Impaired speed encoding and grid cell periodicity in a mouse model of tauopathy
Figures
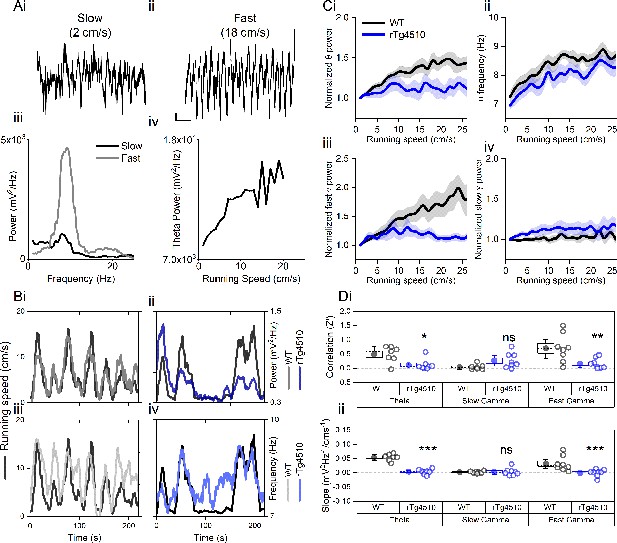
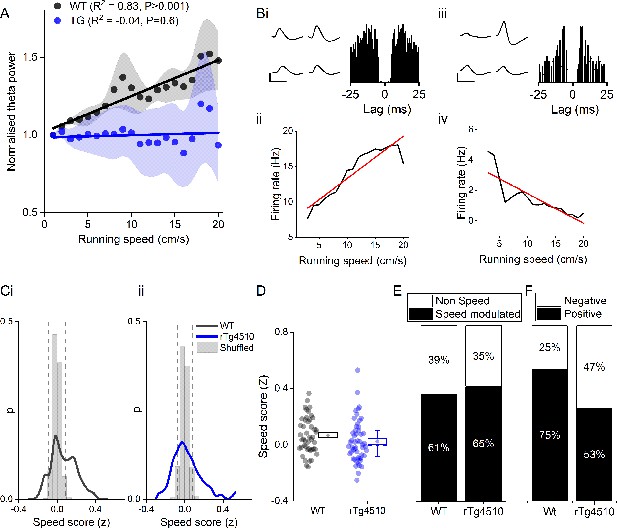
Oscillation-running speed relationship is impaired in rTg4510 mice.
(A) Local field potential from WT mice showing periods of slow (i) and fast (ii) running speed showing faster and larger theta oscillations during locomotor activity. (iii) Power spectra of data shown in i and ii for slow (black) and fast (grey) running periods. (iv) Example relationship between running speed and average theta oscillation power across recording session. (B) Example plots showing animals running speed on linear track (black, left Y axis) showing high correlation with theta oscillation (grey, right Y axis) power (i) and frequency (iii) over several minutes of recording. Corresponding example from rTg4510 mouse showing with theta oscillation amplitude (ii) and frequency (iv) with decreased association with running speed. (C) Running speed-theta oscillation relationships for power (i; normalised to 1–2 cm/s bin) and frequency (ii). Also shown are fast (iii) and slow (iv) gamma power-running speed relationships. (D) Pooled data for each animal showing Z-transformed correlation coefficients (i) and slopes (ii) of running speed-oscillatory power relationships for different frequency bands (*p<0.05, **p<0.01, ***p<0.001, ns = not significant, Bonferroni-corrected pairwise-multiple comparisons; for 2-way repeated measures ANOVA main effects and interactions see Table 1).
