PLK-1 promotes the merger of the parental genome into a single nucleus by triggering lamina disassembly
Figures
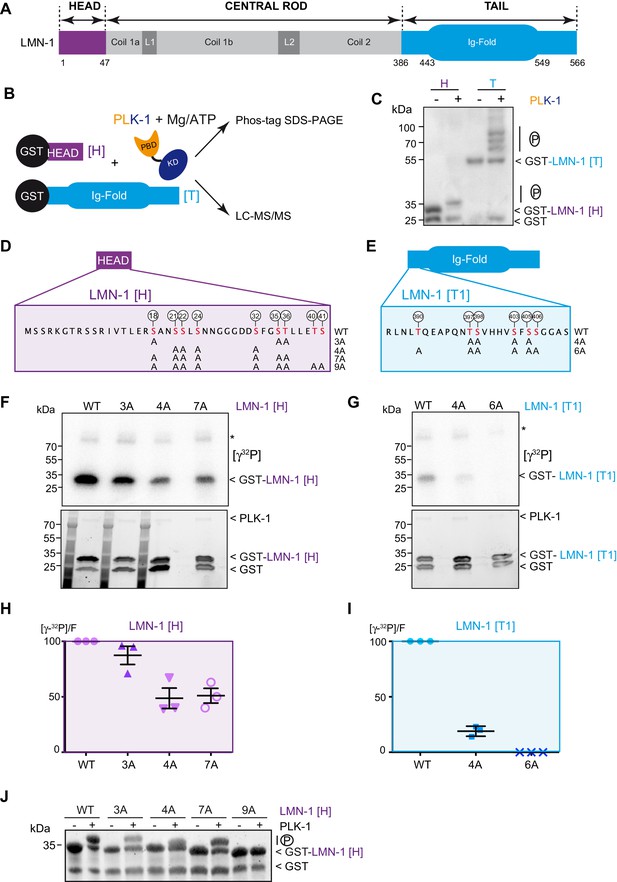
PLK-1 phosphorylates LMN-1 head and tail domains at multiple sites in vitro.
(A) Schematic of LMN-1 domain structure. Similar to lamins from other species, LMN-1 has a tripartite structure consisting of non-α-helical N-terminal head (violet) and C-terminal tail (blue) domains flanking an α-helical central rod domain. The C-terminal tail contains the Ig-fold motif. (B) Schematic of the approach used to investigate whether PLK-1 directly phosphorylates LMN-1 head and tail domains and to map the phosphorylated sites by tandem mass spectrometry (LC-MS/MS). (C) In vitro kinase assay was performed with C. e PLK-1 and the GST-LMN-1 head or tail as substrates. The samples were subjected to Phos-Tag SDS-PAGE followed by a western blot analysis using GST antibody. (D–E) Protein sequence of the head and tail domains with the residues phosphorylated by PLK-1 in red. The position of the phosphorylated residues is also indicated by white circles. The different non-phosphorylatable versions of LMN-1[H] 3A, 4A, 7A, 9A and LMN-1[T1] 4A, 6A with the positions of the alanine substitutions are presented below the protein sequence. (F–G) In vitro kinase assays using PLK-1 and LMN-1 WT or mutated fragments (LMN-1[H] and [T1]) as substrates. An autoradiograph of the SDS-PAGE showing γ-[32P] incorporation in LMN-1 (upper panel). Coomassie brilliant blue (CBB) staining of the same SDS-PAGE (bottom panel). Asterisk marks autophosphorylated PLK-1. GST is present as an impurity from the production of GST-LMN-1 fragments. (H–I) The graphs correspond to the quantification of the radioactivity incorporated into LMN-1 [H] and [T1] fragments divided by the total amount of LMN-1 fragments quantified using tryptophan fluorescence (Stain-Free; Bio-Rad). The ratio obtained for the WT fragments was arbitrary defined as 100. The quantification of three independent experiments is presented. Error bars represent the standard deviation. (J) In vitro kinase assay was performed with C. e PLK-1 and the GST-LMN-1 [H] WT or mutated fragments as substrates. The samples were subjected to SDS-PAGE and the gel was revealed using tryptophan fluorescence (Stain-Free; Bio-Rad).
-
Figure 1—source data 1
Quantification of the in vitro kinase assays.
- https://cdn.elifesciences.org/articles/59510/elife-59510-fig1-data1-v1.xlsx
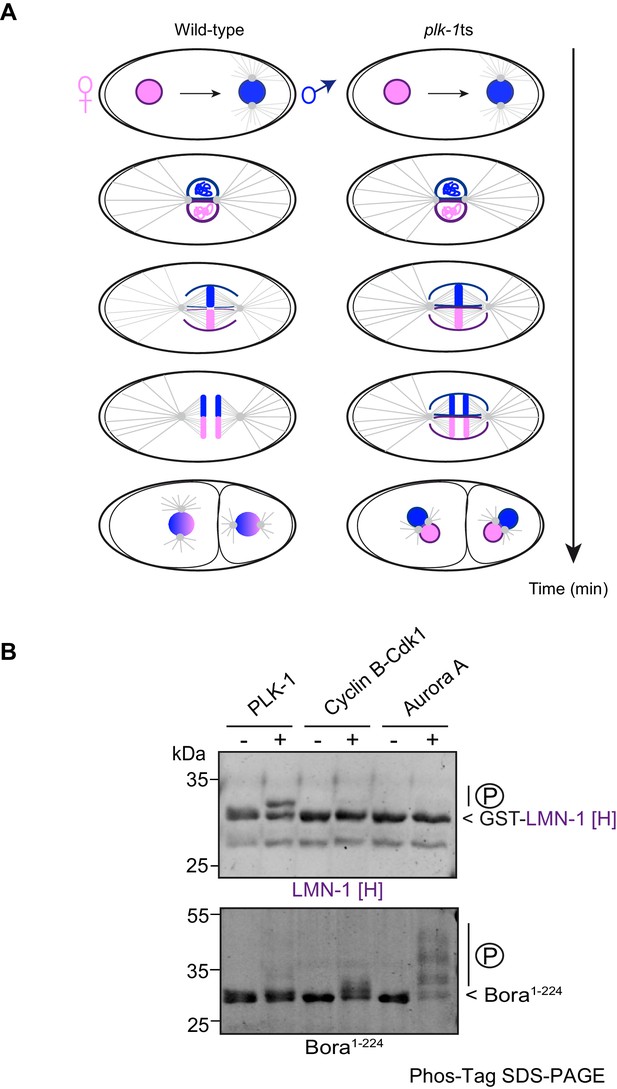
PLK-1 but not Aurora A or Cyclin B/Cdk1 phosphorylates LMN-1[H] in vitro.
