Cell types and neuronal circuitry underlying female aggression in Drosophila
Figures
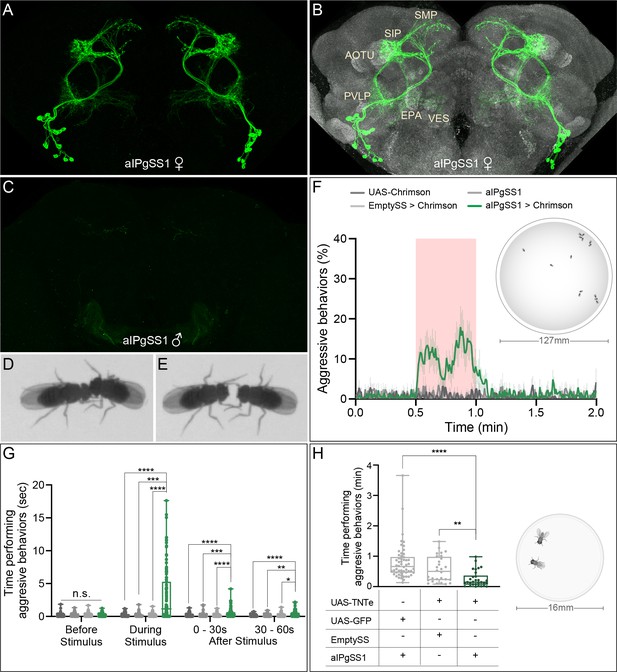
Activation of 11 aIPg neurons increases, while inactivation decreases, female aggressive behaviors.
(A, B) Maximum intensity projection (MIP; 63x) image of the central brain of a female from the aIPgSS1 line crossed with 20xUAS-CsChrimson::mVenus and stained with anti-GFP antibody. Major neuropils innervated are indicated in (B) along with the reference stain (nc82) in gray. (C) A male brain of the same genotype (MIP; 20x) without the reference stain. Images of the complete brain and ventral nerve cord of a female and male are shown in Figure 1—figure supplement 1A–C. Images of individual aIPg neurons, generated by stochastic labeling are shown in Figure 1—figure supplement 2. (D,E) Images of female flies displaying head butting (D) and fencing (E) behaviors. (F) Percentage of flies engaging in aggressive behaviors over the course of a 2 min trial during which a 30 s 0.4 mW/mm2 continuous light stimulus (pink shading) was delivered. The mean is represented as a solid line and shaded bars represent standard error between experiments. The timeseries shows the percentage of flies performing aggression displayed as the mean of 0.5 s (15-frame) bins. See Figure 1—figure supplement 6H for per experiment quantification. See Supplementary file 1 and methods for a description of the JAABA classifier. Data was pooled from two independent biological replicates, which included separate parental crosses and were collected on different days. Inset image shows the arena size for F and G, for more detailed view see Figure 1—figure supplement 3B,C. (G) Total time an individual spent performing aggressive behaviors during each of four 30 s periods: prior to, during, immediately following, and 30–60 s after the stimulus. Individuals were pooled over two independent testing days from two separate parental crosses. Points represent individual flies. (H) Total time an individual spent performing aggressive behaviors over a 30 min trial. Individuals were pooled over two independent testing days during the same week that were from the same parental cross. Inset image shows arena size, for more details see Figure 1—figure supplement 10E and Methods. Points indicate individual flies. Data supporting the plots shown in panels F-H were as follows: F: 20xUAS-CsChrimson, n = 5 experiments; aIPgSS1, n = 5 experiments; EmptySS > 20xUAS-CsChrimson, n = 5 experiments; aIPgSS1 > 20xUAS-CsChrimson, n = 7 experiments. G: 20xUAS-CsChrimson, n = 71 flies; aIPgSS1, n = 65 flies; EmptySS > 20xUAS-CsChrimson, n = 78 flies; aIPgSS1 > 20xUAS-CsChrimson, n = 100 flies. H: aIPgSS1 > UAS-GFP, n = 54 flies; EmptySS > UAS-TNTe, n = 28 flies; aIPgSS1 > UAS-TNTe, n = 30 flies. Data are representative of at least two independent biological repeats, one of which is shown here; see Supplementary file 3 for exact p-values for each figure. For biological repeats of H, see Figure 1—figure supplement 11. Box-and-whisker plots show median and IQR; whiskers show range. A Kruskal-Wallis and Dunn’s post hoc test (G, H) was used for statistical analysis on each time point tested. Asterisk indicates significance from 0: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
-
Figure 1—source data 1
Source data for behavioural experiments in Figure 1.
- https://cdn.elifesciences.org/articles/58942/elife-58942-fig1-data1-v2.xlsx
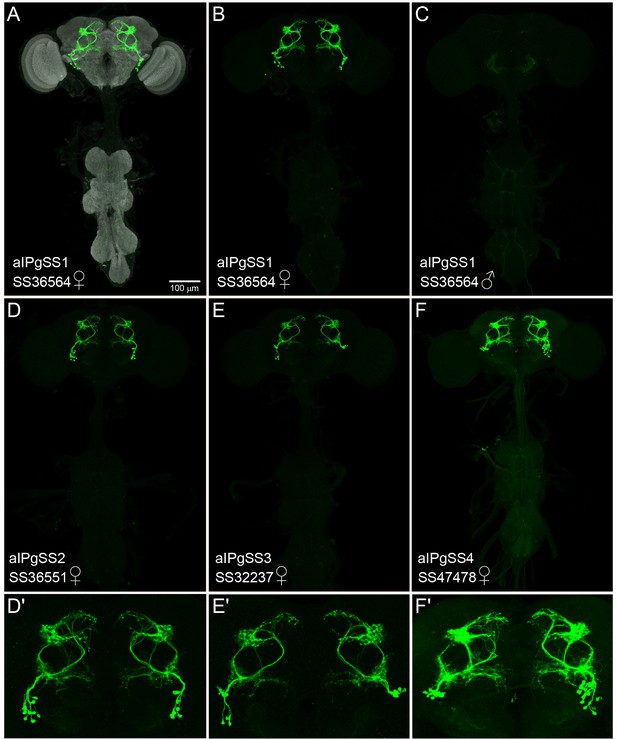
Expression patterns of aIPg split-GAL4 lines.
(A–F) MIP images (20x) of the brains and ventral nerve cords of the indicated split-GAL4 lines crossed to 20xUAS-CsChrimson::mVenus and stained with anti-GFP antibody are shown. Images indicate the relative expression levels of the effector in the different split-GAL4 lines. The scale bar shown in A applies to panels A-F. (D'–F') Enlargements of the central brain of the images shown in D-F. (A–C) SS36564 (aIPgSS1) expresses in 11.3 ± 1.5 cells per female brain hemisphere (n = 8). Note the lack of expression in aIPg neurons in the male central nervous system (C) imaged under identical conditions to that of the female (B). (D,D') SS36551 (aIPgSS2) expresses in 8.9 ± 1.4 cells per female brain hemisphere (n = 8). A few cells with distinct morphology are observed in males. (E,E') SS32237 (aIPgSS3) expresses in 7.8 ± 0.89 cells per female brain hemisphere (n = 8). A few cells with distinct morphology are observed in males. (F,F'). SS47478 (aIPgSS4) expresses in 12.8 ± 1.8 cells per female brain hemisphere (n = 8). No expression was seen in males.
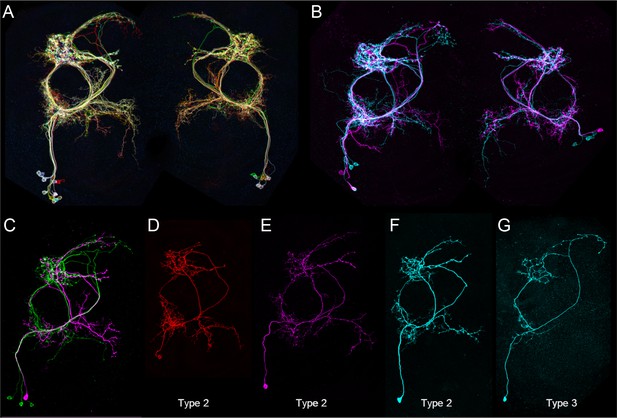
Morphologies of individual aIPg1 - 3 neurons.
The images were generated by stochastic labeling of aIPg split-GAL4 lines using the MultiColor FlipOut method (Nern et al., 2015). (A,B) Images from SS36564 (aIPgSS1) with both hemispheres shown. (C,E,F,G) Images from SS36551 (aIPgSS2), with one brain hemisphere shown. (D) Image from SS47478 (aIPgSS4), with one brain hemisphere shown. (D,E,F) Images are examples of type 2 aIPg cells, while (G) shows a type 3 aIPg cell.
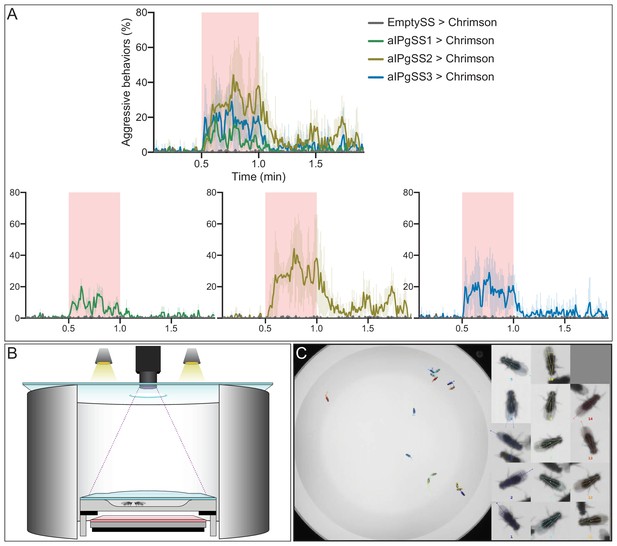
Additional aIPg split-GAL4 lines also induce aggressive behavior.
(A) Percentage of flies engaging in aggressive behaviors over a trial during which a 30 s 0.1 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. The expression pattern of these three aIPgSS lines is shown in Figure 1—figure supplement 1. For clarity, the individual plots for each line and the EmptySS > Chrimson are also shown. EmptySS > Chrimson, n = 7 experiments; aIPgSS1 > Chrimson, n = 5 experiments; aIPgSS2 > Chrimson, n = 3 experiments; aIPgSS3 > Chrimson, n = 4 experiments. Data are representative of at least three independent biological repeats using different parental crosses, one of which is shown here. (B,C) Schematic (B) and an example image from the analysis (C) taken from Video 2 of the 127 mm arena used for the activation experiments detailed in Figure 1.
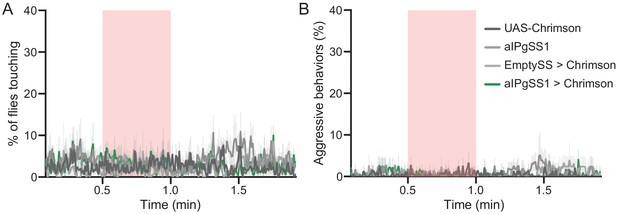
Optogenetic stimulation of aIPgSS1 > Chrimson males does not result in aggressive behavior.
(A,B) Percentage of male flies touching (A) or performing aggressive behaviors (fencing and head butting) (B) over the course of a trial during which a 30 s 0.4 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. Data was pooled from two independent biological replicates, which included separate parental crosses and were collected on different days. 20xUAS-CsChrimson, n = 5 experiments; aIPgSS1, n = 4 experiments; EmptySS > 20xUAS-CsChrimson, n = 3 experiments; aIPgSS1 > 20xUAS-CsChrimson, n = 6 experiments. Data are representative of at least two independent biological repeats using different parental crosses.
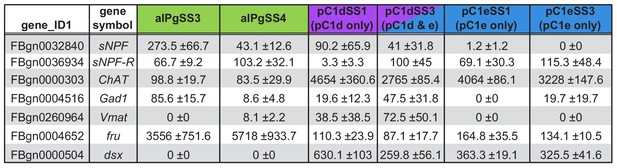
aIPg neurons are cholinergic, fru+ and sNPF+; pC1 neurons are cholinergic and dsx+.
Expression (CPM) of genes related to neurotransmitters (sNPF, sNPF-R, ChAT, Gad1, Vmat) and sex determination pathways (dsx and fru). Cells were dissociated from dissected brains and sorted on the basis of their expression of the indicated split-GAL4 driver: aIPgSS3 (n = 2 samples), aIPgSS4 (n = 3 samples), pC1dSS1 (n = 3 samples), pC1dSS3 (n = 6 samples), pC1eSS1 (n = 3 samples), and pC1eSS3 (n = 3 samples). The mean and standard deviation of the indicated number of samples are shown. Cell sorting and RNA profiling was done as described in Methods. Data were deposited to NCBI Gene Expression Omnibus (Acc No. GSE158748).
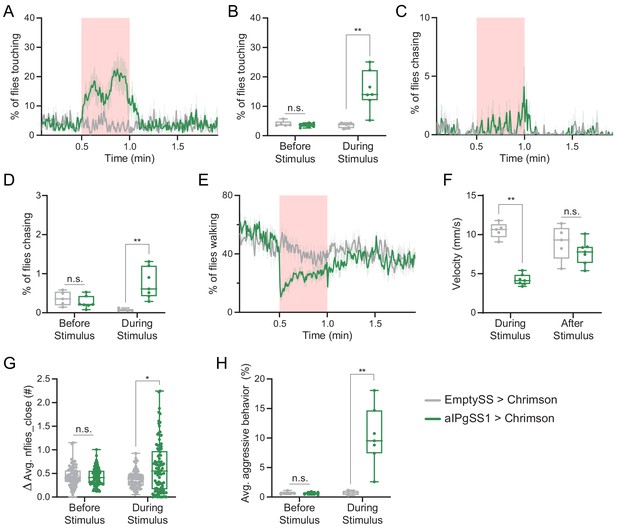
Changes in behavioral metrics in females following activation of aIPgSS1 neurons.