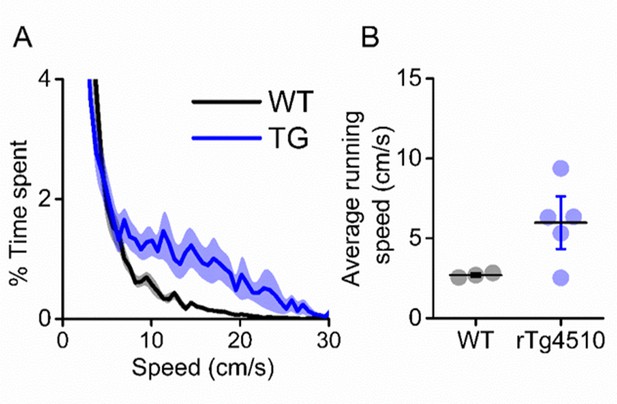
rTg4510 mice display hyperactive phenotype.
(A) Breakdown of animals time spent at eat running speed, showing rTg4510 mice spend more time moving at higher velocities. (B) Average running speed across (mean ± SEM) entire recording session is greater in rTg4510 mice (P=0.04, Unpaired T-test).
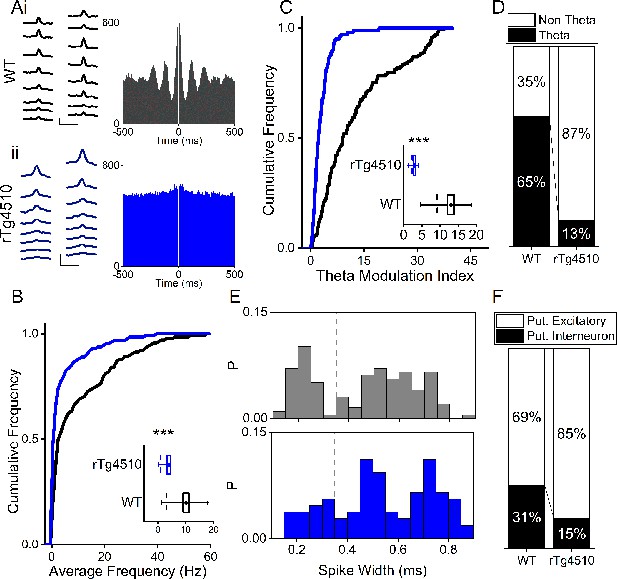
Firing properties of mEC single units.
(A) Average waveforms from an example cell recorded from a 16-channel silicone probe shank for WT (i) and rTg4510 (ii) mouse, with firing autocorrelations. Scale bars: 0.4 ms, 50 µV. (B) Cumulative frequency plot of theta modulation index for all recorded mEC single units, with average modulation for WT (black) and rTg4510 (blue) mice inset. (C) Proportion of cells displaying theta modulation (threshold: TMI>5). (D) Average firing frequency across entire recording session of mEC neurons for WT (black) and rTg4510 (blue) mice, average inset. (E) Spike-width probability histogram for WT (black) and rTg4510 (blue) units. (F) Proportion of cells classified as putative interneurons (spike-width <0.35 ms, dotted line in (E) and putative excitatory. Box plots: dotted line: median, diamond: mean ± SEM, whiskers: 25th/75th centile), *** p <0.001 Mann-Whitney U test.
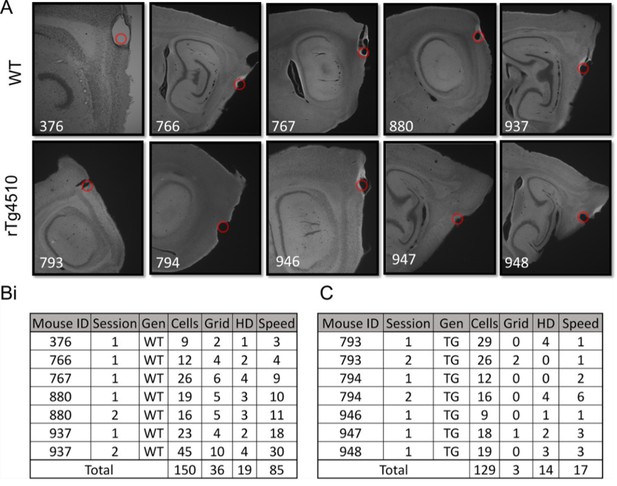
Recording of mEC single units.