(A) Context of the study: schematics of the first mitotic division of wild-type and plk-1(-) embryos. In the wild type after fertilization, the female (pink) and the male (blue) pronuclei, which are surrounded by a nuclear envelope, meet at the posterior pole of the embryo. After rotation and centration of the nucleo-centrosomal complex, the nuclear envelope breaks down in the vicinity of the centrosomes and between the juxtaposed pronuclei. This allows the capture of the chromosomes by the microtubules and the merging of the parental chromosomes into a single nucleus. In plk-1(-) embryos, the nuclear envelope persists and physically separates the parental genomes during DNA segregation resulting in the formation of paired nuclei in each blastomere at the two-cell stage embryo. (B) In vitro kinase assays using PLK-1, Cyclin B-Cdk1 or Aurora A kinases and GST-LMN-1 [H] (top panel) or Bora1-224 fragments (bottom panel) as substrates. The samples were subjected to Phos-Tag SDS-PAGE followed by a Coomassie blue staining.
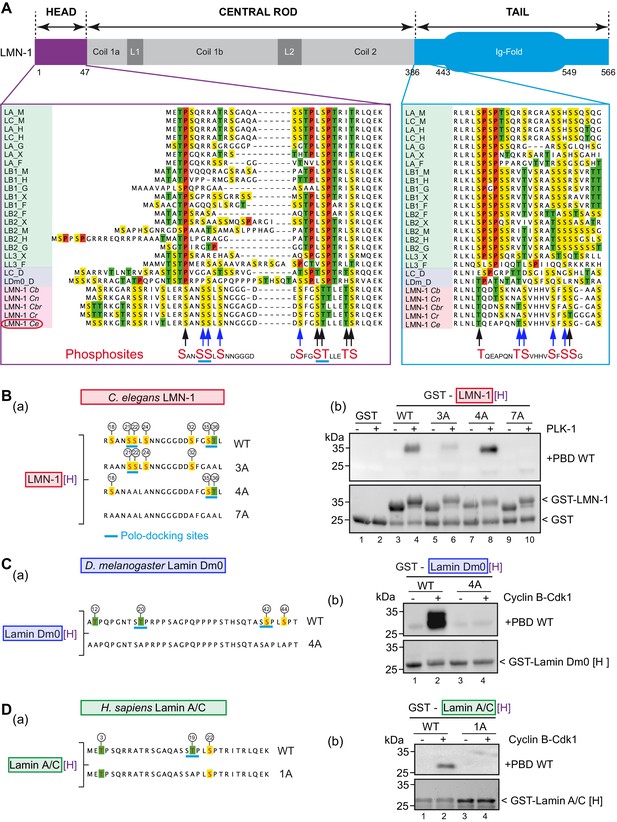
PLK-1 interacts with lamins from several species via self and non-self-priming and binding mechanisms.
(A) Schematics of LMN-1 and multiple protein alignments of the head (violet) and the tail (blue) domains of Ce LMN-1 (surrounded in red, bottom lane) with lamins from vertebrates (light green), Drosophila melanogaster (light blue) and other nematodes (light red). Serine residues are highlighted in yellow, Threonine in green and Proline in red. The sequence correspondence and accession numbers are provided in the Supplementary file 3. The black and blue arrows point the C. e LMN-1 residues phosphorylated by PLK-1 in red at the bottom of the alignment. The blue arrows point residues previously shown to be phosphorylated in the C. elegans germline. Putative polo-docking sites are underlined in light blue. (B) (a) Schematics and sequence of the GST-LMN-1 [H] fragment WT or with the indicated substitution of serine and threonine residues by non-phosphorylatable alanine (3A, 4A, and 7A). Polo-docking sites are underlined in light blue. (b) In vitro kinase assay was performed with PLK-1 and the GST-LMN-1 [H] fragments as substrate. The samples were subjected to SDS-PAGE, followed by a Far-Western ligand-binding assay using the Polo-box domain fused to GST (+PBD, upper panel). The bottom panel shows the Stain-Free Blot (Chemidoc, Bio-Rad) of the same membrane. The full experiment is presented in Figure 2—figure supplement 1B. (C) (a) Schematics and sequence of Drosophila melanogaster lamin Dm0 head domain WT or 4A fused to GST. The sites phosphorylated by Cyclin-Cdk1 are circled (Mehsen et al., 2018). These four sites are substituted by alanine in the 4A construct. The canonical polo-docking sites are underlined in light blue. (b) In vitro kinase assay was performed with Cyclin-Cdk1 and the GST-lamin Dm0 [H] fragments as substrate. The samples were subjected to SDS-PAGE, followed by a Far-Western ligand-binding assay using the Polo-box domain fused to GST (+GST-PBD, upper panel). The bottom panel shows the Stain-Free Blot (Chemidoc, Bio-Rad) of the same membrane. The full experiment is presented in Figure 2—figure supplement 1C. (D) (a) Schematics and sequence of human lamin A/C head domain. This fragment contains three S/T-P sites phosphorylatable by Cyclin-Cdk1 kinase including one polo-docking site (underlined in light blue). This specific site is substituted by alanine in the 1A version. (b) In vitro kinase assay was performed with cyclin-cdk1 and the GST-lamin A/C [H] WT or 1A fragments as substrate. The samples were subjected to SDS-PAGE, followed by a Far-Western ligand-binding assay using the Polo-box domain fused to GST (+GST-PBD, upper panel). The bottom panel shows the Stain-Free Blot (Chemidoc, Bio-Rad) of the same membrane. The full experiment is presented in Figure 2—figure supplement 1D.