(A,C,E) Percentage of female flies touching (A), chasing (C), or walking (E) over the course of a trial during which a 30 s 0.4 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. (B,D) Average percentage of flies touching (B) or chasing (D) over the 30 s period prior to or during the stimulus delivery. (F) Velocity during the stimulus period and in the 30–60 s following the stimulus. (G) Difference in the average number of flies within two body lengths over the 30 s prior to and during the stimulus; points represent individual flies. (H) Average percentage of flies per experiment engaging in aggressive behaviors over the 30 s period prior to or during the stimulus delivery. Points in panels B, D, F and H represent separate experiments consisting of approximately 15 flies. Analyses were performed on the same data set used in Figure 1. Data supporting the plots shown in the individual panels were as follows: A-F,H: EmptySS > 20xUAS-CsChrimson, n = 5 experiments; aIPgSS1 > 20xUAS-CsChrimson, n = 7 experiments. G: EmptySS > 20xUAS-CsChrimson, n = 78 flies; aIPgSS1 > 20xUAS-CsChrimson, n = 100 flies. Data are representative of at least two independent biological repeats; see Supplementary file 3 for exact p-values for each figure. Box-and-whisker plots show median and IQR; whiskers show range. Mann-Whitney U-tests were used for statistical analysis. Asterisk indicates significance from 0: *p<0.05; **p<0.01; n.s, not significant.
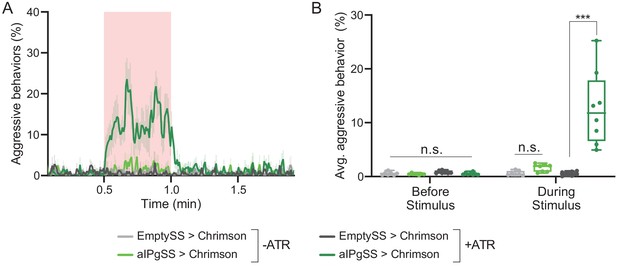
Optogenetic activation of aggression with Chrimson requires feeding all trans-retinal.
(A) Percentage of flies that engaged in aggressive behaviors during a trial in which a 30 s 0.1 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. Flies were raised with food supplemented with all trans-retinal (+ATR) or not (-ATR). (B) Average percentage of flies engaging in aggressive behaviors over the 30 s period prior to or during the stimulus delivery in A. Points represent separate experiments consisting of approximately 15 flies. +ATR: EmptySS > Chrimson, n = 7 experiments; aIPgSS1 > Chrimson, n = 8 experiments; -ATR: EmptySS > Chrimson, n = 7 experiments; aIPgSS1 > Chrimson, n = 7 experiments. Data was pooled from two independent biological replicates, which included separate parental crosses and were collected on different days; see Supplementary file 3 for exact p-values for each figure. Box-and-whisker plots show median and IQR; whiskers show range. A Kruskal-Wallis and Dunn’s post hoc test was used for statistical analysis. Asterisk indicates significance from 0: ***p<0.001; n.s, not significant.
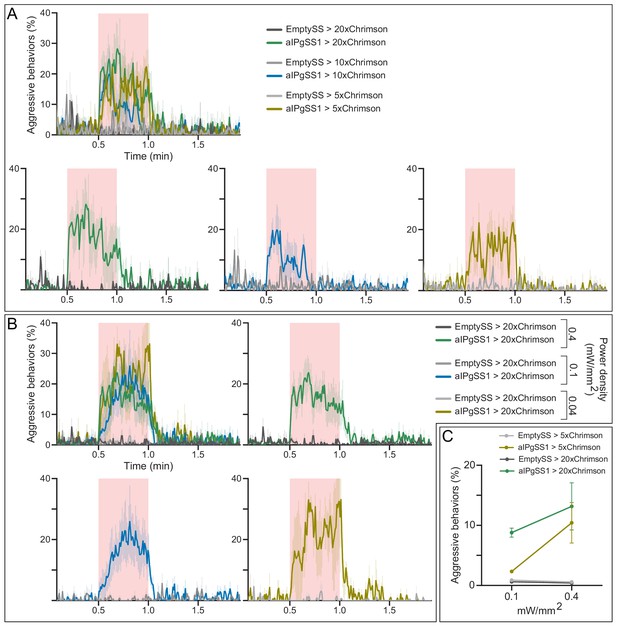
Behavioral effects of effector strength and stimulus delivery.
(A) Percentage of flies that carry 5x, 10x, or 20xUAS-Chrimson effectors that engage in aggressive behaviors over the course of a trial during which a 30 s 0.1 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. Individual curves are shown for comparison. (B) Percentage of flies engaging in aggressive behaviors over the course of a trial during which a 30 s 0.4, 0.1, or 0.04 mW/mm2 continuous light stimulus was delivered, plotted as in Figure 1F. Individual curves are shown for comparison. Data are representative of at least two independent biological repeats using different parental crosses, one of which is shown here. (C) Average percentage of flies with either the 5x or 20xUAS-Chrimson effector that perform aggressive behaviors over the 30 s period during the delivery of either a 0.1 or 0.4 mW/mm2 continuous light stimulus. Data supporting the plots shown in the individual panels were as follows: A: EmptySS > 20xUAS-Chrimson, n = 2 experiments; aIPgSS1 > 20xUAS-Chrimson, n = 3 experiments; EmptySS > 10xUAS-Chrimson, n = 3 experiments; aIPgSS1 > 10xUAS-Chrimson, n = 4 experiments; EmptySS > 5xUAS-Chrimson, n = 3 experiments; aIPgSS1 > 5xUAS-Chrimson, n = 3 experiments. B: 0.04 mW/mm2: EmptySS > Chrimson, n = 3 experiments; aIPgSS1 > Chrimson, n = 3 experiments; 0.1 mW/mm2: EmptySS > Chrimson, n = 4 experiments; aIPgSS1 > Chrimson, n = 4 experiments; 0.4 mW/mm2: EmptySS > Chrimson, n = 5 experiments; aIPgSS1 > Chrimson, n = 6 experiments. C: 0.1 mW/mm2: EmptySS > 5xUAS-Chrimson, n = 6 experiments; aIPgSS1 > 5xUAS-Chrimson, n = 5 experiments; EmptySS > 20xUAS-Chrimson, n = 6 experiments; aIPgSS1 > 20xUAS-Chrimson, n = 8 experiments; 0.4 mW/mm2: EmptySS > 5xUAS-Chrimson, n = 6 experiments; aIPgSS1 > 5xUAS-Chrimson, n = 9 experiments; EmptySS > 20xUAS-Chrimson, n = 4 experiments; aIPgSS1 > 20xUAS-Chrimson, n = 5 experiments. Bars are mean ± S.E.M.
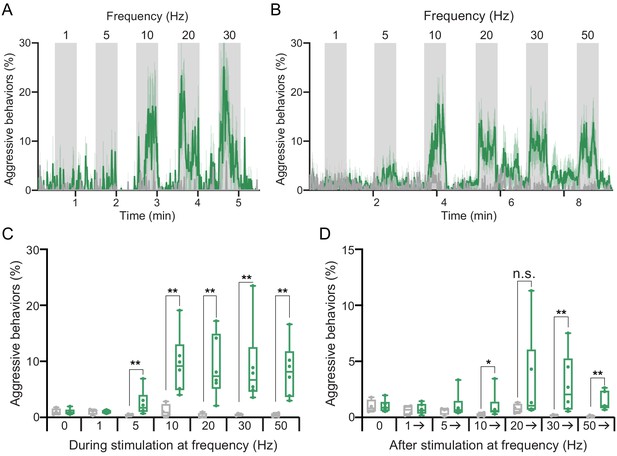
Higher frequency optogenetic stimulation increases the persistence of aggressive behaviors.
(A,B) Blocks of 30 s photostimulation (gray bars) with increasing stimulation frequency separated by 30 s (A) or 60 s (B) sequential intervals. Light was delivered at 0.1 mW/mm2 with at a 10 ms pulse width, with increasing pulse frequency. The pulse period and pulse number during each period was as follows: 1000 ms, 30 (1 Hz); 200 ms, 150 (5 Hz); 100 ms, 300 (10 Hz); 50 ms, 600 (20 Hz); 33 ms, 909 (30 Hz); 20 ms, 1500 (50 Hz). aIPgSS1 > 20xUAS-Chrimson is shown in green; EmptySS > 20xUAS-Chrimson is shown in gray. (C) Average percentage of flies performing aggressive behaviors over the 30 s period during stimulus delivery in B. (D) Average percentage of flies performing aggressive behaviors over the 30 s period before 1 Hz, the 60 s periods between subsequent stimuli, and the 30 s after 50 Hz stimulation in B. Data was pooled from two independent biological replicates, which included separate parental crosses and were collected on different days. Points indicate separate experiments. A: EmptySS > 20xUAS-Chrimson, n = 3 experiments; aIPgSS1 > 20xUAS-Chrimson, n = 3 experiments; B: EmptySS > 20xUAS-Chrimson, n = 4 experiments; aIPgSS1 > 20xUAS-Chrimson, n = 6 experiments. Data are representative of at least two independent biological repeats; see Supplementary file 3 for exact p-values for each figure. Box-and-whisker plots show median and IQR; whiskers show range. Mann-Whitney U-tests were used for statistical analysis. Asterisk indicates significance from 0: *p<0.05; **p<0.01; n.s., not significant.
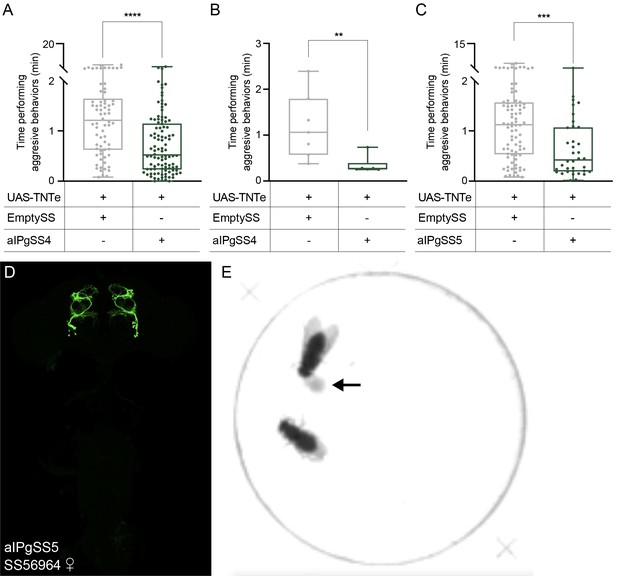
Inactivation of aIPg neurons using additional split-GAL4 lines also decreases aggressive behaviors.
(A–C) Total time an individual spent performing aggressive behaviors over a 30 min trial. Experiments in A - C were pooled from three, two, and two testing days, respectively. The experiments in A–C used separate parental crosses. Experiments in B were manually scored with JWatcher software, whereas the experiments in A and C were scored using an automated classifier generated with JAABA (see Methods). Points in A and C indicate individual flies, while in B points represent pairs of flies. (D) Expression pattern of SS56964 (aIPgSS5); this line expresses in 15.8 ± 0.4 cells per female brain hemisphere (n = 6). (E) Image of the 16 mm arena used for inactivation experiments. Arrow points to the 1 mm yeast spot. Data supporting the plots shown in the individual panels were as follows: A: EmptySS > UAS-TNTe, n = 70 flies; aIPgSS4 > UAS-TNTe, n = 102 flies. B: EmptySS > UAS-TNTe, n = 7 pairs; aIPgSS4 > UAS-TNTe, n = 6 pairs. C: EmptySS > UAS-TNTe, n = 84 flies; aIPgSS5 > UAS-TNTe, n = 36 flies. Box-and-whisker plots show median and IQR; whiskers show range. Kruskal-Wallis and Dunn’s post hoc tests were used for statistical analysis. Asterisk indicates significance from 0: **p<0.01; ***p<0.001; ****p<0.0001.
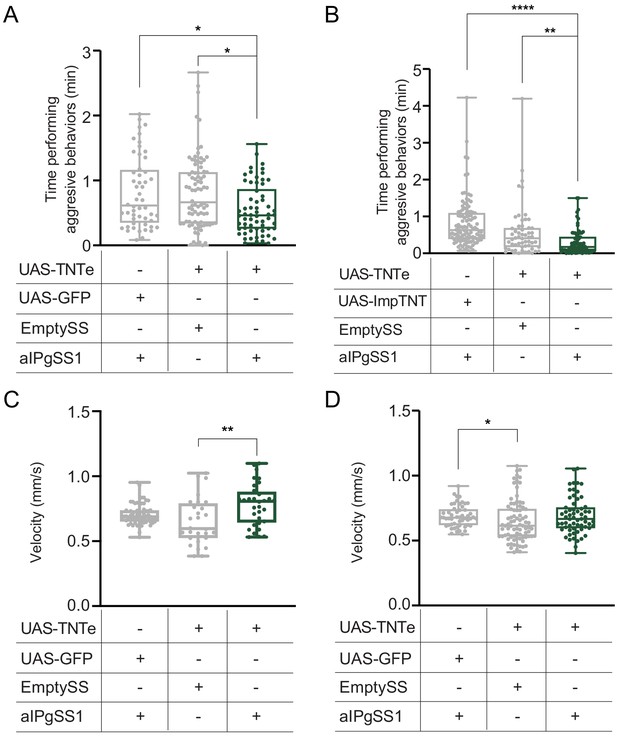
aIPg inactivation reproducibly decreases aggressive behaviors but not velocity.