(A) Example sections showing electrode lesions (red dot) of final probe location in mEC of WT and rTg4510 mice. (B) Breakdown of cells recorded from each animal, for each recording location for wildtype (WT) mice. (C) Breakdown of cells from rTg4510 (TG) mice.
-
Figure 2—figure supplement 1—source data 1
Example images of electrolytic lesions for each mouse.
- https://cdn.elifesciences.org/articles/59045/elife-59045-fig2-figsupp1-data1-v2.zip
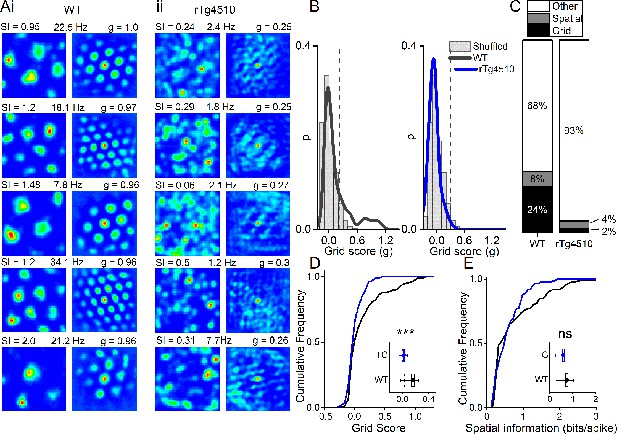
Breakdown of grid cell periodicity in rTg4510 mice.
(A) Example spatial firing patterns (left) and 2D autocorrelations (right) of cells from WT (i) and rTg4510 (ii) mice in a 0.8 x 0.8 m square arena, displayed with grid score (g), spatial information content (SI) and peak firing rate across recording environment. Five cells with the highest grid score are displayed for each genotype, showing irregular firing patterns even in the rTg4510 cells with the highest grid scores. (B) Histograms of grid scores for all single units recorded from WT and rTg4510 mice. Observed data plotted as a solid line, shuffled data shown as grey bars. The 95th centile of shuffled distributions is plotted as a dashed line. (C) Cumulative frequency plots for grid score; inset box plot illustrating average values for each genotype (dotted line: median, diamond: mean ± SEM, whiskers: 25 th /75 th centile), *** p<0.001, Mann-Whitney U test. (D) Proportions of grid and spatial non-grid cells greater than threshold in WT and rTg4510 mice. (E) Cumulative frequency plots for spatial information score; inset box plot illustrating average values for each genotype (dotted line: median, diamond: mean ± SEM, whiskers: 25th/75th centile), ns p>0.05, Mann-Whitney U test.
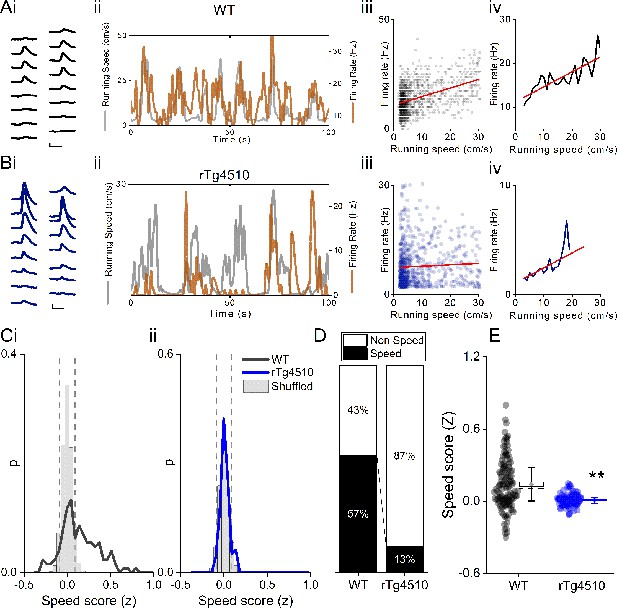
Decreased speed modulation of MEC single units in rTg4510 mice.