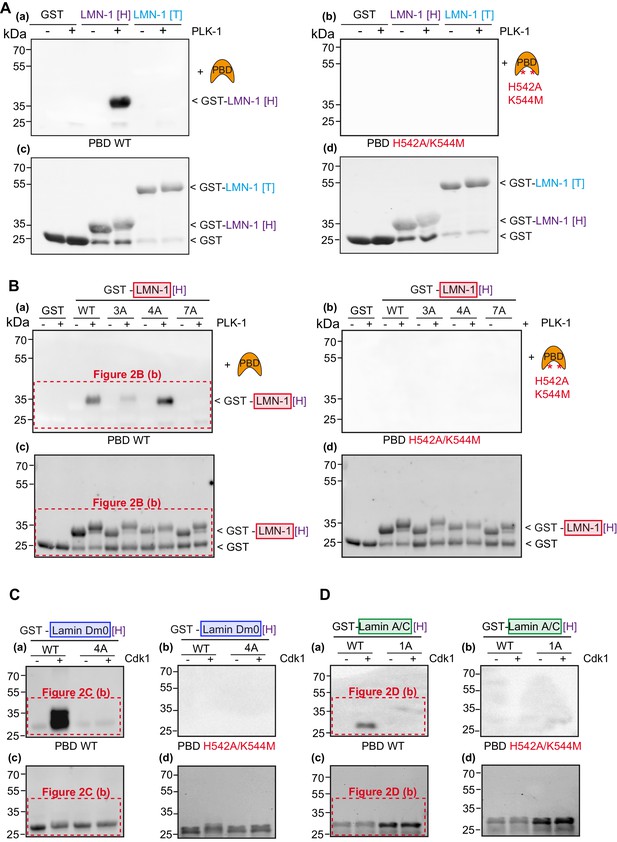
Lamins from several species interact with the Plk1 PBD in a phospho-dependent manner.
(A) In vitro kinase assay was performed with C.e PLK-1 and the GST-LMN-1 head [H] or tail [T] fragments as substrates. The samples were subjected to SDS-PAGE, followed by a Far-Western ligand-binding assay using GST-PBD wild type (a) or the corresponding phosphate pincer (GST-PBD H538A/K540M) mutant (b). The bottom panel shows the Stain-Free Blot (Chemidoc, Bio-Rad) of the same membrane (c, d). GST is present as an impurity from the production of GST-LMN-1 fragments. (B) Full scans of Western blots corresponding to Figure 2B (panels b and d) and including the Far-Western blot using the PBD mutated on the phosphate pincer as control (panels a and c). In vitro kinase assay was performed with PLK-1 and the GST-LMN-1 [H] WT and mutated fragments as substrate. The samples were subjected to SDS-PAGE, followed by a Far-Western ligand-binding assay using GST-PBD wild type (a) or the corresponding phosphate pincer (GST-PBD H538A/K540M) mutant (b). The bottom panel shows the Stain-Free Blot (Chemidoc, Bio-Rad) of the same membrane (c, d). GST is present as an impurity from the production of GST-LMN-1 fragments. (C) Full scans of Western blots corresponding to Figure 2C (panels b and d) and including the Far-Western blot using the PBD mutated on the phosphate pincer as control (panels a and c). In vitro kinase assay was performed with Cyclin-Cdk1 and the GST-lamin Dm0 [H] fragments as substrate. The samples were subjected to SDS-PAGE, followed by a Far-Western ligand-binding assay using GST-PBD wild-type (a) or the corresponding phosphate pincer (GST-PBD H538A/K540M) mutant (b). The bottom panel shows the Stain-Free Blot (Chemidoc, Bio-Rad) of the same membrane (c, d). (D) Full scans of Western blots corresponding to Figure 2D. In vitro kinase assay was performed with Cyclin-Cdk1 and the GST-lamin A/C [H] WT or 1A fragments as substrate. The samples were subjected to SDS-PAGE, followed by a Far-Western ligand-binding assay using GST-PBD wild type (a) or the corresponding phosphate pincer (GST-PBD H538A/K540M) mutant (b). The bottom panel shows the Stain-Free Blot (Chemidoc, Bio-Rad) of the same membrane (c, d).
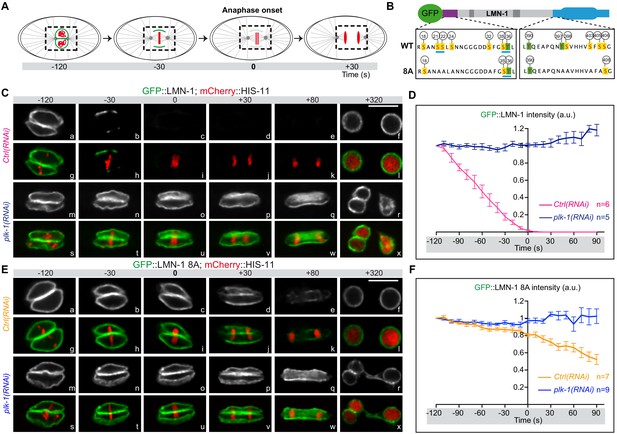
LMN-1-mediated PLK-1 phosphorylation promotes lamina disassembly during mitosis.
(A) Schematics of the approach used to monitor and quantify lamina disassembly during mitosis using spinning disk confocal microscopy in embryos expressing GFP::LMN1 WT or 8A (green) and mCherry-HIS-11 (red). GFP::LMN-1 intensity was quantified over time in the boxed region relative to anaphase onset (time 0). (B) Schematics of GFP::LMN-1 wild-type and 8A with the position of the serine (boxed in yellow) and threonine (boxed in green) residues phosphorylated by PLK-1 in vitro. The eight serine residues substituted by non-phosphorylatable alanine in the head and the tail domains of GFP::LMN-1 8A are S21, S24, S22, S32, T397, T398, S403 and S405. The polo-docking sites are underlined in light blue. (C) Spinning disk confocal micrographs of lmn-1∆ mutant embryos expressing wild-type GFP::LMN-1 (shown alone, and in green in the merged images) and mCherry::HIS-11 (red, in the merged image) exposed to control or plk-1(RNAi). Timings in second are relative to anaphase onset. All panels are at the same magnification. Scale Bar, 10 μm. D- Quantification of GFP::LMN-1 signal intensity above background at the NE in embryos of the indicated genotype during mitosis. The mean +/- SEM is presented for n = 6 embryos for control and n = 5 plk-1(RNAi) embryos. Data were collected from three independent experiments. (E) Spinning disk confocal micrographs of lmn-1∆ mutant embryos expressing GFP::LMN-1 8A (shown alone, and in green in the merged images) and mCherry::HIS-11 (red, in the merged image) exposed to control or plk-1(RNAi). Timings, in seconds, are relative to anaphase onset. All panels are at the same magnification. Scale Bar, 10 μm. (F) Quantification of GFP::LMN-1 8A signal intensity above background at the NE in embryos of the indicated genotype during mitosis. The mean +/- SEM is presented for n = 7 embryos for control and n = 9 for plk-1(RNAi). Data were collected from three independent experiments.
-
Figure 3—source data 1
Quantification of GFP::LMN-1 WT and 8A signal intensity during mitosis in control and plk-1(RNAi) embryos (related to Figure 3D and F).