(A,B) Total time an individual spent performing aggressive behaviors over a 30 min trial. Plots in A and B each represent the pooled data from four and five testing days, respectively, of separate biological repeats using different parental crosses. The experiments in A and B differ in that a different negative control UAS line was used: UAS-GFP in A and UAS-ImpTNT in B; ImpTNT is an inactive form of TNT (see Methods). Panel A represents an independent biological repeat of the experiment shown in Figure 1H. Experiments were scored using an automated classifier generated with JAABA (see Methods). Points indicate individual flies. (C) Average velocity over the 30 min trial shown in Figure 2. Points indicate individual flies. (D) Average velocity over the 30 min trial in A. Data supporting the plots shown in the individual panels were as follows: A: aIPgSS1 > UAS-GFP, n = 52 flies; EmptySS > UAS-TNTe, n = 82 flies; aIPgSS1 > UAS-TNTe, n = 62 flies. B: aIPgSS1 > UAS-ImpTNTe, n = 110 flies; EmptySS > UAS-TNTe, n = 56 flies; aIPgSS1 > UAS-TNTe, n = 74 flies. C: aIPgSS1 > UAS-GFP, n = 54 flies; EmptySS > UAS-TNTe, n = 28 flies; aIPgSS1 > UAS-TNTe, n = 30 flies. See Supplementary file 3 for exact p-values for each figure. Box-and-whisker plots show median and IQR; whiskers show range. Kruskal-Wallis and Dunn’s post hoc tests were used for statistical analysis. Asterisk indicates significance from 0: *p<0.05; **p<0.01; ****p<0.0001.
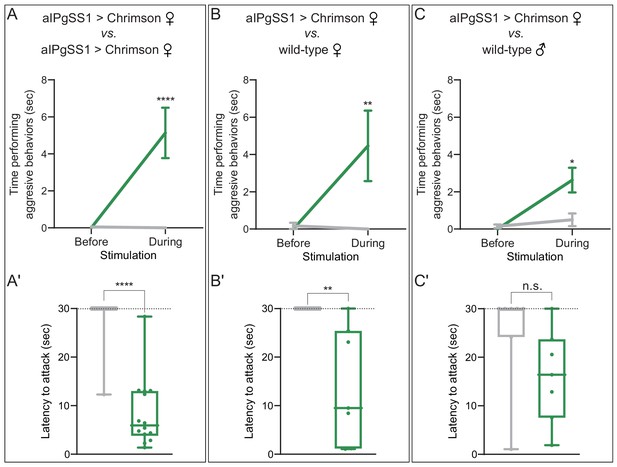
aIPg activation increases aggression against wild-type females and males.
(A–C) Total time spent performing aggressive behaviors in a 16 mm arena over the 30 s period prior to or during a 0.1 mW/mm2 stimulation. The plots refer only to the behavior of aIPgSS1 > Chrimson females and each arena contained only two flies: (A) two aIPgSS1 > Chrimson females; (B) an aIPgSS1 > Chrimson female and a wild-type (Canton-S) female; and (C) an aIPgSS1 > Chrimson female and a wild-type (Canton-S) male. The green line shows the stated genotype; the gray line shows the results when EmptySS > Chrimson was used instead of aIPgSS1 > Chrimson. Note that the difference observed between aggression against wild-type females (B) and males (C) was not significant (p=0.65). (A'–C') Amount of time during a 30 s 0.1 mW/mm2 continuous stimulation period until first aggressive encounter. Points indicate individual flies. Dotted lines indicate the end of the trial and error bars in A–C are mean ± S.E.M. Box-and-whisker plots show median and IQR; whiskers show range. Data supporting the plots shown in the individual panels were as follows: (A') EmptySS > 20xUAS-Chrimson, n = 22 flies; aIPgSS1 > 20xUAS-Chrimson, n = 14 flies. (B') EmptySS > 20xUAS-Chrimson, n = 8 flies; aIPgSS1 > 20xUAS-Chrimson, n = 7 flies. (C') EmptySS > 20xUAS-Chrimson, n = 7 flies; aIPgSS1 > 20xUAS-Chrimson, n = 7 flies. We performed at least two biological repeats that confirmed aggression against all three types of target flies; results for a typical repeat had the following p-values during stimulation: A: p=0.0035; A’: p=0.0067; B: p=0.0006; B’: p=0.0157; C: p=0.0123; C’: p=0.0030. A Mann-Whitney U post hoc test was used for statistical analysis. Asterisk indicates significance from 0: *p<0.05; **p<0.01; ****p<0.0001; n.s., not significant.
-
Figure 2—source data 1
Source data for behavioural experiments in Figure 2.
- https://cdn.elifesciences.org/articles/58942/elife-58942-fig2-data1-v2.xlsx
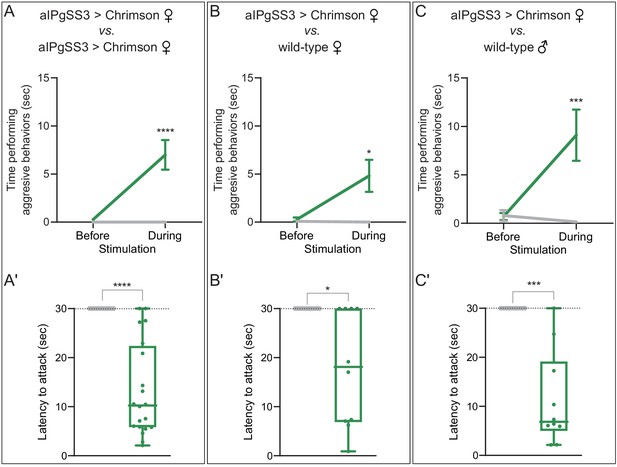
Increased aggression against wild-type females and males reproduced with a second aIPgSS line.
(A–C) Total time spent performing aggressive behaviors in a 16 mm arena over the 30 s period prior to or during a 0.1 mW/mm2 stimulation. The plots refer only to the behavior of aIPgSS3 > Chrimson females and each arena contained only two flies: (A) two aIPgSS3 > Chrimson females; (B) an aIPgSS3 > Chrimson female and a wild-type (Canton-S) female; and (C) a aIPgSS3 > Chrimson female and a wild-type (Canton-S) male. The green line shows the stated genotype; the gray line shows the results when EmptySS > Chrimson was used instead of aIPgSS3 > Chrimson. Note that the difference observed between aggression against wild-type females (B) and males (C) was not significant (p=0.16). (A'–C') Amount of time during a 30 s 0.1 mW/mm2 continuous stimulation period until first aggressive encounter. Points indicate individual flies. Dotted lines indicate the end of the trial and error bars in A—C are mean ± S.E.M. Box-and-whisker plots show median and IQR; whiskers show range. (A') EmptySS > 20xUAS-Chrimson, n = 8 flies; aIPgSS3 > 20xUAS-Chrimson, n = 20 flies; (B') EmptySS > 20xUAS-Chrimson, n = 7 flies; aIPgSS3 > 20xUAS-Chrimson, n = 10 flies; (C') EmptySS > 20xUAS-Chrimson, n = 8 flies; aIPgSS3 > 20xUAS-Chrimson, n = 10 flies. Data are representative of two independent biological repeats, one of which is shown here; see Supplementary file 3 for exact p-values. A Mann-Whitney U post hoc test was used for statistical analysis. Asterisk indicates significance from 0: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; n.s., not significant.
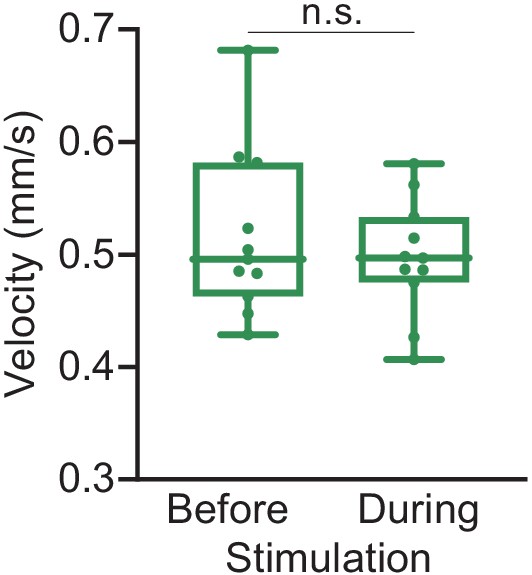
aIPg activation in the absence of a target fly does not alter velocity.
Average velocity over the 30 s either prior to or during the stimulus. Points represent individual flies and data are representative of two independent biological repeats, one of which is shown here. aIPgSS1 > 20xUAS-Chrimson (Before), n = 11 flies; aIPgSS1 > 20xUAS-Chrimson (During), n = 11 flies. See Supplementary file 3 for exact p-values. Box-and-whisker plots show median and IQR; whiskers show range. A Mann-Whitney U post hoc test was used for statistical analysis. n.s., not significant. See also Video 5.
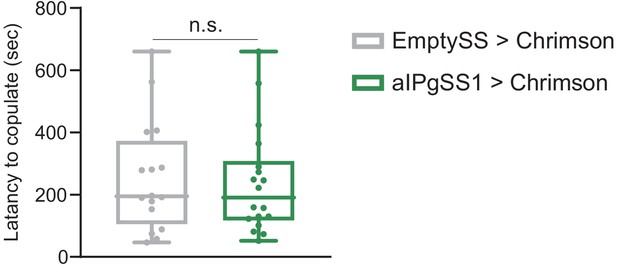
aIPg activation does not significantly alter copulation latency.
Amount of time until first copulation event following a 0.1 mW/mm2 continuous stimulus. Virgin females were added into a 10 mm arena containing virgin wild type (Canton-S) males immediately prior to the stimulus onset. Points indicate individual flies. EmptySS > 20xUAS-Chrimson, n = 16 flies; aIPgSS1 > 20xUAS-Chrimson, n = 16 flies. Data are representative of at least two independent biological repeats, one of which is shown here; see Supplementary file 3 for exact p-values. Box-and-whisker plots show median and IQR; whiskers show range. A Mann-Whitney U post hoc test was used for statistical analysis. n.s., not significant.

An unbiased screen suggests that aIPg1-3 and pC1d are key nodes for gating female-female aggression.
(A) The fourteen top hits for female-female chasing from an unbiased activation screen of 2204 generation 1 GAL4 lines are listed along with their Z-scores, the signed number of standard deviations from the mean behavior of the control, as determined by Robie et al., 2017. Also shown is the relevant cell type we concluded from our intersectional analysis (Figure 4—figure supplement 1) to be present in each line. N.D., no cell type reproducibly detected, suggesting that R48F12 may not share a common cell type with any of the other 13 lines. The final column in the table refers to the panels in Figure 3—figure supplement 1 where results supporting the stated conclusion are shown. (B,C) The expression patterns of the two indicated GAL4 lines (Jenett et al., 2012). The images shown were taken from the database at http://www.janelia.org/gal4-gen1, where the expression patterns of the other lines listed in A can also be found. (D) The expression pattern of a split-GAL4 line made by intersecting these two enhancers; a cell with the morphology of pC1d can be seen.
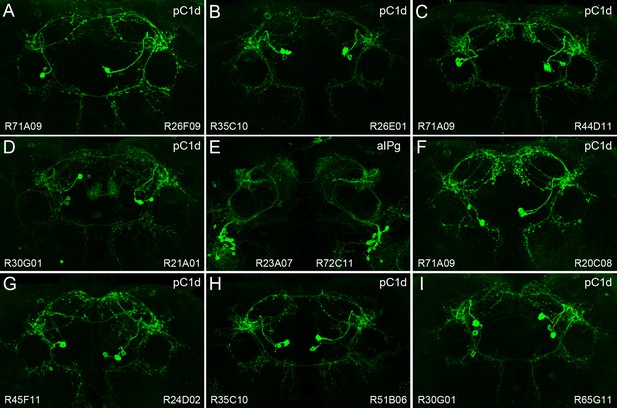
Expression patterns resulting from split-GAL4 intersections of hits from the unbiased screen.
(A–I) Each panel shows the intersection of the indicated enhancers; the enhancer in the lower left was used to drive the split-GAL4 AD domain and the enhancer in the lower right drove the DBD domain. We observed either the pC1d or aIPg cell type in each intersection. Nearly all of the pC1d containing intersections also show expression in pC1e or other members of the pC1 cell-type group.
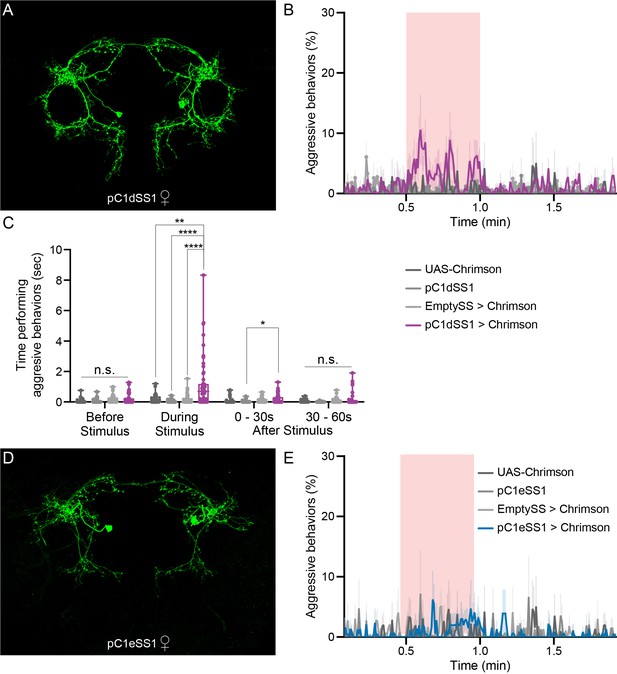
pC1d, but not pC1e, significantly increases aggressive social interactions in female flies.