Average waveforms of a single unit recorded from a 16-channel shank silicone probe, from a WT (Ai; black) and rTg4510 (Bi; blue) mouse. The running speed (grey) and cell firing rate (orange) (ii), correlation between running speed and firing rate for each time bin (40 ms) (iii), and average for each speed bin (1 cm/s) (iv) are shown for each of these example cells. Red line: linear fit for each. Scale bars: 0.3 ms, 25 µV. (C) Distribution of speed scores for WT (i) and rTg4510 units (ii) with shuffled distribution of scores (grey bars); 5th/95th centile threshold: dotted lines. (D) Proportion of cells classified as speed modulated (>95th or <5th centile of shuffled distribution). (E) Average speed score for each recorded MEC unit. Box plots: dotted line: median, diamond: mean ± SEM, whiskers: 25th/75th centile, **p<0.01, Mann-Whitney U test.
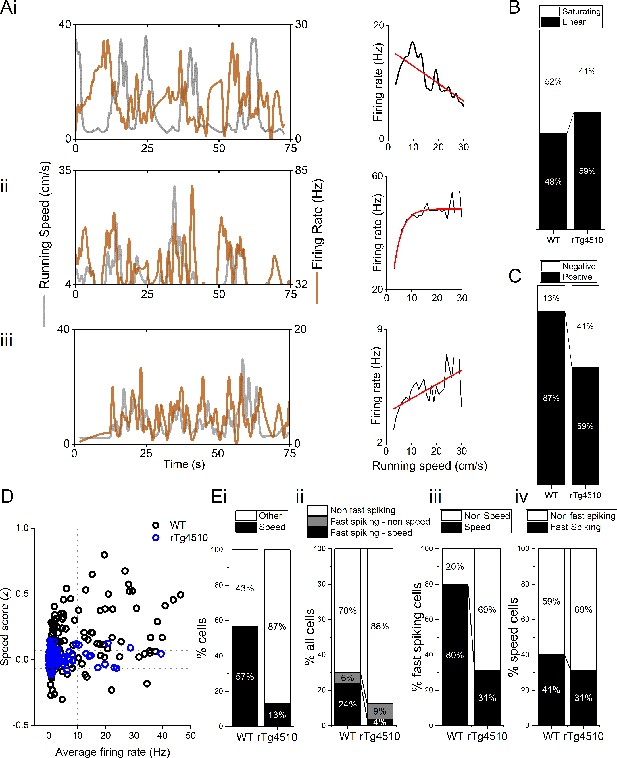
Properties of MEC speed-modulated cells.
(A) Example negative (i), saturating (ii), and linear (iii) speed-modulated cells, with running speed (grey) and cell firing rate (orange), with average for each speed bin (1 cm/s) (right). Red line: linear/exponential fit for each. (B) Proportion of speed-modulated mEC cells that are best described by linear and saturating fits for WT and rTg4510 mice. (C) Proportion of speed-modulated units that show positive (>95th centile of shuffled distribution) and negative (<5th centile of shuffled distribution) speed modulation. (D) Plot of speed score vs average firing rate, showing faster spiking cells are more likely to have high-speed modulation in WT, but not rTg4510 mice. (E)(i) Proportion of speed-modulated cells in WT and rTg4510 mice (replotted from Figure 3E). (ii) Proportions of cells classified as fast-spiking (>10 Hz), with breakdown into speed and non-speed modulators for each genotype. (iii) Proportion of fast-spiking cells which display speed modulation, which shows decreased numbers in rTg4510 mice compared to WT. (iv) Proportion of speed-modulated cells which are classified as fast spiking shows only small decrease in rTg4510 mice.
-
Figure 4—figure supplement 1—source data 1
Example images of electrolytic lesions for each mouse.
- https://cdn.elifesciences.org/articles/59045/elife-59045-fig4-figsupp1-data1-v2.zip
Speed modulation of single units in hippocampal CA1 pyramidal cell layer also shows similar increase in negatively speed-modulated firing.