- https://cdn.elifesciences.org/articles/59510/elife-59510-fig3-data1-v1.xlsx
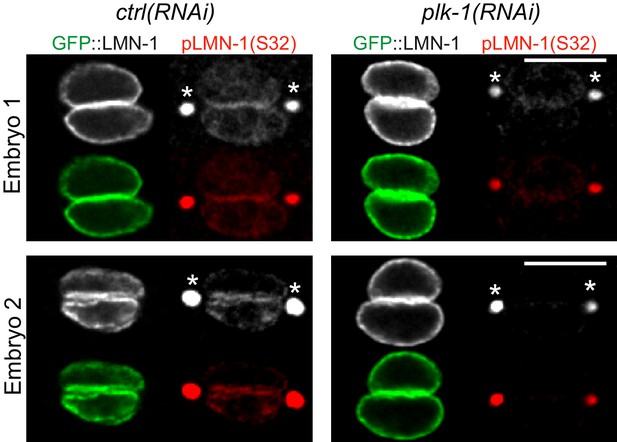
LMN-1 Serine 32 is phosphorylated in a PLK-1-dependent manner in one-cell C. elegans embryos.
Immunostaining of ctrl(RNAi) and plk-1(RNAi) one-cell embryos using phosphospecific [pS32] antibody (shown alone and in red) and GFP::LMN-1 (green). The asterisk denotes non-specific stainings at the centrosomes. Two embryos are presented.
Lamina disassembly in early embryos expressing GFP::LMN-1 and mCherry::HIS-11 exposed to control (video 1) or plk-1(RNAi) (video 2).
Lamina disassembly in early embryos expressing GFP::LMN-1 and mCherry::HIS-11 exposed to control (video 1) or plk-1(RNAi) (video 2).
Lamina disassembly in early embryos expressing GFP::LMN-1 8A and mCherry::HIS-11 exposed to control (video 3) or plk-1(RNAi) (video 4).
Lamina disassembly in early embryos expressing GFP::LMN-1 8A and mCherry::HIS-11 exposed to control (video 3) or plk-1(RNAi) (video 4).
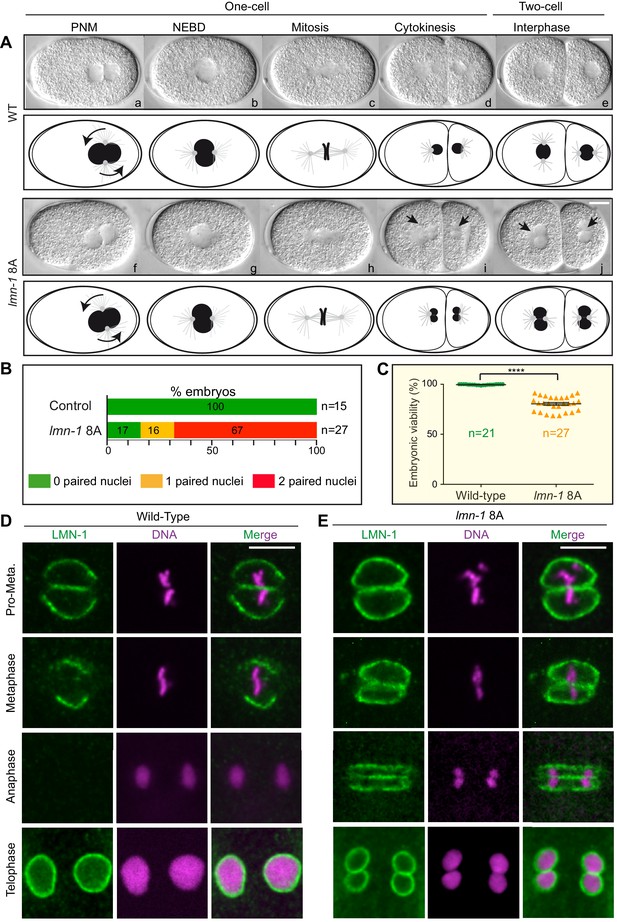
Non-phosphorylatable LMN-1 8A is sufficient to cause the appearance of embryos with a paired nuclei phenotype.
(A) Differential interference contrast micrographs and corresponding schematics of the first two divisions in wild type (WT) (a–e) and lmn-1 8A (f–j) embryos at 23°C. Black arrowheads highlight paired nuclei in lmn-1 8A mutant embryos. PNM: Pronuclear meeting, NEBD: nuclear envelope breakdown. Scale Bar, 10 μm. (B) Percentage of WT and lmn-1 8A two-cell stage embryos presenting 0 (green bars), 1 (orange bars) or 2 (red bars) paired nuclei at the two-cell stage at 23°C. The number of embryos analyzed (n) is indicated on the right and was generated by aggregation over more than three independent experiments. The values inside the bars represent the percentage of embryos of a given phenotype. (C) Graph presenting the embryonic viability (%) determined from wild-type (n = 21) and lmn-1 8A (n = 27) mutant animals. Data were collected from two independent experiments containing each two replicates. The results are presented as means ± SEM. **** indicates p<0.0001. (D–E) Confocal images of fixed wild-type and lmn-1 8A mutant one-cell embryos in pro-metaphase, metaphase, anaphase and telophase, stained with LMN-1 antibodies (green) and counterstained with DAPI (magenta). All panels are at same magnification. Scale Bar, 10 μm.
-
Figure 4—source data 1
Progeny test analysis of WT versus lmn-1 8A mutants (related to Figure 4C).
- https://cdn.elifesciences.org/articles/59510/elife-59510-fig4-data1-v1.xlsx
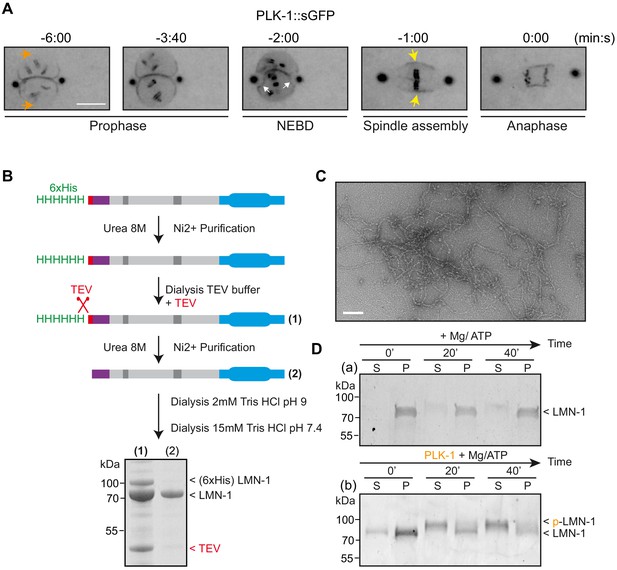
PLK-1 localizes at sites of lamina depolymerization in vivo and disassembles C.e lamin filaments in vitro.