(A) MIP (63x) image of the central brain of a female from the pC1dSS1 split-GAL4 line crossed with 20xUAS-CsChrimson::mVenus and stained with anti-GFP antibody. Images of the complete brain and ventral nerve cord of a female and male of the same genotype are shown in Figure 4—figure supplement 1A–C. (B) Percentage of flies engaging in aggressive behaviors over the course of a trial during which a 30 s 0.4 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. (C) Total time an individual spent performing aggressive behaviors during each of four 30 s periods: prior to, during, immediately following, and 30–60 s after the stimulus. Points represent individual flies. Note that we used 20xUAS-CsChrimson for these experiments to be consistent with the experiments done with aIPg split-GAL4 lines, but the levels of aggression observed with pC1dSS1 are actually higher when a weaker effector line (5xUAS-CsChrimson) is used (Figure 4—figure supplement 9A). Box-and-whisker plots show median and IQR; whiskers show range. Kruskal-Wallis and Dunn’s post hoc tests were used for statistical analysis. Asterisk indicates significance from 0: *p<0.05; **p<0.01; ****p<0.0001; n.s., not significant. (D) MIP (63x) image of the central brain of a female from the pC1eSS1 line crossed with 20xUAS-CsChrimson::mVenus and stained with anti-GFP antibody. Images of the complete brain and ventral nerve cord of a female and male of the same genotype are shown in Figure 4—figure supplement 2A–C. (E) Percentage of flies engaging in aggressive behaviors over the course of a trial during which a 30 s 0.4 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. No obvious differences between genotypes were observed. Data supporting the plots shown in the individual panels were as follows: B: 20xUAS-CsChrimson, n = 2 experiments; pC1dSS1, n = 3 experiments; EmptySS > 20xUAS-CsChrimson, n = 5 experiments; pC1dSS1 > 20xUAS-CsChrimson, n = 5 experiments. C: 20xUAS-CsChrimson, n = 29 flies; pC1dSS1, n = 48 flies; EmptySS > 20xUAS-CsChrimson, n = 46 flies; pC1dSS1 > 20xUAS-CsChrimson, n = 53 flies. E: 20xUAS-CsChrimson, n = 2 experiments; pC1eSS1, n = 2 experiments; EmptySS > 20xUAS-CsChrimson, n = 3 experiments; pC1eSS1 > 20xUAS-CsChrimson, n = 3 experiments. For all panels, data are representative of at least three independent biological repeats, one of which is shown here; see Supplementary file 3 for exact p-values.
-
Figure 4—source data 1
Source data for behavioural experiments in Figure 4.
- https://cdn.elifesciences.org/articles/58942/elife-58942-fig4-data1-v2.xlsx
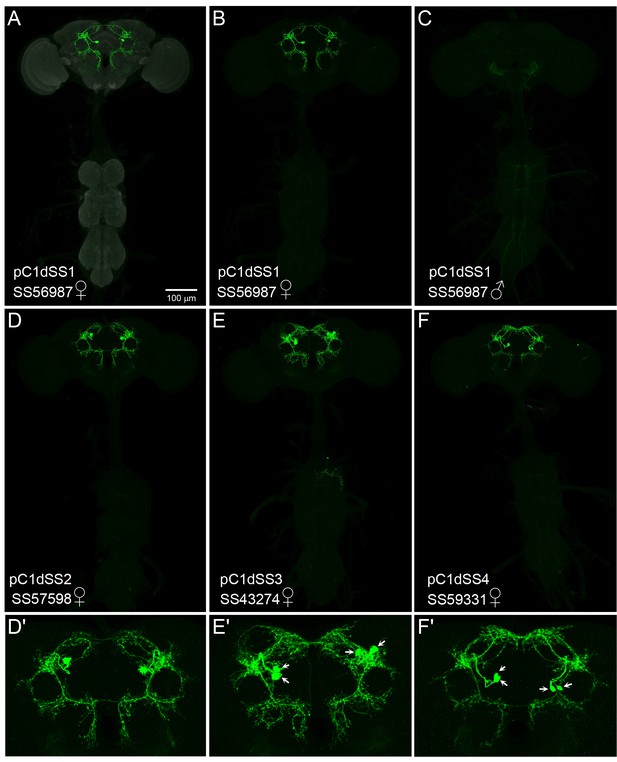
Expression patterns of pC1d split-GAL4 lines.
(A–F) 20X MIP images of the brains and ventral nerve cords of the indicated split-GAL4 lines crossed to 20xUAS-CsChrimson::mVenus. The scale bar shown in A applies to panels A - F. (D'–F') Enlargements of the central brain of the images shown in D - F. (A–C) The SS56987 (pC1dSS1) expression pattern is shown; the neuropil reference channel shown in A (gray). Note that no expression in pC1d neurons is seen in the male nervous system which was imaged under identical conditions to the female nervous system shown in B. (D,D') Expression pattern of SS57598 (pC1dSS2). (E,E') Expression pattern of SS43274 (pC1dSS3). Note that this line expresses in both pC1d and pC1e. This line shows expression in males in a cell type clearly distinct from pC1d. Arrows indicate cell bodies. (F,F'). Expression pattern of SS59331 (pC1dSS4). Note that this line expresses in both pC1d and pC1e. Arrows indicate cell bodies.
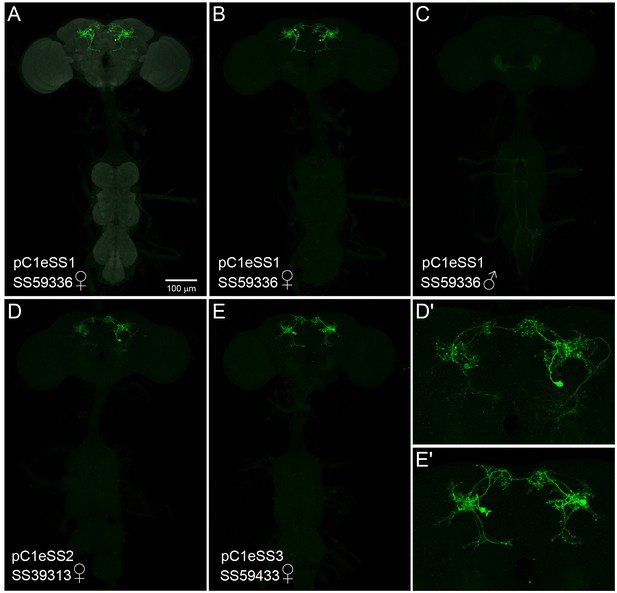
Expression patterns of pC1e split-GAL4 lines.
(A–E) 20X MIP images of the brains and ventral nerve cords of the indicated split-GAL4 lines crossed to 20xUAS-CsChrimson::mVenus. The scale bar shown in A applies to panels A-E. (D',E') Enlargements of the central brain of the images shown in D and E. (A–C) The SS59336 (pC1eSS1) expression pattern is shown; the neuropil reference channel is shown in A (gray). Note that no expression in aIPg neurons is seen in the male nervous system (C) which was imaged under identical conditions to the female nervous system shown in B. (D,D') Expression pattern of SS39313 (pC1eSS2). This line shows expression in males in a cell type clearly distinct from pC1e (not shown). (E,E') Expression pattern of SS59433 (pC1eSS3).
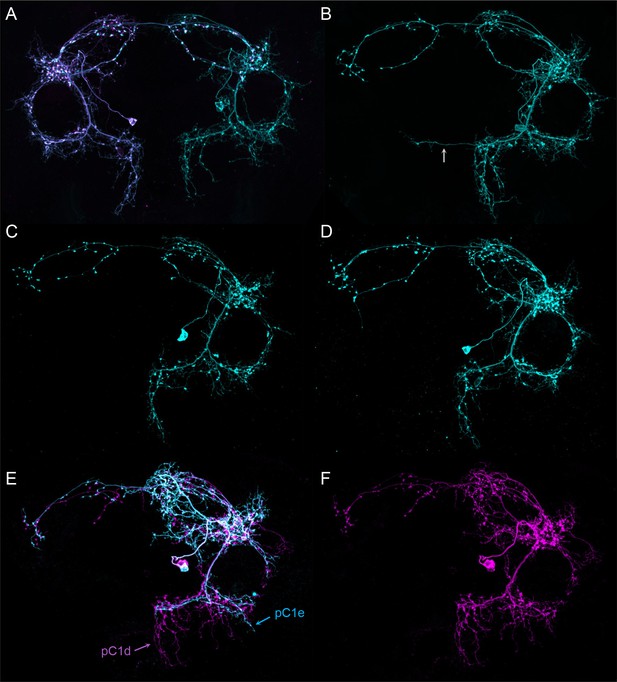
Morphologies of individual pC1d neurons.
The images in all panels were generated by stochastic labeling of pC1d split-GAL4 lines using the MultiColor FlipOut method (Nern et al., 2015). Both brain hemispheres are shown. (A–D) Images from SS56987 (pC1dSS1). The arrow in (B) highlights a process that crosses the midline which is often seen in pC1d cells; see (D') and (F') in Figure 4—figure supplement 1 for additional examples. (A) The right and left hemisphere pC1d cells are shown. (E,F) Images from SS43274 (pC1dSS3).
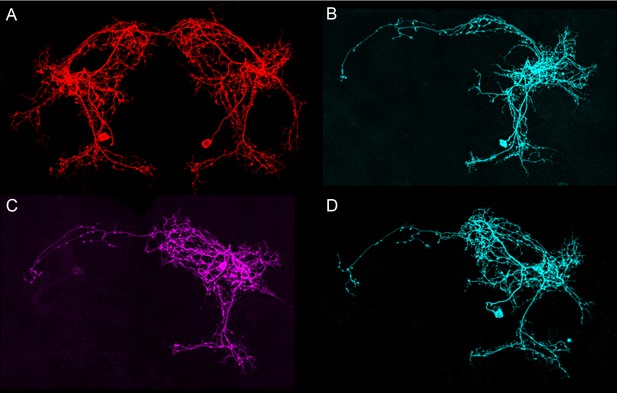
Morphologies of individual pC1e neurons.
The images in all panels were generated by stochastic labeling of pC1e split-GAL4 lines using the MultiColor FlipOut method (Nern et al., 2015). (A,B) Images from SS59336 (pC1eSS1). (A) The right and left hemisphere pC1e cells are shown. (C,D) Images from SS43274 (pC1dSS3). Note this line contains both pC1d and pC1e cells. See Figure 4—figure supplement 3 for pC1d cells.
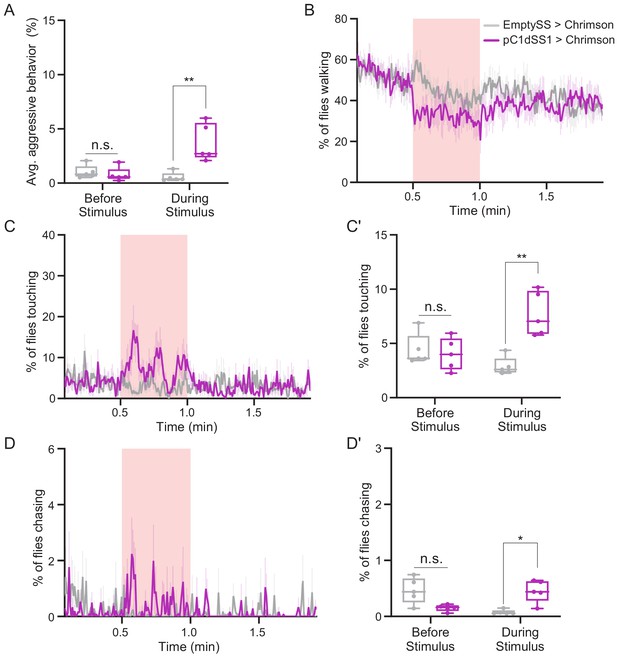
Behavioral characterization of female flies after pC1d activation.
Behaviors were measured over the course of a trial during which a 30 s 0.4 mW/mm2 continuous light stimulus (pink shading) was delivered. (A) Average percentage of flies per experiment engaging in aggressive behaviors over the 30 s period prior to or during stimulus delivery. (B,C,D) Percentage of female flies walking (B), touching (C), or chasing (D), plotted as in Figure 1F. (C', D') Average percentage of flies touching (C') or chasing (D') over the 30 s period prior to or during the stimulus delivery in C or D, respectively. Points represent separate experiments consisting of approximately 15 flies. EmptySS > 20xUAS-CsChrimson, n = 5 experiments; pC1dSS1 > 20xUAS-CsChrimson, n = 5 experiments. Data are representative of at least three independent biological repeats, one of which is shown here; see Supplementary file 3 for exact p-values. Box-and-whisker plots show median and IQR; whiskers show range. Mann-Whitney U-tests were used for statistical analysis. Asterisk indicates significance from 0: *p<0.05; **p<0.01; n.s., not significant.
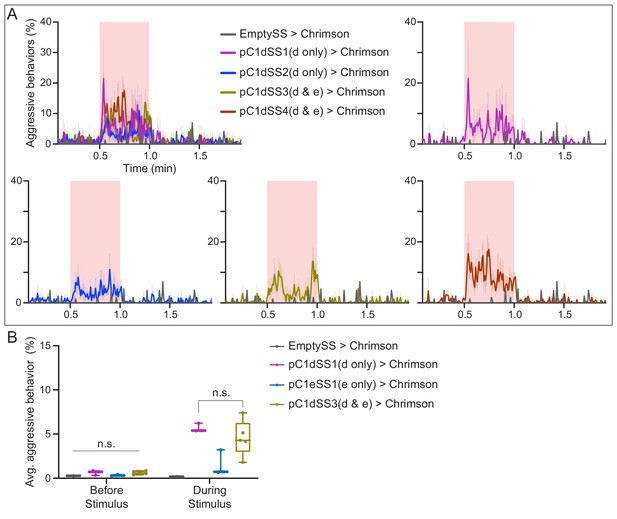
Optogenetic activation of additional lines labeling pC1d split-GAL4 lines display similar behavioral results to pC1dSS1.