(A) Normalised theta oscillation amplitude from WT and rTg4510 mice with increasing running speed, with average linear regression above (WT: R2 = 0.83, p < 0.001, n = 6 mice; rTg4510: R2 = -0.04, p = 0.6, n = 4 mice). (B) Example speed-modulated cells recorded from the CA1 pyramidal cell layer. The cell on the left (i and ii) was positively speed-modulated, whereas the cell on the right (iii and iv) was negatively speed-modulated. (C) Histograms of speed scores for WT and rTg4510 mice. Fifth and 95th centiles of shuffled data shown as dotted lines. (D) Box plot comparing speed scores between the two genotypes (E) Proportion of cells passed criteria for speed-modulated firing is approximately even between genotypes (WT: 25/46 units, rTg4510: 27/52 units). (F) Increased proportion of negatively speed-modulated units in CA1 in rTg4510 mice, compared to WT controls (WT: 5/25 units, rTg4510:13/27 units; χ² (1) = 4.5, p = 0.03, Chi-Square test).
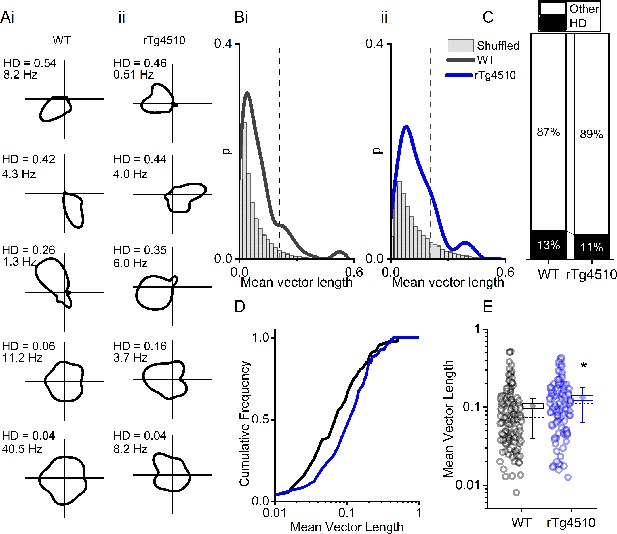
rTg4510 mice retain MEC head-direction (HD) tuning.
(A) Example HD tuning of 5 WT and 5 rTg450 units, displayed with HD score and peak firing rate across HD bins. (B) Distribution of HD scores for WT (i) and rTg4510 mice (ii) with shuffled distribution of scores (grey), 95th-centile threshold: dotted line. (C) Proportions of cells with HD scores over threshold in WT and rTg4510 mice. (D) Cumulative frequency distribution of HD scores (mean vector length). (E) Mean vector length for all cells, showing a small but significant increase in HD tuning across the population in rTg410 mice (dotted line: median, diamond: mean ± SEM, whiskers: 25th/75th centile), * p<0.05 Mann-Whitney U test.
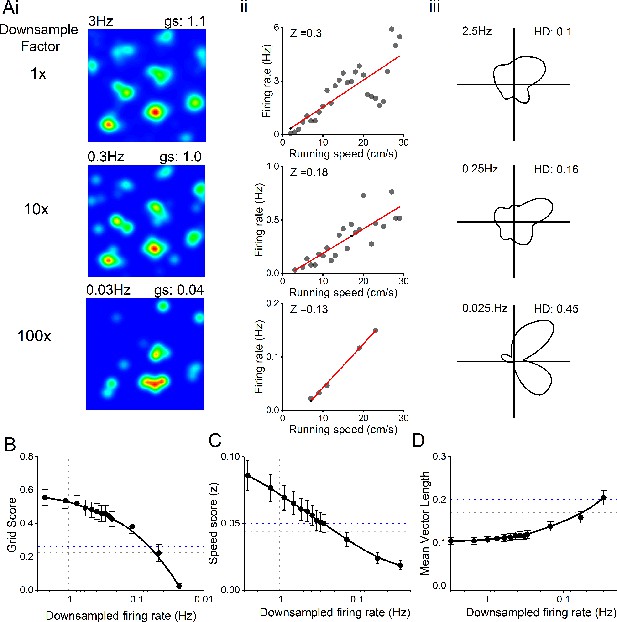
Decreased firing rate does not account for changes in spatial metrics.