(A) Images from a time-lapse spinning disk confocal movie showing the spatiotemporal localization of PLK-1::sGFP to centrosomes, chromosomes and to the NE during the first mitosis of the C. elegans zygote. Orange arrows point to the regions of the NE located in the regions of the pronuclei where PLK-1::sGFP is excluded before NEBD. NEBD is defined as the time point at which the nuclear envelope starts to deform (white arrows). Yellow arrows point to the region of the NE surrounding the mitotic spindle where PLK-1::sGFP accumulates. Times are in minutes:seconds relative to anaphase onset. Scale bar, 5 μm. (B) Flow-chart of the approach used to purify LMN-1 from E. coli and to assemble LMN-1 filaments in vitro. Coomassie blue staining of purified 6xHis-LMN-1 before and after cleavage of the 6xHis tag with TEV protease. As the TEV protease is also tagged with the 6xHis tag (not mentioned in the figure), the protease is removed from the LMN-1 preparation after purification on Ni2+ column (step 2). (C) Negative staining electron microscopy micrograph of C. elegans lamin filaments assembled from purified LMN-1. Scale Bar, 100 nm. D- In vitro kinase assay was performed with Mg/ATP alone as control (a) or with PLK-1 plus Mg/ATP (b) and LMN-1 filaments as substrate. The samples were taken at the indicated time, centrifuged and the pellet (P) and supernatant (S) fractions were subjected to SDS-PAGE and the gel was stained with Coomassie brilliant blue.
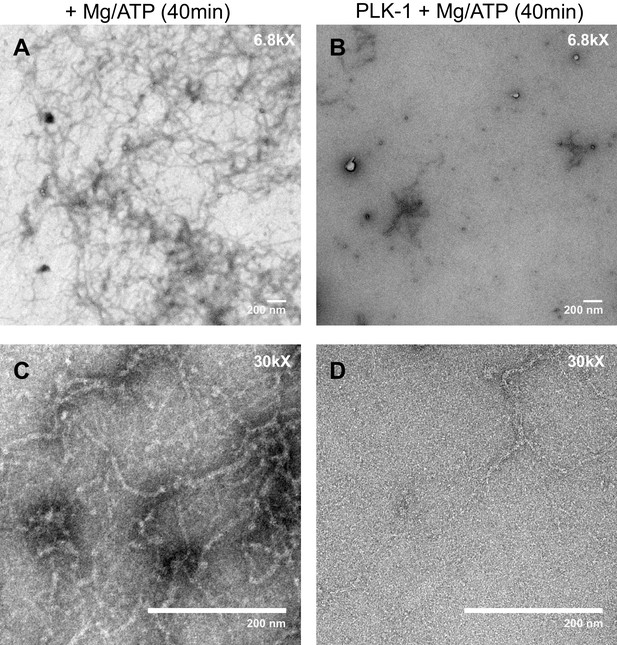
PLK-1 kinase promotes LMN-1 filaments disassembly in vitro.
Negative staining electron microscopy micrograph of C. elegans lamin filaments assembled from purified LMN-1 after 40 min incubation with Mg/ATP as control (A, C) or Mg/ATP and PLK-1 kinase (B, D). Scale Bar, 200 nm.
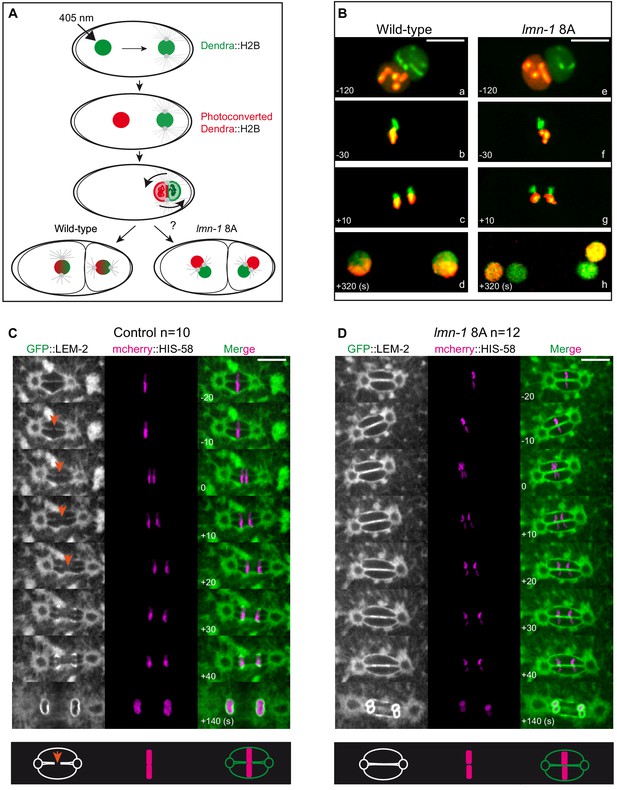
Paternally- and maternally-derived chromatin remain physically separated during mitosis in lmn-1 8A mutant one-cell embryos.
(A) Schematics of the live-imaging approach used to differentially label parental chromosomes. The female pronuclei of the Dendra2-H2B-expressing strain are photoconverted (405 nm laser beam) in the early zygote changing the fluorescence of the female pronucleus from green-to-red and the embryos are imaged until the 2 cell stage. (B) Representative spinning disk confocal images are taken from different wild-type (a, d) and lmn-1 8A (e, f) mutant embryos. Shown are maximum projections of image stacks. Nuclei are in mitotic prophase (a, e), metaphase (b, f) anaphase (c, g) and telophase (d, h). Scale Bar, 10 μm. (C-D) Spinning disk confocal micrographs of WT and lmn-1 8A mutant one-cell stage embryos expressing the INM protein LEM-2 fused to GFP (GFP::LEM-2, shown alone on the right of the panels and in green in the merged images) and mCherry::HIS-58 (magenta). In the control embryo (left), the nuclear envelope breaks as the chromosomes align at the metaphase plate (orange arrows), allowing the chromosomes from the sperm and oocyte pronuclei to merge on the spindle before their segregation. In the lmn-1 8A mutant embryos, the nuclear envelope remains intact and acts as a physical barrier that separates the chromosomes from the two pronuclei during their segregation. On the bottom panels, schematics illustrate the presence of a membrane gap in wild-type as opposed to lmn-1 8A embryos. Scale Bar, 10 μm. Times are in seconds relative to anaphase onset.