(A) Percentage of flies engaging in aggressive behaviors over the course of a trial during which a 30 s 0.1 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. Note that two pC1d split-GAL4 lines label just pC1d and two others label both pC1d and pC1e, as indicated. Individual curves are shown for comparison. (B) Average percentage of flies engaging in aggressive behaviors over the 30 s period prior to or during the delivery of a 0.1 mW/mm2 stimulus. Note that the amount of aggression seen in lines expressing either in just pC1d or both pC1d and pC1e are not significantly different. Points represent separate experiments consisting of approximately 15 flies. Data supporting the plots shown in the individual panels were as follows: (A) EmptySS > Chrimson, n = 1 experiment; pC1dSS1 > Chrimson, n = 3 experiments; pC1dSS2 > Chrimson, n = 6 experiments; pC1dSS3 > Chrimson, n = 5 experiments; pC1dSS4 > Chrimson, n = 5 experiments. B: EmptySS > Chrimson, n = 1 experiment; pC1dSS1 > Chrimson, n = 3 experiments; pC1eSS1 > Chrimson, n = 3 experiments; pC1dSS3 > Chrimson, n = 5 experiments. Data was pooled from two independent biological replicates, which included separate parental crosses and were collected on different days. Data are representative of at least two independent biological repeats; see Supplementary file 3 for exact p-values. Box-and-whisker plots show median and IQR; whiskers show range. Kruskal-Wallis and Dunn’s post hoc tests were used for statistical analysis. Asterisk indicates significance from 0: n.s., not significant.
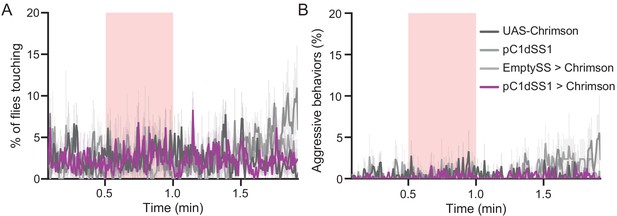
Optogenetic stimulation of pC1dSS1 > Chrimson males does not result in aggressive behavior.
(A–B) Percentage of male flies touching (A) or performing aggressive behaviors (B) over the course of a trial during which a 30 s 0.4 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. 20xUAS-CsChrimson, n = 5 experiments; pC1dSS1, n = 4 experiments; EmptySS > 20xUAS-CsChrimson, n = 3 experiments; pC1dSS1 > 20xUAS-CsChrimson, n = 5 experiments. Data are representative of at least two independent biological repeats, one of which is shown here.
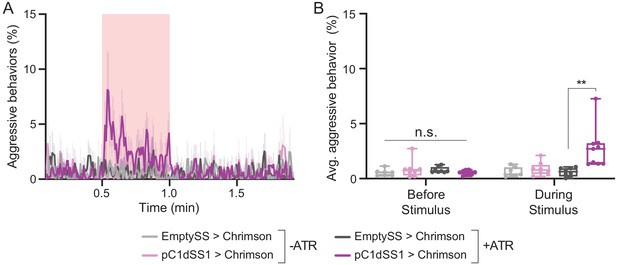
Optogenetic activation of aggression depends on feeding all trans-retinal.
(A) Percentage of flies that engaged in aggressive behaviors during a trial in which a 30 s 0.1 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. Flies were raised with food supplemented with all trans-retinal (+ATR) or not (-ATR). (B) Average percentage of flies engaging in aggressive behaviors over the 30 s period prior to or during the stimulus delivery in A. Points represent separate experiments consisting of approximately 15 flies. +ATR: EmptySS > Chrimson, n = 8 experiments; pC1dSS1 > Chrimson, n = 9 experiments; -ATR: EmptySS > Chrimson, n = 7 experiments; pC1dSS1 > Chrimson, n = 8 experiments. Data was pooled from two independent biological replicates, which included separate parental crosses and were collected on different days. Data are representative of at least two independent biological repeats; see Supplementary file 3 for exact p-values. Box-and-whisker plots show median and IQR; whiskers show range. Kruskal-Wallis and Dunn’s post hoc tests were used for statistical analysis. Asterisk indicates significance from 0: **p<0.01; n.s., not significant.
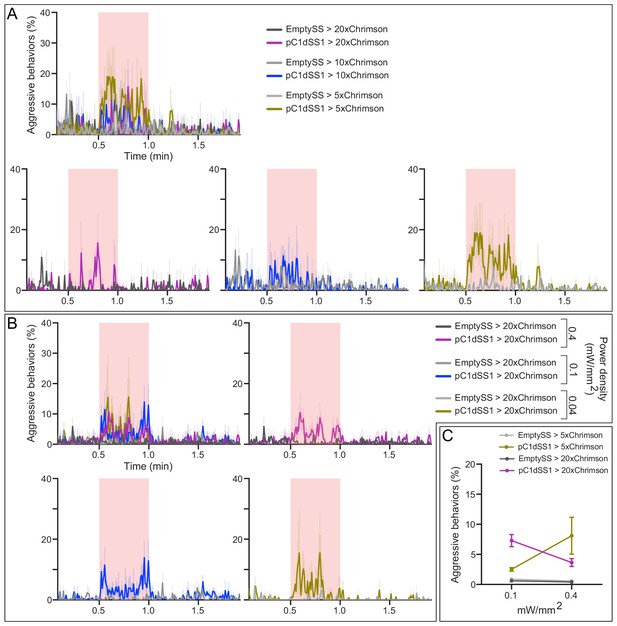
Behavioral effects of stimulus delivery and effector strength.
(A) Percentage of flies engaging in aggressive behaviors over the course of a trial during which a 30 s 0.1 mW/mm2 continuous light stimulus (pink shading) was delivered to flies carrying 5XUAS, 10xUAS, and 20xUAS effectors, data plotted as in Figure 1F. Individual curves are shown for comparison. (B) Percentage of flies engaging in aggressive behaviors over the course of a trial during which a 30 s 0.4, 0.1, or 0.04 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. Individual curves are shown for comparison. (C) Average percentage of flies engaging in aggressive behaviors over the 30 s period during stimulus delivery. Data supporting the plots shown in the individual panels were as follows: (A) EmptySS > 20xUAS-Chrimson, n = 2 experiments; pC1dSS1 > 20xUAS-Chrimson, n = 2 experiments; EmptySS > 10xUAS-Chrimson, n = 3 experiments; pC1dSS1 > 10xUAS-Chrimson, n = 2 experiments; EmptySS > 5xUAS-Chrimson, n = 3 experiments; pC1dSS1 > 5xUAS-Chrimson, n = 3 experiments. (B) 0.04 mW/mm2: EmptySS > Chrimson, n = 3 experiments; pC1dSS1 > Chrimson, n = 3 experiments; 0.1 mW/mm2: EmptySS > Chrimson, n = 4 experiments; pC1dSS1 > Chrimson, n = 4 experiments; 0.4 mW/mm2: EmptySS > Chrimson, n = 5 experiments; pC1dSS1 > Chrimson, n = 5 experiments. (C) 0.1 mW/mm2: EmptySS > 5xUAS-Chrimson, n = 6 experiments; pC1dSS1 > 5xUAS-Chrimson, n = 6 experiments; EmptySS > 20xUAS-Chrimson, n = 6 experiments; pC1dSS1 > 20xUAS-Chrimson, n = 7 experiments; 0.4 mW/mm2: EmptySS > 5xUAS-Chrimson, n = 6 experiments; pC1dSS1 > 5xUAS-Chrimson, n = 2 experiments; EmptySS > 20xUAS-Chrimson, n = 4 experiments; pC1dSS1 > 20xUAS-Chrimson, n = 6 experiments. Bars are mean ± S.E.M. Data was pooled from two independent biological replicates, which included separate parental crosses and were collected on different days. Data are representative of at least two independent biological repeats.
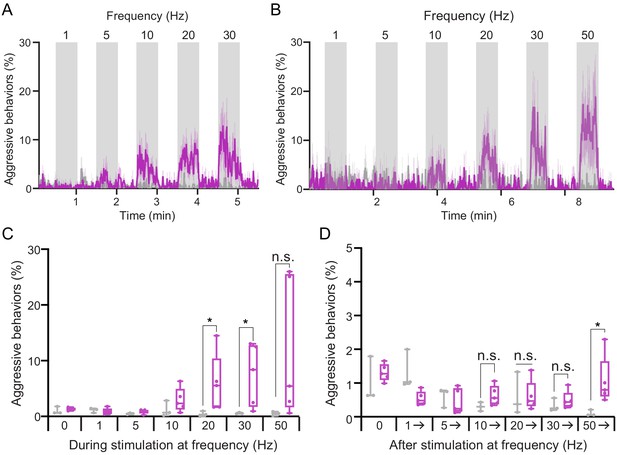
Behavioral effects of the frequency of optogenetic stimulation.
(A,B) Blocks of 30 s photostimulation (gray bars) with increasing stimulation frequency separated by 30 (A) or 60 (B) second intervals were delivered sequentially to pC1dSS1females. Light was delivered at 0.1 mW/mm2 with at a 10 ms pulse width, but with increasing pulse frequency. The pulse period and pulse number during each period was as follows: 1000 ms, 30 (1 Hz); 200 ms, 150 (5 Hz); 100 ms, 300 (10 Hz); 50 ms, 600 (20 Hz); 33 ms, 909 (30 Hz); 20 ms, 1500 (50 Hz). pC1dSS1 > 20xUAS-Chrimson is shown in purple; EmptySS > 20xUAS-Chrimson is shown in gray. The mean is represented as a solid line and shaded error bars represent variation between experiments. (C) Average percentage of flies performing aggressive behaviors over the 30 s period during stimulus delivery in B. (D) Average percentage of flies performing aggressive behaviors over the following periods: the 30 s before the onset of the first stimulus (1 Hz), the 60 s periods between subsequent stimuli, and the 30 s after the last stimulus (50 Hz) in B. Points indicate separate experiments. Data supporting the plot shown in the panel A were as follows: EmptySS > 20xUAS-Chrimson, n = 8 experiments; pC1dSS1 > 20xUAS-Chrimson, n = 8 experiments; (B) EmptySS > 20xUAS-Chrimson, n = 3 experiments; pC1dSS1 > 20xUAS-Chrimson, n = 5 experiments. Data was pooled from two independent biological replicates, which included separate parental crosses and were collected on different days. Data are representative of at least two independent biological repeats; see Supplementary file 3 for exact p-values. Box-and-whisker plots show median and IQR; whiskers show range. Mann-Whitney U-tests were used for statistical analysis. Asterisk indicates significance from 0: *p<0.05; n.s., not significant.
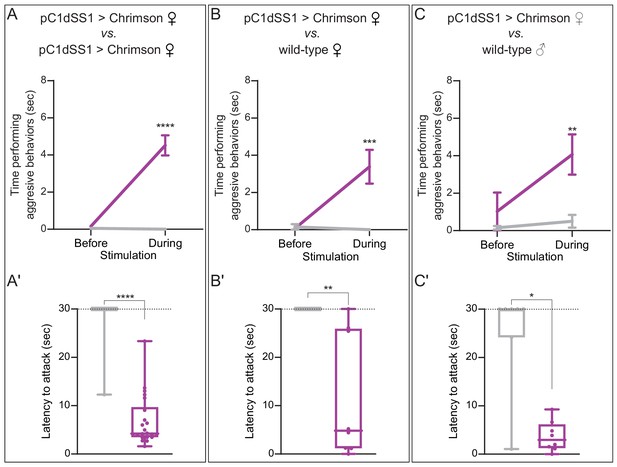
pC1d activation also increases aggression against wild-type females and males.
(A–C) Total time an individual spent performing aggressive behaviors over the 30 s period prior to or during stimulation, see Figure 2 legend for detail. (A'–C') Amount of time during a 30 s 0.1 mW/mm2 continuous stimulation period until first aggressive encounter. Points indicate individual flies. Dotted lines at 30 s indicate the end of the trial and error bars are mean ± S.E.M. Data supporting the plots shown in the individual panels were as follows: (A) EmptySS > 20xUAS-Chrimson, n = 22 flies; pC1dSS1 > 20xUAS-Chrimson, n = 22 flies; (B) EmptySS > 20xUAS-Chrimson, n = 8 flies; pC1dSS1 > 20xUAS-Chrimson, n = 8 flies; (C) EmptySS > 20xUAS-Chrimson, n = 7 flies; pC1dSS1 > 20xUAS-Chrimson, n = 8 flies. Data are representative of at least two independent biological repeats, one of which is shown here; see Supplementary file 3 for exact p-values. Box-and-whisker plots show median and IQR; whiskers show range. A Mann-Whitney U post hoc test was used for statistical analysis. Asterisk indicates significance from 0: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
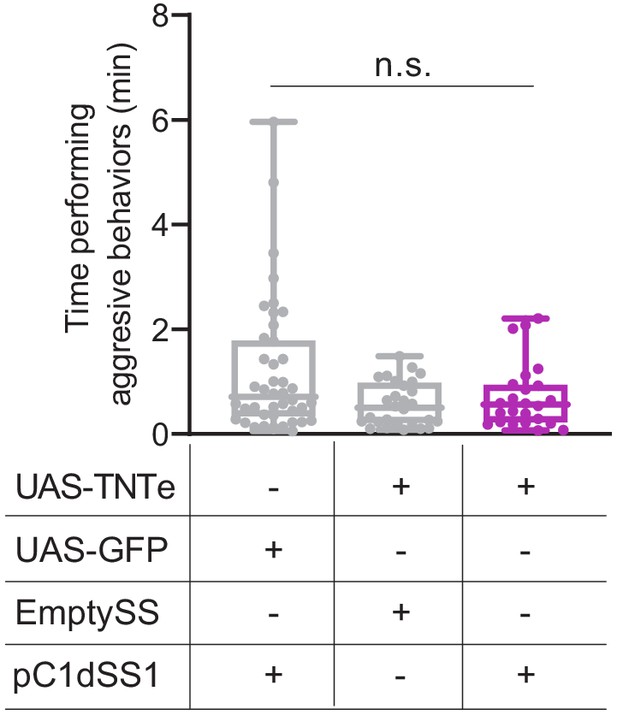
pC1d inactivation did not significantly diminish aggressive behavior.