(A) Example grid (i), speed (ii), and head-direction (iii) cell firing rate maps, with progressive down sampling of spikes. (B) Effect of down-sampled spike trains on grid score output on WT neurons. Horizontal line: grid score threshold from 95th centile of shuffled distributions for WT (grey) and rTg4510 (blue) mice. Vertical line: Average firing of rTg4510 MEC neurons. (C) Effect of down sampled spike trains on speed score output on WT neurons. Horizontal line: grid score threshold from 95th centile of shuffled distributions. Vertical line: Average firing of rTg4510 MEC neurons. (D) Effect of down sampled spike trains on head-direction score output on WT neurons. Horizontal line: grid score threshold from 95th centile of shuffled distributions. Vertical line: Average firing of rTg4510 MEC neurons.
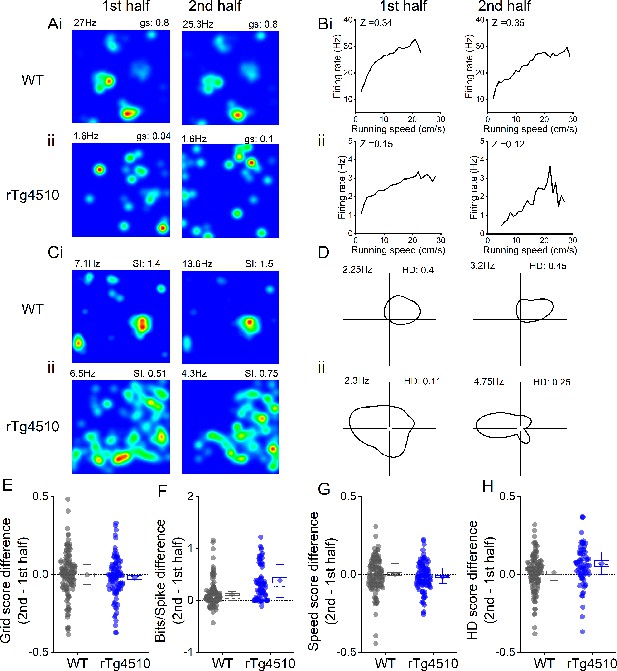
Rate coding stability across recording session.
Example grid (A), speed (B), spatial (C), and head direction (HD) (D) cell firing rate maps displayed for the first (left) and second (right) half of the recording session for WT (i) and rTg4510 (ii) mice. (E) Difference between grid score in first and second halves of the recording session (dotted line at zero). (F) Difference between spatial information score in first and second halves of the recording session (dotted line at zero). (G) Difference between speed score in first and second halves of the recording session (dotted line at zero). (H) Difference between HD score in first and second halves of the recording session (dotted line at zero) All box plots: dotted line: median, diamond: mean ± SEM, whiskers: 25th/75th centile.
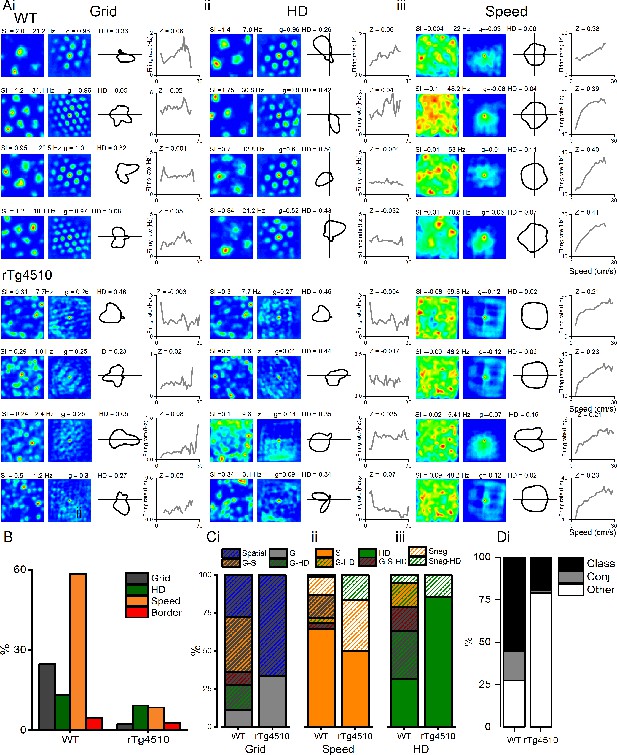
Conjunctive representation of grid, head direction, and running speed in WT and rTg4510 mice.