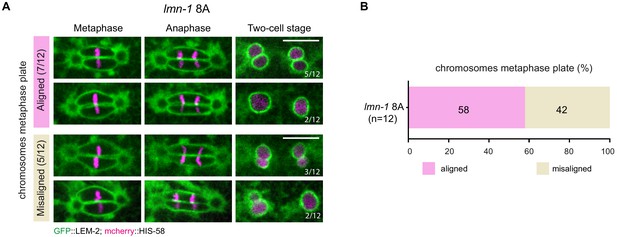
lmn-1 8A mutant one-cell embryos are defective in membrane gap formation but properly align parental chromosomes.
(A) Spinning disk confocal micrographs of representative early lmn-1 8A mutant embryos expressing GFP::LEM-2 (green) and mCherry::HIS-58 (magenta) in metaphase, anaphase and telophase. 7/12 early embryos properly aligned chromosomes on the metaphase plate. Among these seven embryos, five presented a double paired-nuclei phenotype and two did not present paired nuclei. Scale Bar, 10 μm. (B) Percentage of lmn-1 8A early embryos with aligned (pink) or misaligned chromosomes (brown).
Control (video 1) or lmn-1 8A (video 2) mutant embryos expressing GFP::LEM-2 and mCherry::HIS-58.
Control (video 1) or lmn-1 8A (video 2) mutant embryos expressing GFP::LEM-2 and mCherry::HIS-58.
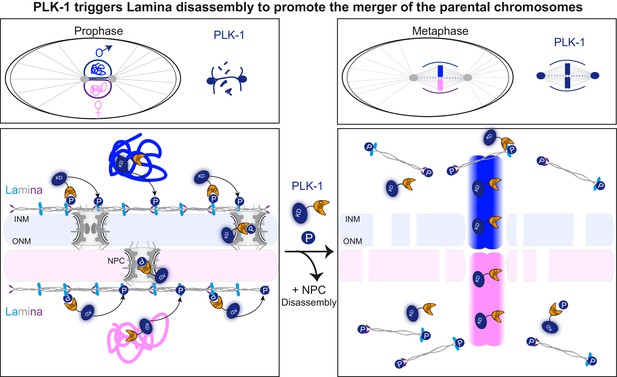
PLK-1 dynamics at the nuclear envelope dictates the timing of lamina depolymerization during mitosis The maternal and paternal chromosomes are colored in pink and blue respectively.
Polo-like kinase1 with the kinase domain (KD) in dark blue and the polo-box domain (PBD) in orange. Phosphorylated residues (P) circled in dark blue are modified by PLK-1.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Caenorhabditis elegans) | C. elegans N2 Bristol | CGC | http://www.cgc.cbs.umn.edu/strain.php?id=10570 | |
| Strain, strain background (C. elegans) | [gfp::lmn-1] MosSCI: lmn-1(tm1502) I; jfSi68[Plmn-1::gfp cb-unc-119(+)] II | Link et al., 2018, #49005 | UV120 | |
| Strain, strain background (C. elegans) | [gfp::lmn-1S8A] MosSCI: lmn-1(tm1502) I; jfSi89[Plmn-1S(21,22,24,32,397,398,403,405)A::gfp cb-unc-119(+)] II | Link et al., 2018, #49005 | UV122 | |
| Strain, strain background (C. elegans) | lmn-1(jf140[S21,22,24,32,397,398, 403,405A]) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III) | This study | UV2059 | V. Jantsch Lab |
| Strain, strain background (C. elegans) | tonSi1[mex-5p::Dendra2::his-66::tbb-2 3’UTR + Cbr-unc-119(+)]II | Bolková and Lanctôt, 2016, #90027 | JBL1 | |
| Strain, strain background (C. elegans) | lmn-1(jf140[S21,22,24,32,397,398, 403,405A]) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III);tonSi1[mex-5p::Dendra2::his-66::tbb-2 3’UTR + Cbr-unc-119(+)]II | This study | WLP831 | L. Pintard Lab |
| Strain, strain background (C. elegans) | Itls37 [(pAA64) pie-1p::mCherry::his-58 + unc-119(+)]IV qals3507 [pie-1::GFP::LEM-2 + unc-119(+)] | CGC | OD83 | |
| Strain, strain background (C. elegans) | lmn-1S8AS(21,22,24,32,397,398,403, 405)A Itls37 [(pAA64) pie-1p::mCherry::his-58 + unc-119(+)]IV qals3507 [pie-1::GFP::LEM-2 + unc-119(+)] | This study | WLP833 | L. Pintard Lab |
| Strain, strain background (C. elegans) | plk-1(lt17) ([plk-1::sgfp] loxp)III | Martino et al., 2017, #66043 | OD2425 | |
| Strain, strain background (Escherichia coli) | BL21(DE3) | Sigma-Aldrich | CMC0016 | |
| Antibody | LMN-1 (Rabbit polyclonal) | Novus Biologicals | Cat#38530002 RRID:AB_10005072 | (1:100 IF) (1:1,000 WB) |
| Antibody | LMN-1Ser32pi (Rabbit polyclonal) | Link et al., 2018, #49005 | V. Jantsch Lab (Home made) | (1:10 IF) |
| Antibody | Plk1 (human) (Mouse monoclonal) | Merck Millipore | Cat#05–844 RRID:AB_310836 | (1:1,000 WB) |
| Antibody | GST (Rabbit polyclonal) | This study | L. Pintard Lab (Home made) | (1:1,000 WB) |
| Antibody | Anti-Mouse IgG (Fab specific) -Peroxidase antibody (produced in goat) | Sigma | Cat#A9917 RRID:AB_258476 | (1:5,000 WB) |
| Antibody | Anti-Rabbit IgG (whole molecule) -Peroxidase antibody (Produced in goat) | Sigma | Cat#A0545 RRID:AB_257896 | (1:3,000 WB) |
| Antibody | Anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 (Produced in goat) Fluor 568 | Invitrogen | Cat#A-11011 RRID:AB_143157 | (1:1,000 IF) |
| Antibody | Anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Produced in goat) | Invitrogen | Cat#A-11001 RRID:AB_2534069 | (1:1,000 IF) |
| Chemical compound, drug | Coomassie R250 | Sigma | Cat#B014925G | |
| Chemical compound, drug | Ponceau Red | Sigma | Cat#A1405 | |
| Chemical compound, drug | VECTASHIELD Mounting Medium with DAPI | Eurobio | Cat#H-1200 | |
| Chemical compound, drug | IPTG | Euromedex | Cat#EU0008-B | |
| Chemical compound, drug | Adenosine TriPhosphate (ATP) | Sigma | Cat#A2383 | |
| Chemical compound, drug | γ-[32P] Adenosine TriPhosphate (ATP)500 μCi 3000 Ci/mmol | PerkinElmer | Cat#NEG002A500U | |
| Chemical compound, drug | Phos-TagAcrylamide AAL-107 | WAKO-SOBIODA | Cat#W1W304-93521 | |