Total time an individual spent performing aggressive behaviors over a 30 min trial; for more experimental detail see Figure 1—figure supplement 10E and Methods. Points indicate individual flies. pC1dSS1 > UAS-GFP, n = 42 flies; EmptySS > UAS-TNTe, n = 28 flies; pC1dSS1 > UAS-TNTe, n = 24 flies. Data are representative of at least two independent biological repeats, one of which is shown here; see Supplementary file 3 for exact p-values. Box-and-whisker plots show median and IQR; whiskers show range. Kruskal-Wallis and Dunn’s post hoc tests were used for statistical analysis. Asterisk indicates significance from 0: n.s., not significant.
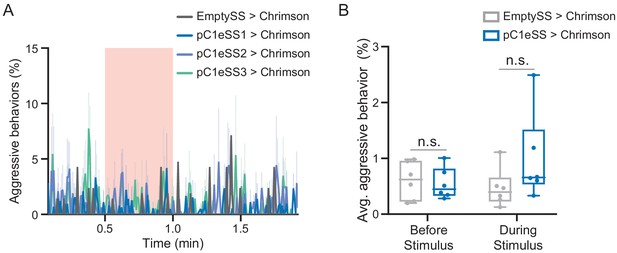
Optogenetic activation of additional lines labeling pC1e.
(A) Percentage of flies engaging in aggressive behaviors over the course of a trial during which a 30 s 0.1 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. (B) Average percentage of flies engaging in aggressive behaviors over the 30 s period prior to or during the delivery of a 0.1 mW/mm2 stimulus. Data supporting the plots shown in the individual panels were as follows: (A) EmptySS > Chrimson, n = 1 experiment; pC1eSS1 > Chrimson, n = 3 experiments; pC1eSS2 > Chrimson, n = 3 experiments; pC1eSS3 > Chrimson, n = 3 experiments. (B) EmptySS > 20xUAS-CsChrimson, n = 6 experiments; pC1eSS1 > 20xUAS-CsChrimson, n = 6 experiments. Data are representative of at least two independent biological repeats, one of which is shown here; see Supplementary file 3 for exact p-values. Box-and-whisker plots show median and IQR; whiskers show range. A Mann-Whitney U post hoc test was used for statistical analysis. Asterisk indicates significance from 0: n.s., not significant.
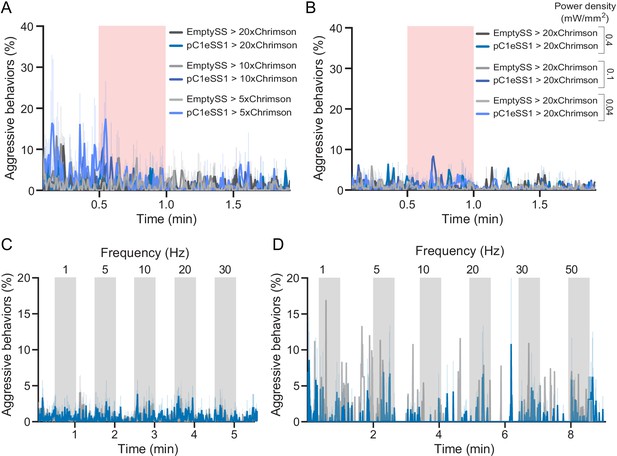
Behavioral effects of stimulus delivery and effector copy number.
(A) Percentage of flies engaging in aggressive behaviors over the course of a trial during which a 30 s 0.1 mW/mm2 continuous light stimulus (pink shading) was delivered to flies carrying either 5xUAS, 10XUAS or 20xUAS effectors, time course plotted as in Figure 1F. (B) Percentage of flies engaging in aggressive behaviors over the course of a 2 min trial during which a 30 s 0.4, 0.1, or 0.04 mW/mm2 continuous light stimulus (pink shading) was delivered, plotted as in Figure 1F. (C,D) Blocks of 30 s photostimulation (gray bars) with increasing stimulation frequency separated by 30 (C) or 60 (D) second intervals were delivered sequentially to females. Light was delivered at 0.1 mW/mm2 with a 10 ms pulse width and increasing frequencies resulting in higher numbers of pulses given over the 30 s stimulus on period. The pulse period and pulse number during each period was as follows: 1000 ms, 30 (1 Hz); 200 ms, 150 (5 Hz); 100 ms, 300 (10 Hz); 50 ms, 600 (20 Hz); 33 ms, 909 (30 Hz); 20 ms, 1500 (50 Hz). Data supporting the plots shown in the individual panels were as follows: (A) EmptySS > 20xUAS-Chrimson, n = 2 experiments; pC1eSS1 > 20xUAS-Chrimson, n = 2 experiments; EmptySS > 10xUAS-Chrimson, n = 3 experiments; pC1eSS1 > 10xUAS-Chrimson, n = 2 experiments; EmptySS > 5xUAS-Chrimson, n = 3 experiments; pC1eSS1 > 5xUAS-Chrimson, n = 2 experiments. (B) 0.04 mW/mm2: EmptySS > Chrimson, n = 3 experiments; pC1eSS1 > Chrimson, n = 3 experiments; 0.1 mW/mm2: EmptySS > Chrimson, n = 4 experiments; pC1eSS1 > Chrimson, n = 3 experiments; 0.4 mW/mm2: EmptySS > Chrimson, n = 5 experiments; pC1eSS1 > Chrimson, n = 5 experiments. (C) EmptySS > 20xUAS-Chrimson, n = 8 experiments; pC1eSS1 > 20xUAS-Chrimson, n = 7 experiments; (D) EmptySS1 > 20xUAS-Chrimson, n = 1 experiment; pC1eSS1 > 20xUAS-Chrimson, n = 2 experiments. Data are representative of at least two independent biological repeats, one of which is shown here.

Comparison of activation phenotypes of pC1d, pC1e and pC1a-c.
(A–C) Percentage of female flies engaged in aggressive behaviors (A) touching (B) or chasing (C) over the course of a trial during which a 30 s 0.1 mW/mm2 continuous light stimulus was delivered (pink shading), time courses plotted as in Figure 1F. EmptySS > 20xUAS-Chrimson, n = 4 experiments; pC1a-cSS > 20xUAS-Chrimson, n = 4 experiments; pC1dSS1 > 20xUAS-Chrimson, n = 5 experiments; pC1eSS1 > 20xUAS-Chrimson, n = 6 experiments. Data was pooled from two independent biological replicates, which included separate parental crosses and were collected on different days. Data are representative of at least two independent biological repeats. The pC1a-c line was provided by K. Wang and Barry Dickson.
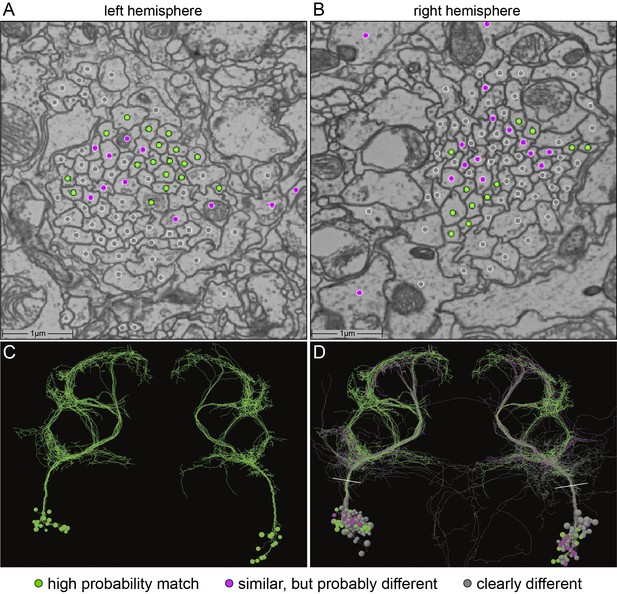
Identification of aIPg neurons in the FAFB dataset.
(A,B) Images of an area of the left and right hemisphere of an EM section from the FAFB dataset containing the fiber tracts of the putative aIP-g neurons described by Cachero et al., 2010. A dot has been placed in each axon, color-coded to reflect the degree of similarity of its morphology, revealed by manual tracing and visual inspection, to the aIPg neurons contained in our split-GAL4 lines. Gray represents neurons whose morphology clearly differed, magenta represents neurons whose morphology were similar but differed in one or more branches, and green represents neurons that we judged to correspond to those in our split-GAL4 lines. Note that, as we often observe in our split-GAL4 lines (see Figure 1—figure supplement 1) and has been reported for many cell types in connectomic studies (see for example, Bates et al., 2020), the number of neurons often differs between hemispheres. In this brain, the left hemisphere had 17 green cells, while the right hemisphere had only 12. (C,D) Skeleton rendering of the traced aIPg neurons, colored based on their similarity to the aIPg neurons identified in our split-GAL4 lines. Panel C shows only cells we judged to correspond to those in our split-GAL4 lines, while D shows all traced cells. White lines in D indicate the approximate plane of the images shown in A and B. Tracing of gray neurons was stopped when it was clear they did not match our split-GAL4 lines; therefore, their arbors are likely to be incomplete in these images.
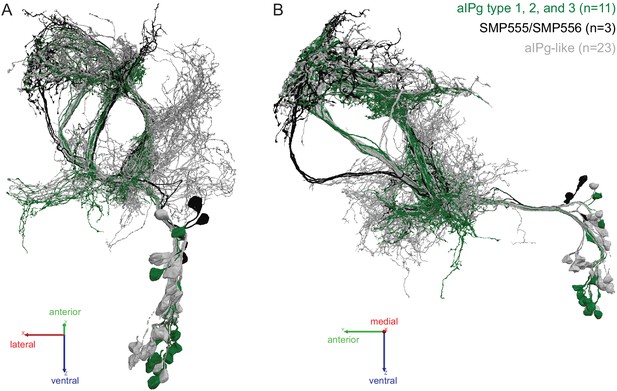
Identification of aIPg neurons in the hemibrain dataset.
(A,B) Skeleton renderings of the 37 putative aIP-g neurons described in Cachero et al., 2010 identified in the hemibrain dataset; two views are shown. The 11 aIPg type 1–3 neurons are displayed in green, the three SMP555/SMP556 neurons are displayed in black, and the remaining 23 putative aIPg neurons are displayed in gray. Note that the aIP-g cluster described in Cachero et al., 2010 included neurons with cell bodies in the anterior of the brain, none of which were traced for this project in the FAFB dataset or included in this grouping of neurons traced in the hemibrain as the neurons in our split-GAL4 lines only contained posterior located cell bodies.

Major inputs to and outputs from pC1d.
(A) Pre-synaptic inputs to the right-hemisphere pC1d neuron. All neurons making more than 100 synapses are shown, along with other neurons selected based on biological interest. Numbers in the arrows represent synapse number. The size of the circles representing input neurons indicates the estimated percentage of their output (number of synapses to pC1d/total output synapse number in the hemibrain volume to all ‘traced’ neurons) that goes to pC1d; note that vpoDN is a descending interneuron with outputs in the ventral nerve cord, which lies outside the hemibrain volume. Numbers in the circles indicate the number of neurons of that cell type present in the hemibrain, if greater than one. Note that, while there is only one pC1a neuron per hemisphere, both the left and right-hemisphere pC1a neurons make synapses onto the right-hemisphere pC1d. (B) Post-synaptic outputs of the single right-hemisphere pC1d. All neurons receiving more than 100 synapses from pC1d are shown, along with other neurons selected based on biological interest. Numbers in the arrows represent synapse number. The size of the circles representing output neurons indicates the percentage of their input (estimated by synapse number) that comes from pC1d. (C) Positions on the pC1d arbor of post-synaptic sites, color-coded to match diagram in (A). (D) Positions on the pC1d arbors of the presynaptic sites where the connections diagrammed in (B) occur, color-coded. (E) Positions of pC1d’s pre- (yellow) and post-synaptic (blue) connections to all neurons are shown; relevant brain areas are indicated. See Video 10 for better visualization of the inputs to pC1d and Video 12 for better visualization of pC1d’s outputs.
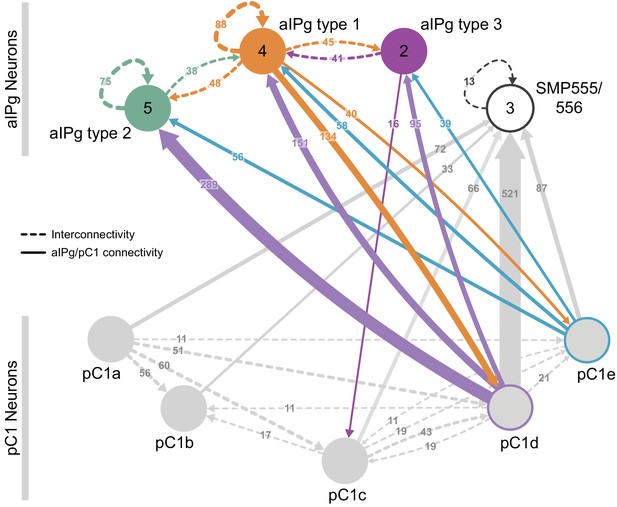
Reciprocal connections between aIPg and pC1 neurons.