(A) Example grid (i), head direction (ii), and speed (iii) cells taken from the highest modulation scores from WT (top) and rTg4510 (bottom) mice, showing spatial firing patterns with 2D spatial autocorrelation, head-direction tuning and running speed-firing rate relationship and corresponding score (SI: spatial information, HD: mean vector length, Z: speed score). (B) Proportions of grid, head-direction, and speed-modulated cells recorded in mEC, showing reduced number of grid and speed, but not head-direction, cells passing threshold (95% centile of shuffled distribution). (C) Conjunctive proportions of grid cells (i), head-direction cells (ii), and speed-modulated cells (iii) recorded from WT and rTg4510 mice scaled to 100%. (D) Breakdown of units from WT and rTg4510 mice that satisfied a single criteria (class), multiple criteria (conj) or no discernible firing pattern (other). Key, G: grid, S: speed, HD: head direction, G-S: grid-speed, G-HD: grid-head direction, G-S-HD: grid-speed-head direction, SnegHD: negative speed–head-direction, Sneg: negative speed, Class: classed as cell type, Conj: conjunctive representation (more than 1 classification), other: not classified as grid, speed or HD.
Tables
Results of two-way repeated measure ANOVA for oscillation-running speed interaction.
Results of relevant post-hoc pairwise-comparisons displayed on Figure 1.
| Correlation (Z) | |||||
|---|---|---|---|---|---|
| Source of variation | Df | Sum-of-squares | Mean square | F | p |
| Frequency | 2 | 0.470 | 0.235 | 4.341 | 0.051 |
| Genotype | 1 | 0.602 | 0.602 | 5.927 | 0.038 |
| Interaction | 2 | 1.330 | 0.665 | 8.379 | 0.005 |
| Slope (mV2/Hz.cm-1 ) | |||||
| Source of variation | Df | Sum-of-squares | Mean square | F | p |
| Frequency | 2 | 0.004 | 0.002 | 8.973 | 0.004 |
| Genotype | 1 | 0.008 | 0.008 | 86.224 | >0.001 |
| Interaction | 2 | 0.005 | 0.003 | 16.623 | >0.001 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (M. musculus, Male) | rTg4510, Tg(Camk2a-tTA)1Mmay Fgf14 | ENVIGO | RRID:MGI:4819951 | Gift from Eli Lilly. (Ramsden et al., 2005) |
| Software, algorithm | Klusta suite | Rossant et al., 2016 | RRID:SCR_014480 | https://klusta.readthedocs.io/ |
| Software, algorithm | MATLAB R2019b | Mathworks https://uk.mathworks.com/ | RRID:SCR_001622 Chronux toolbox: RRID:SCR_005547 | |
| Software, algorithm | OriginPro 2019b | OriginLab https://www.originlab.com/ | RRID:SCR_014212 | |
| Other | Cresyl violet stain | Sigma aldrich | ID: C5042 | |
| Other | High-density silicon probe electrode array | Cambridge NeuroTech https://www.cambridgeneurotech.com/ | P2 | two × 16 channel shanks |
| Other | Silicon Probe | Neuronexus https://neuronexus.com/ | A1 × 16–5 mm-150-703 | 16 channel linear array |
| Other | Digital Lynx 10S recording system | Neuralynx https://neuralynx.com/ | HS-18 or HS-36 | Cheetah five data acquisition software |