| Chemical compound, drug | Imidazole | Sigma | Cat#I202 | |
| Chemical compound, drug | Glutathione | Sigma | Cat#G4251 | |
| Chemical compound, drug | HiTrap Chelating HP 5 × 1 mL | GE Healthcare | Cat#17-0408-01 | |
| Chemical compound, drug | Glutathion Sepharose 4B Fast Flow | GE Healthcare | Cat#17-0756-01 | |
| Chemical compound, drug | Pfu | Promega | Cat#M7741 | |
| Chemical compound, drug | DpnI | Biolabs | Cat#R0176S | |
| Commercial assay or kit | ECL reagent | Millipore | Cat#WBKLS0500 | |
| Commercial assay or kit | BP Clonase II Enzyme Mix (Gateway cloning) | Invitrogen | Cat#11789–020 | |
| Commercial assay or kit | LR Clonase II Enzyme Mix (Gateway cloning) | Invitrogen | Cat#11791–020 | |
| Recombinant protein | Human Cyclin B-Cdk1 kinase | New England Biolabs | Cat#P6020L | |
| Recombinant DNA reagent | L4440 (RNAi Feeding vector) | Kamath et al., 2001, #2201 | N/A | |
| Recombinant DNA reagent | plk-1 cloned into L4440 | Kamath et al., 2003, #46063 | Arhinger Library | |
| Recombinant DNA reagent | pDESTttTi5605[R4-R3] for MOS insertion on Chromosome II | Frøkjaer-Jensen et al., 2008, #79702 | pCFJ150 Addgene plasmid # 19329 | |
| Recombinant DNA reagent | MOS transposase Pglh-2::MosTase::glh-2utr | Frøkjaer-Jensen et al., 2008, #79702 | pJL43.1 Addgene plasmid # 19332 | |
| Recombinant DNA reagent | Prab-3::mCherry | Frøkjaer-Jensen et al., 2008, #79702 | pGH8 | |
| Recombinant DNA reagent | Pmyo-2::mCherry::unc-54 | Frøkjaer-Jensen et al., 2008, #79702 | pCFJ90 | |
| Recombinant DNA reagent | Entry clone for Gateway pDONR221 (lmn-1 CDS was cloned in this vector) | Multistite Gateway Kit, Thermo Fischer Scientific | Cat# 12537–023 | |
| Recombinant DNA reagent | pDONR P4-P1R (the lmn-1 5’ UTR with and without GFP was cloned in this vector) | Multistite Gateway Kit, Thermo Fischer Scientific | Cat# 12537–023 | |
| Recombinant DNA reagent | pDONRTM P2r-P3 (thelmn-1 3’UTR was cloned in this vector) | Multistite Gateway Kit, Thermo Fischer Scientific | Cat# 12537–023 | |
| Recombinant DNA reagent | Minigene from IDT for generating the lmn-18A containing the whole CDS of lmn-1(S21,22,24,32,397, 398,403,405A) | Link et al., 2018, #49005 | N/A | |
| Recombinant DNA reagent | Plmn-1_gfp::lmn-1_lmn-1 3’UTR in pCFJ150 | Link et al., 2018, #49005 | N/A | |
| Recombinant DNA reagent | Plmn-1_gfp::lmn-1(S8A) lmn-1 3’UTR in pCFJ150 | Link et al., 2018, #49005 | N/A | |
| Recombinant DNA reagent | Gal4 pDEST DB | Noatynska et al., 2010, #23221 | pMG97 | |
| Recombinant DNA reagent | Gal4 pDEST DB-PLK-1 PBD | Noatynska et al., 2010, #23221 | pMG477 | |
| Recombinant DNA reagent | Gal4 pDEST DB-PLK-1 PBD H542A, K544M | Noatynska et al., 2010, #23221 | pMG538 | |
| Recombinant DNA reagent | pGEX-4T (GST) | GE Healthcare | Cat#GE28-9545-49 | |
| Recombinant DNA reagent | pFasTBAC Hta PLK-1 C. elegans | Tavernier et al., 2015, #22388 | pLP871 | |
| Recombinant DNA reagent | pGEX-6p1 GST-PLK-1 PBD H. s | Gift I. Sumara | N/A | |
| Recombinant DNA reagent | pGEX-6p1 GST-PLK-1 PBD H538A/K540M H. s | Gift I. Sumara | N/A | |
| Recombinant DNA reagent | pDONR201 TEV-Aurora A H. s | This study | pLP1884 | |
| Recombinant DNA reagent | His-GST-TEV-Aurora A H. s | This study | pLP2067 | |
| Recombinant DNA reagent | pBABE-puro-GFP-wt-lamin A | Gift B. Cisneros | Addgene #17662 | |
| Recombinant DNA reagent | cDNA LMN-1 13A (S18A, S21A, S22A, S24A, S32A, S35A, T36A, T390A, T397A, S398A, S403A, S405A, S406A) Generate by Thermofisher | ThermoFisher | N/A | |
| Recombinant DNA reagent | pDONR201 | ThermoFisher | N/A | |
| Recombinant DNA reagent | pDONR201 LMN-1 [H] (aa1-47) | This study | pLP2234 | |
| Recombinant DNA reagent | pDONR201 LMN-1 [H] (aa1-47) 3A S18A S35A T36A | This study | pLP2277 | |
| Recombinant DNA reagent | pDONR201 LMN-1 [H] (aa1-47) 4A S21A S22A S24A S32A | This study | pLP2236 | |
| Recombinant DNA reagent | pDONR201 LMN-1 [H] (aa1-47) 7A S18A S21A S22A S24A S32A S35A T36A | This study | pLP2276 | |
| Recombinant DNA reagent | pDONR201 LMN-1 [T] (aa387-548) | This study | pLP2270 | |
| Recombinant DNA reagent | pDONR201 LMN-1 [T1] (aa387-436) | This study | pLP2249 | |
| Recombinant DNA reagent | pDONR201 LMN-1 [T1] 4A (aa387-436) T397A S398A S403A S405A | This study | pLP2250 | |
| Recombinant DNA reagent | pDonR201 LMN-1 [T1] 6A (aa387-436) T390A, T397A, S398A, S403A, S405A, S406A | This study | pLP2269 | |
| Recombinant DNA reagent | pDONR201 cDNA of lamin Dm0 from Drosophila melanogaster | Mehsen et al., 2018, #44350 | N/A | |