(A) Interconnectivity between the right-hemisphere pC1a-e, aIPg type 1–3, and SMP555/556 neurons; only connections with 10 or more synapses are shown. SMP555/556 neurons also appear to derive from the aIP-g lineage (see Figure 5—figure supplement 1). Synapse number is noted on each arrow and dashed lines represent interconnectivity within the aIPg type 1, type 2 or type 3, SMP555/556 or pC1a – e neurons. Numbers in the circles indicate the number of neurons within the cell type present in the right brain hemisphere, if greater than one. Arrows are color-coded to correspond to the presynaptic cell type. See Video 8 for additional morphological detail on interconnectivity among aIPg neuron types and Video 11 for additional morphological detail on the connections between aIPg and pC1 neurons.
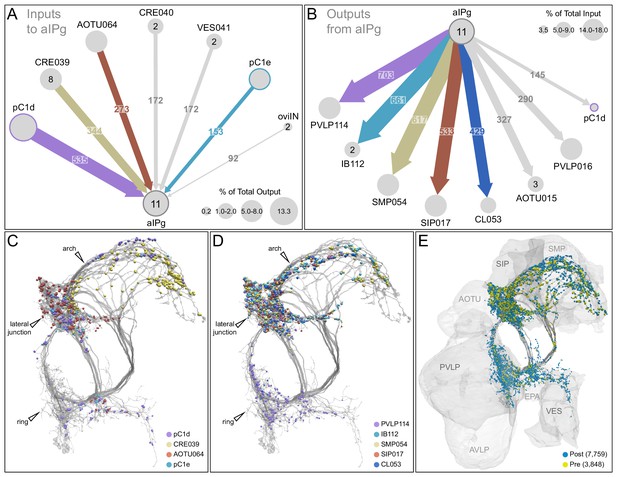
Major inputs to and outputs from aIPg neurons.
(A) Inputs to the 11 aIPg type 1–3 neurons in the right hemisphere; only connections with 150 or more synapses are shown and inputs to all 11 aIPg neurons have been pooled. OviIN was included, even though it did not meet this threshold, due to its involvement in other female behaviors. The size of the circles representing input neurons indicates the percentage of their output (estimated by synapse number) that goes to aIPg neurons. Numbers in the circles indicate the number of neurons of that cell type present, if greater than one. The number of cells and synapses given for a cell type include neurons in the left hemisphere if they are connected to the right-hemisphere aIPgs, with the exception of pC1d where only the right-hemisphere pC1d is shown; pC1d_L makes 71 synapses to aIPgs. (B) Post-synaptic outputs of aIPg neurons; only connections with 290 or more synapses are shown except for pC1d (145 synapses), which has been included to point out reciprocal connections. The size of the circles representing the downstream targets of aIPg indicates the percentage of their input (estimated by synapse number) that comes from aIPg neurons. Only one of the top downstream targets of the aIPg neurons, SMP054, also receives strong input from pC1d, although the connectivity strengths differ considerably between the two, with 14.4% and 1.1% of its inputs provided by aIPg and pC1d neurons, respectively. (C) Positions on the aIPg arbors of post-synaptic sites where the connections diagrammed in (A) occur, color-coded. (D) Positions on the aIPg arbors of the presynaptic sites where the connections diagrammed in (B) occur, color-coded. (E) Positions of aIPg’s pre- (yellow) and post-synaptic (blue) connections to all neurons are shown; relevant brain areas are indicated. See Video 13 for better visualization of the inputs to aIPg neurons and Video 14 for better visualization of their outputs.
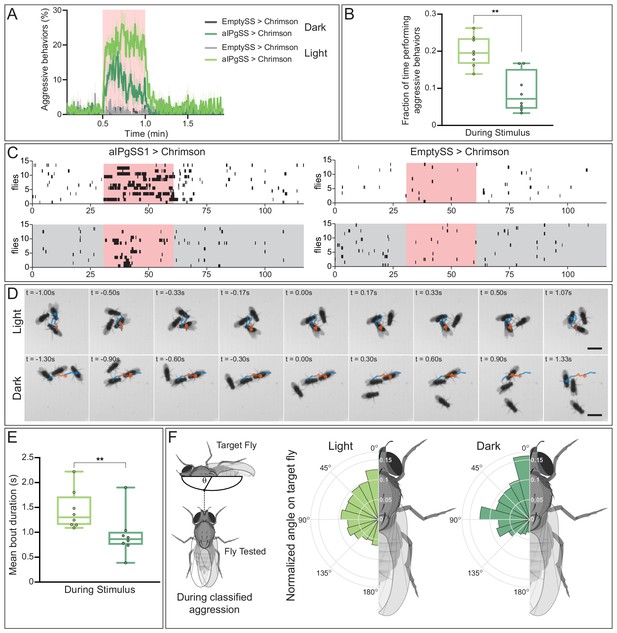
Absence of visual information alters aggressive behaviors during aIPgSS1 stimulation.
(A) Percentage of individual female flies engaging in aggressive behaviors over the course of a 2 min trial during which a 30 s 0.1 mW/mm2 continuous light stimulus was delivered (pink shading). Trials were conducted either in the presence or absence of visible light, as indicated. (B) Fraction of time individuals spent performing aggressive behaviors during the stimulus period. Each dot represents the mean of the individuals within each experiment. (C) Ethogram plots of the aggressive events in light (white) and dark (gray) conditions. Each row represents one individual in a trial and black bars represent classified aggressive events. The stimulus period is indicated in pink and the ethograms were chosen to represent a typical trial for each condition. (D) Time course of example aggressive events during a single bout in light (top) and dark (bottom) conditions. The trajectory of one fly is shown both prior to (blue), during (orange), and immediately following (blue) an aggressive bout. The time before (-) and after (+) a central frame of the behavior is shown in the upper left corner of each image. Note that although average bout length in longer in the light, we selected bouts of similar length for display to facilitate comparison. The scale bar (3 mm) applies throughout each condition. See Video 15 for examples of multiple bouts in the light and dark. (E) Mean length of classified aggressive behavioral events during the stimulus period. Event lengths were calculated from the per-frame classifications. Each dot represents the mean of the individuals within each experiment. (F) Polar plots of the angle on the targeted fly (θ), measured as shown in the diagram at the left, during classified aggressive events. Each wedge represents 15 degrees and plots were normalized within each condition. Light: EmptySS > Chrimson, n = 89 flies in eight experiments; aIPgSS1 > Chrimson, n = 136 flies in eight experiments; Dark: EmptySS > Chrimson, n = 90 flies in eight experiments; aIPgSS1 > Chrimson, n = 126 flies in eight experiments. Box-and-whisker plots in panels B and E show median and IQR; whiskers show range. A Mann-Whitney U test was used for statistical analysis. Asterisk indicates significance: **p<0.01.
-
Figure 9—source data 1
Source data for behavioural experiments in Figure 9.
- https://cdn.elifesciences.org/articles/58942/elife-58942-fig9-data1-v2.xlsx
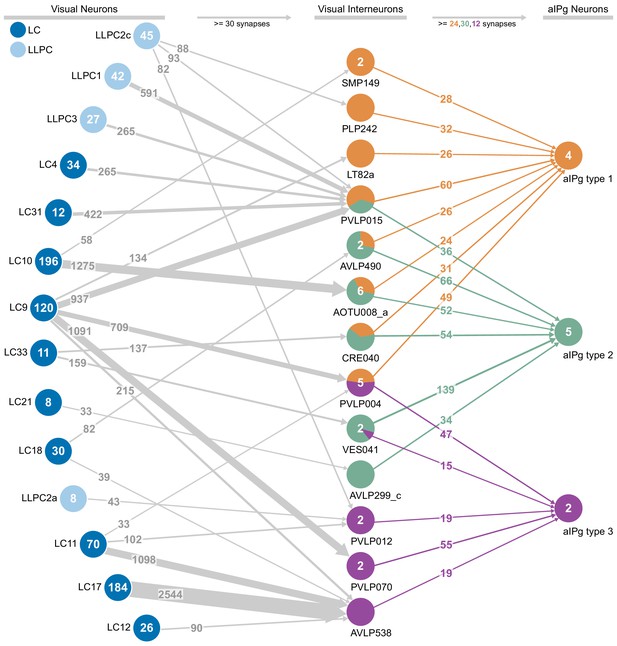
Connections between visual projection neurons, interneurons, and aIPg type 1, type 2 and type 3.
Connections with 30 or more combined synapses for each population of visual projection neurons to interneurons are shown. Inputs to the aIPg neurons were thresholded in proportion to the number of cells in each aIPg type resulting in thresholds of 24, 30, and 12 synapses to type 1, type 2, and type 3, respectively. The color-coding of the circles representing interneurons indicates the fraction of their outputs going to each aIPg type.

Connection paths of aIPg and pC1d neurons to descending neurons.
(A) The numbers within the arrows represent synapse numbers; only connections with greater than 50 synapses between cell types are shown. Cell types are labelled and the number of neurons for cell types with more than one cell that participate in the diagrammed connections are given inside the circle representing the cell. The proportion of inputs from type 1 and type 2 aIPg neurons is indicated by the color-coding of the circle. AOTU-innervating neurons that also get input of more than 500 synapses from LC10 neurons are highlighted with a dark green circumference (see panel B). Some of the neurons downstream of aIPg (AOTU023 and VES041) make bilateral connections to DNs and others (AOTU019 and SMP148) make contralateral connections. For this reason, the right (RHS, above the line representing the midline) and left (LHS, below the line representing the midline) brain hemispheres are diagrammed separately to show all connections to right-hemisphere DNs. DNa03 is strongly connected to DNa02 in the LAL and VES. (B) AOTU-innervating cell types that make more than 100 synapses onto one or more DN cell types and that also get strong input (more than 500 combined synapses) from LC10 neurons are shown. The aIPg type 1 and type 2 cell types make inputs onto the majority of these AOTU neurons, as shown; the relative proportion of inputs from type 1 and type two neurons is indicated by color-coding of the circle representing the AOTU neuron. The numbers within the arrows represent synapse number. The numbers in the circles indicate the number of cells within that cell type in the right brain hemisphere that participate in the indicated connections. For example, 386 of the 449 LC10 cells identified in the hemibrain volume make connections to at least one of the shown AOTU neurons. Only about half the known DN cell types have been definitively identified in the hemibrain v1.1 dataset and thus there are likely to be additional DNs that meet our criteria that are not shown here.
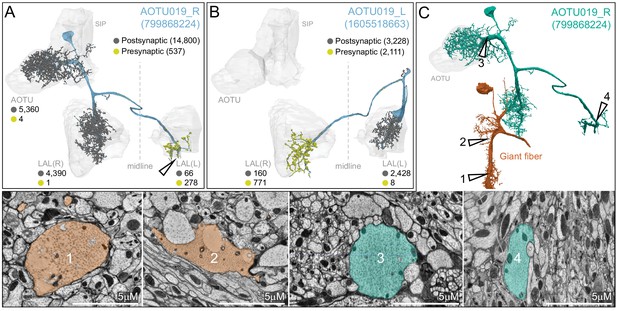
Morphology and connectivity of AOTU019.
(A,B) The right (A) and left (B) hemisphere AOTU019 cells are shown in order to illustrate the complete morphology of AOTU019 which makes bilateral connections and extends outside of the hemibrain volume. AOTU019 gets ipsilateral inputs (gray dots) in the AOTU, SIP and LAL and makes most of its output synapses (yellow dots) in the contralateral LAL and VES. Synapse numbers are indicated for reconstructed portion of the entire neuron as well as separately for each brain region. (C) A comparison of the morphologies of the giant fiber and AOTU neurons to illustrate the large diameter of the AOTU19 axons. Cross-sections of the axons of each neuron at the positions of the arrowheads are shown below.
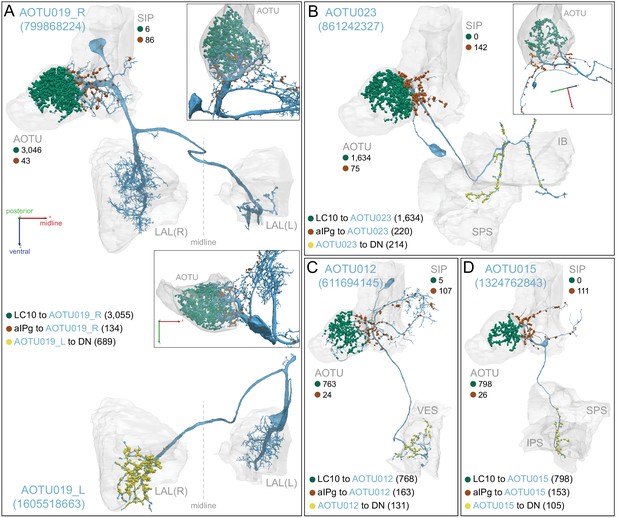
Synaptic inputs from LC10 and aIPg neurons are segregated in different dendritic arbors of AOTU neurons.
The synapses from LC10 neurons (green dots) and aIPg neurons (brown dots) occur in distinct arbors on AOTU019 (A), AOTU023 (B), AOTU012 (C) and AOTU015 (D). Neuronal morphologies of the indicated neurons and synapse numbers are shown; selected brain areas are indicated. Only one of the three right-hemisphere AOTU15 neurons (neuPrint ID: 1324762843) is shown. Output synapses to DNs are indicated by yellow dots. The inserts in panels A and B present rotated views at the orientation shown in the panel B inset.