| Recombinant DNA reagent | pDONR201 cDNA of lamin with 7 mutations of the CDK motifs from Drosophila melanogaster T12A, T20A, S42A, S45A, T413A, T435A, T440A | Mehsen et al., 2018, #44350 | N/A | |
| Recombinant DNA reagent | pDONR201 Lamin Dm0 [H] (aa1-57) | This study | pLP2314 | |
| Recombinant DNA reagent | pDONR201 Lamin Dm0 [H] (aa1-57) 4A T12A T20A S42A S44A | This study | pLP2315 | |
| Recombinant DNA reagent | pDONR201 Lamin A/C [H] (aa1-34) | This study | pLP2316 | |
| Recombinant DNA reagent | pDONR201 TEV-Bora 1–224 Hs | This study | pLP1848 | |
| Recombinant DNA reagent | pDEST17 His-TEV-Bora 1–224 Hs | This study | pLP1850 | |
| Recombinant DNA reagent | pDEST15 | ThermoFisher | Cat#11802014 | |
| Recombinant DNA reagent | pDEST15 GST- LMN-1 [H] (aa 1–47) | This study | pLP2235 | |
| Recombinant DNA reagent | pDEST15 GST- LMN-1 [H] (aa 1–47) 3A S18A S35A T36A | This study | pLP2282 | |
| Recombinant DNA reagent | pDEST15 GST- LMN-1 [H] (aa 1–47) 4A S21A S22A S24A S32A | This study | pLP2237 | |
| Recombinant DNA reagent | pDEST15 GST- LMN-1 [H] (aa1-47) 7A S18A S21A S22A S24A S32A S35A T36A | This study | pLP2281 | |
| Recombinant DNA reagent | pDEST15 GST- LMN-1 [H] (aa1-47) 9A S18A S21A S22A S24A S32A S35A T36A T40A S41A | This study | pLP2393 | |
| Recombinant DNA reagent | pDEST15 GST- LMN-1 [T] (aa387-548) | This study | pLP2271 | |
| Recombinant DNA reagent | pDEST15 GST- LMN-1 [T1] (aa387-436) | This study | pLP2253 | |
| Recombinant DNA reagent | pDEST15 GST- LMN-1 [T1] 4A (aa387-436) T397A S398A S403A S405A | This study | pLP2254 | |
| Recombinant DNA reagent | pDEST15 GST- LMN-1 [T1] 6A (aa387-436) T390A, T397A, S398A, S403A, S405A, S406A | This study | pLP2298 | |
| Recombinant DNA reagent | pDEST15 GST- Lamin Dm0 [H] (aa1-57) | This study | pLP2322 | |
| Recombinant DNA reagent | pDEST15 GST- Lamin Dm0 [H] (aa1-57) 4A T12A T20A S42A S44A | This study | pLP2323 | |
| Recombinant DNA reagent | pDEST15 GST- Lamin A/C [H] (aa1-34) | This study | pLP2324 | |
| Recombinant DNA reagent | pDEST15 GST- Lamin A/C [H] (aa1-34)1A T18A | This study | pLP2362 | |
| Recombinant DNA reagent | pET28b | Novagen | Cat#69864 | |
| Recombinant DNA reagent | pET28b (His linker-TEV-linker Nter) LMN-1 | This study | pLP2231 | |
| Recombinant DNA reagent | Alt-R CRISPR-Cas9 tracrRNA, 100 nmol | IDT | Cat#1072534 | |
| Sequence-based reagent | Primers for cloning and site-directed mutagenesis (see oligonucleotide sequences table S4 source) | This study | N/A | |
| Software, algorithm | Clustal Omega | EMBL-EBI | https://www.ebi.ac.uk/Tools/msa/clustalo/ | |
| Software, algorithm | Jalview | Waterhouse et al., 2009, #45561 | https://www.jalview.org/ | |
| Software, algorithm | Adobe Illustrator CS6 | Adobe | https://www.adobe.com/products/illustrator.html | |
| Software, algorithm | Adobe Photoshop CS4 | Adobe | https://www.adobe.com/products/photoshop.html | |
| Software, algorithm | Image J | NIH; Schneider et al., 2012, #74926 | https://imagej.nih.gov/ij/ | |
| Software, algorithm | ZEN | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html | |
| Software, algorithm | PRISM | Graphpad | https://www.graphpad.com/ | |
| Software, algorithm | Metamorph | Molecular Devices | https://www.metamorph.com/ | |
| Software, algorithm | Proteome Discoverer 2.2 | Thermo Scientific | https://www.thermofisher.com/store/products/OPTON-30945#/OPTON-30945 | |
| Software, algorithm | Matrix Science 5.1 | Mascot Server | https://www.matrixscience.com/ |
Additional files
-
Supplementary file 1
LC-MS/MS analysis of phosphorylated LMN-1 in vitro.
- https://cdn.elifesciences.org/articles/59510/elife-59510-supp1-v1.xlsx
-
Supplementary file 2
Sequence analysis of LMN-1 phospho-sites.
Phosphorylated serine and threonine residues (pS and pT) are highlighted in yellow and green respectively. Most phosphosites match the previously described Plk1 consensus motifs [L(Φ)-D/E/N/Q-X-pS/pT-L(Φ) or p[S/T]-F] (Santamaria et al., 2011; Kettenbach et al., 2011). D/E/N/Q residues are highlighted in blue, hydrophobic (Φ) in orange and F. in red.
- https://cdn.elifesciences.org/articles/59510/elife-59510-supp2-v1.xlsx
-
Supplementary file 3
Accession numbers of lamin protein sequences.
- https://cdn.elifesciences.org/articles/59510/elife-59510-supp3-v1.docx
-
Supplementary file 4
Oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/59510/elife-59510-supp4-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59510/elife-59510-transrepform-v1.docx