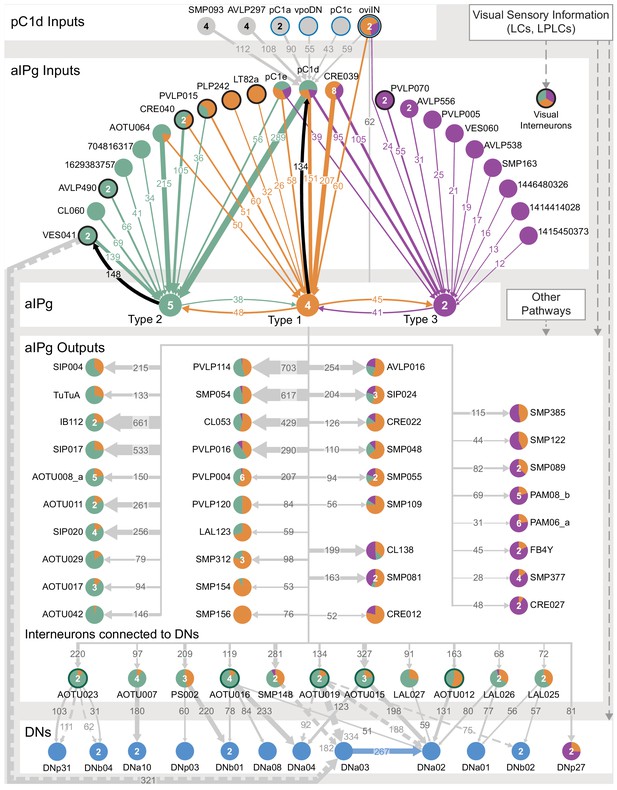
Proposed circuit underlying female aggressive behaviors.
A diagram of key neuronal pathways centered on pC1d and aIPg type1, type 2 and type 3 neurons is shown. The numbers in arrows represent synapse number. We propose that pC1d facilitates female aggressive behaviors by acting through aIPg type 1 (orange), type 2 (green), and type 3 (purple) neurons. The top four inputs to pC1d and three other neurons of interest are shown. These include neurons implicated in other female behaviors (highlighted with a blue circumference), including oviposition (oviIN, pC1a) and mating (vpoDN), suggesting communication between the circuits underlying these behaviors and aggression. We propose that aIPg neurons integrate diverse upstream signals and then alter brain state in a way that increases the likelihood of engaging in aggressive social interactions. Upstream neuronal populations are color-coded by the proportion of their synapses they make onto the aIPg type 1, aIPg type 2, and aIPg type 3 subtypes. The post-synaptic targets of aIPg type 1, aIPg type 2, and aIPg type 3 neurons are shown color-coded by the relative proportion of their synaptic input they receive from each aIPg type. AOTU neurons that also get input from LC10 neurons (see Figure 10B) are highlighted with a dark green circumference. Inputs to the aIPg neurons were thresholded in proportion to the number of cells in each aIPg type resulting in thresholds of 24, 30, and 12 synapses to type 1, type 2, and type 3, respectively. Downstream outputs receiving more than 50 synapses from aIPg neurons (outputs from all 11 aIPg neurons pooled) are shown, with neurons receiving fewer synapses included when a majority came from aIPg type 3. For cell types that get input from multiple types of aIPg neurons, the relative proportions of inputs from different types are indicated by the color-coding of the circle. Connections with DNs with more than 50 synapses are shown. VES041 neurons have strong reciprocal connections with aIPg type 2 neurons; both the right and left hemisphere VES041 neurons are connected to the right-hemisphere DNa03 (321 total synapses; see Figure 9A) and are shown as solid and dotted arrows, respectively, in this diagram. Connections from AOTU019, SMP148, and AOTU023 from the contralateral hemisphere to DNs are similarly shown as dotted arrows (see Figure 10A).

Alternative models for how the saliency of visual input from LC10 neurons is increased by arousal during two different social behaviors.
(A) A conceptual model for gating of visual information by aIPg neurons in females during aggression. Several types of projection neurons (represented by the gray neuron) connect the AOTU with ventral brain regions where they then innervate descending neurons (blue) to drive motor behavior (Figure 10B). These projection neurons get input from both aIPg neurons and LC10 neurons, a population of visual projection neurons that are tuned to be activated by a nearby moving fly. The combined activation of these excitatory cholinergic inputs might act synergistically to drive AOTU firing and signaling to DNs. At this point we do not know if the anatomical separation of the aIPg and LC10 inputs to the AOTU neurons (Figure 10—figure supplement 2) is simply a wiring convenience or has functional significance. The dotted arrow from aIPg to LC10 indicates an indirect path mediated by two types of TuTuA neurons (Figure 12—figure supplement 1). (B) A conceptual model based on the work of Sten et al., 2020 for the gating of visual information by P1 neurons in males during courtship. P1 neurons are proposed to act upstream of LC10 such that LC10 neurons produce more synaptic output when male flies are aroused. The nature of the signal transmission between P1 and LC10 has not been determined. See Sten et al., 2020 for details. We note that in addition to transmission at chemical synapses, signaling by neuropeptides also remains a possibility in both models: RNA profiling of LC10a and LC10b neurons from a mixture of males and females (Davis et al., 2020) has shown that both these cell types express the receptor for sNPF (sNPF-R) and our work showed that aIPg neurons express sNPF (Figure 1—figure supplement 5). Similarly, LC10a neurons express two receptors for Tk (TkR86C and TkR99D), while LC10b neurons appear to express only TkR86C at high levels. Tk is known to be an important regulator of male aggression (Asahina et al., 2014), but a role for Tk in courtship has not been established.
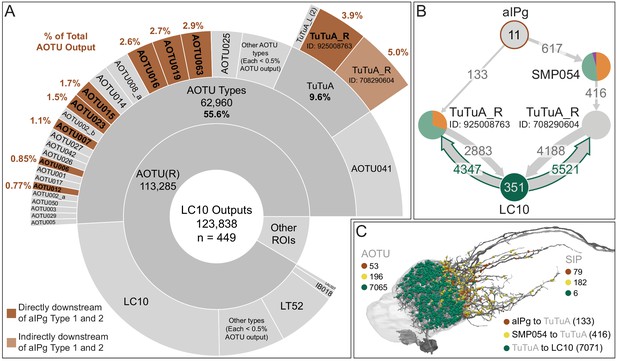
Connectivity of LC10 neurons in the AOTU.
(A) A chart showing the outputs of the 449 LC10 neurons identified in the right brain hemisphere. Cell types that also get aIPg input are shaded brown and the percentage of LC10 output synapses in the AOTU(R) they each receive is indicated. More information about the connectivity of these cells is shown in Figure 10, Figure 10—figure supplement 2 and panel B of this figure. (B) A diagram of two indirect paths from aIPg neurons to LC10 neurons, each mediated by a different TuTuA_R neuron (neuPrint IDs: 925008763 and 708290604). (C) Neuronal morphologies underlying connections diagrammed in panel B. Color-coded synaptic inputs to the two TuTuA neurons (shown in different shades of gray) from aIPg and SMP054 as well as the synapses from the TuTuA_R neurons to LC10 are shown. The AOTU brain region is shown in light gray.
Videos
Behavior of EmptySS > Chrimson flies.
Behavior of aIPgSS1 > Chrimson flies.
High-speed video of two aIPgSS1 > Chrimson female flies.
High-speed video of an aIPgSS1 > Chrimson female fly with a wild-type male.
Behavior of individual aIPgSS1 > Chrimson and EmptySS > Chrimson flies.
Behavior of pC1dSS1 > Chrimson flies.
High-speed video of two pC1dSS1 > Chrimson female flies.
Interconnectivity of aIPg types 1–3 neurons.
Locations of pre- and post-synaptic connections with aIPg type 1–3 neurons.
Top inputs to pC1d.
Interconnectivity of aIPg types 1–3, pC1d and pC1e neurons.
Top outputs from pC1d.
Top inputs to aIPg types 1–3 neurons.
Top outputs from aIPg types 1–3 neurons.
Comparison of aggressive bouts under lighted and dark conditions.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Drosophila melanogaster) | VT064565-x-VT043699 | this paper | splitgal4.janelia.org: SS36564 | split-GAL4 driver line targeting aIPg (aIPgSS1) |
| Genetic reagent (Drosophila melanogaster) | VT043699-x-R11A07 | this paper | splitgal4.janelia.org: SS36551 | split-GAL4 driver line targeting aIPg (aIPgSS2) |
| Genetic reagent (Drosophila melanogaster) | R11A07-x-VT043700 | this paper | splitgal4.janelia.org: SS32237 | split-GAL4 driver line targeting aIPg (aIPgSS3) |
| Genetic reagent (Drosophila melanogaster) | R72C11-x-R11A07 | this paper | splitgal4.janelia.org: SS47478 | split-GAL4 driver line targeting aIPg (aIPgSS4) |
| Genetic reagent (Drosophila melanogaster) | R11A07-x-R72C11 | this paper | splitgal4.janelia.org: SS56964 | split-GAL4 driver line targeting aIPg (aIPgSS5) |
| Genetic reagent (Drosophila melanogaster) | R35C10-x-R71A09 | this paper | splitgal4.janelia.org: SS56987 | split-GAL4 driver line targeting pC1d (pC1dSS1) |
| Genetic reagent (Drosophila melanogaster) | R71A09-x-R26F09 | this paper | splitgal4.janelia.org: SS57598 | split-GAL4 driver line targeting pC1d (pC1dSS2) |
| Genetic reagent (Drosophila melanogaster) | VT25602-x-VT002064 | this paper | splitgal4.janelia.org: SS43274 | split-GAL4 driver line targeting pC1d and pC1e (pC1dSS3) |
| Genetic reagent (Drosophila melanogaster) | R35C10-x-VT002063 | this paper | splitgal4.janelia.org: SS59331 | split-GAL4 driver line targeting pC1d and pC1e (pC1dSS4) |
| Genetic reagent (Drosophila melanogaster) | R35C10-x-R60G08 | this paper | splitgal4.janelia.org: SS59336 | split-GAL4 driver line targeting pC1e (pC1eSS1) |
| Genetic reagent (Drosophila melanogaster) | R60G08-x-VT002063 | this paper | splitgal4.janelia.org: SS39313 | split-GAL4 driver line targeting pC1e (pC1eSS2) |
| Genetic reagent (Drosophila melanogaster) | R60G08-x-R35C10 | this paper | splitgal4.janelia.org: SS59433 | split-GAL4 driver line targeting pC1e (pC1eSS3) |
| Genetic reagent (Drosophila melanogaster) | R18A05-x-dsx-ZpGal4DBD | this paper; gift of K. Wang and B. Dickson | SS75230 | split-GAL4 driver line targeting pC1a-c |
| Genetic reagent (Drosophila melanogaster) | BPp65AD-x-BPZpGal4DBD | Pfeiffer et al., 2010; Aso et al., 2014 | emptySS | split-GAL4 driver line empty brain control |
| Genetic reagent (Drosophila melanogaster) | 20XUAS-CsChrimson-mVenus | Klapoetke et al., 2014; Aso et al., 2014 | ||
| Genetic reagent (Drosophila melanogaster) | 10XUAS-CsChrimson-mVenus | Klapoetke et al., 2014; Aso et al., 2014 | ||
| Genetic reagent (Drosophila melanogaster) | 5XUAS-CsChrimson-mVenus | Klapoetke et al., 2014; Aso et al., 2014 | ||
| Genetic reagent (Drosophila melanogaster) | MCFO-1 | Nern et al., 2015 | ||
| Genetic reagent (Drosophila melanogaster) | pJFRC2-10XUAS-IVS-mCD8::GFP | Pfeiffer et al., 2010 | ||
| Genetic reagent (Drosophila melanogaster) | UAS-impTNT-HA | Sweeney et al., 1995 | ||
| Genetic reagent (Drosophila melanogaster) | UAS-TNT-E | Sweeney et al., 1995 | ||
| Antibody | nc82 supernatant; Mouse α-bruchpilot monoclonal | Developmental Studies Hybridoma Bank | Cat #: nc82-s | https://www.janelia.org/project-team/flylight/protocols Dilution, (1:30) |
| Antibody | Mouse α-V5 Tag monoclonal | AbD Serotec | Cat #: MCA1360D550GA | Dilution (1:500) |
| Antibody | Rabbit α-HA Tag monoclonal | Cell Signaling Technologies | Cat #: 3724S | Dilution (1:300) |
| Antibody | Rat α-FLAG Tag (DYKDDDDK Epitope tag) monoclonal | Novus Biologicals | Cat #: NBP1-06712 | Dilution (1:200) |
| Antibody | rabbit anti-GFP polyclonal | Thermo Fisher Scientific | Cat #: A-11122; RRID:AB_221569 | Dilution, (1:1000) |
| Antibody | Alexa Fluor 488 goat anti-rabbit polyclonal | Thermo Fisher Scientific | Cat #: A-11034; RRID:AB_2576217 | Dilution, (1:800) |
| Antibody | Alexa Fluor 568 goat anti-mouse polyclonal | Thermo Fisher Scientific | Cat #: A-11031; RRID:AB_144696 | Dilution, (1:400) |
| Antibody | ATTO 647N goat anti-rat polycolonal | Rockland Immunochemicals Inc | Cat #: AB_10893386 | Dilution, (1:300) |
| Chemical compound, drug | paraformaldehyde | Electron Microscopy Sciences | 15713 s | https://www.janelia.org/project-team/flylight/protocols |
| Chemical compound, drug | Triton X-100 | Sigma–Aldrich | X100 | https://www.janelia.org/project-team/flylight/protocols |
| Chemical compound, drug | DPX Mountant | Electron Microscopy Sciences | #13512 | https://www.janelia.org/project-team/flylight/protocols |
| Chemical compound, drug | xylene | Fisher Scientific | x5-500 | https://www.janelia.org/project-team/flylight/protocols |
Additional files
-
Supplementary file 1
Framewise performance of automated classifiers.
- https://cdn.elifesciences.org/articles/58942/elife-58942-supp1-v2.xlsx
-
Supplementary file 2
List of hemibrain v1.1 unique neuron identifier numbers and synapse numbers for connected neurons.
- https://cdn.elifesciences.org/articles/58942/elife-58942-supp2-v2.xlsx
-
Supplementary file 3
Sample size and statistics for behavioral analysis.
- https://cdn.elifesciences.org/articles/58942/elife-58942-supp3-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58942/elife-58942-transrepform-v2.docx





