The Natural History of Model Organisms: Insights into mammalian biology from the wild house mouse Mus musculus
Abstract
The house mouse, Mus musculus, was established in the early 1900s as one of the first genetic model organisms owing to its short generation time, comparatively large litters, ease of husbandry, and visible phenotypic variants. For these reasons and because they are mammals, house mice are well suited to serve as models for human phenotypes and disease. House mice in the wild consist of at least three distinct subspecies and harbor extensive genetic and phenotypic variation both within and between these subspecies. Wild mice have been used to study a wide range of biological processes, including immunity, cancer, male sterility, adaptive evolution, and non-Mendelian inheritance. Despite the extensive variation that exists among wild mice, classical laboratory strains are derived from a limited set of founders and thus contain only a small subset of this variation. Continued efforts to study wild house mice and to create new inbred strains from wild populations have the potential to strengthen house mice as a model system.
https://doi.org/10.7554/eLife.05959.001Introduction
Today, house mice (Mus musculus) are widely known as an excellent mammalian model for studying a wide variety of traits and diseases, including those involved in metabolism, development, neurological disorders, immunity, and others (Morse, 2007). At the beginning of the 20th century, however, Mendel's work had only recently been rediscovered and the race was on to develop genetic model systems. In a series of papers from 1902–1905, Lucien Cuénot used mice to demonstrate Mendel's laws for the first time in mammals (Hickman and Cairns, 2003). Around the same time, William E Castle, a pioneer of the use of Drosophila to study genetics, also began investigating the inheritance of coat color in mice (Castle and Allen, 1903). The Castle lab and others launched research programs focused on mouse genetics, and soon realised the need to create inbred strains (see Box 1 for a glossary of specialist terms used in this article) of mice (e.g., Castle and Allen, 1903; Castle and Little, 1910; Russell, 1978; Morse, 1981, 1985). By 1909, the first inbred strain, DBA, was created and the era of modern mouse genetics had begun (Russell, 1978). Since then, hundreds of inbred strains have been developed.
Glossary.
Adaptive introgression and adaptive introgressive hybridization—See ‘Introgression’.
Association mapping—A technique in which genetic variation is surveyed to identify statistical associations with phenotypic variation.
Endogenous retroviral sequences—Sequences within a genome that are similar to and derived from retroviruses.
Human commensals—Organisms that live in close association with humans. The term commonly describes symbiotic relationships where one partner benefits and the other is unaffected. House mice are called commensals despite their potential negative effects on humans.
Inbred strains—A strain created by many generations of brother-sister or parent-offspring mating. Historically, strains were considered inbred after 20 such generations, when residual heterozygosity would be vanishingly small. High-throughput genotyping can now evaluate homozygosity as strains are inbred.
Inbreeding coefficient—A statistic that summarizes the probability that any two genes in a population are identical by descent.
Introgression—The transmission of an allele from one population to another via hybridization and subsequent backcrossing. Adaptive introgression and adaptive introgressive hybridization refer to when an allele is transmitted from one population to another and confers a selective advantage.
Meiotic drive—Also called transmission ratio distortion, where two alleles at a locus carried by a heterozygote are transmitted to a zygote unequally (i.e., non-Mendelian inheritance).
Responder locus—An allele at a locus that rescues the fertility of sperm carrying a t-haplotype but that does not rescue fertility in wild-type sperm.
Retrovirus—A virus that is RNA based and that uses reverse transcriptase to transcribe itself into DNA.
Robertsonian translocation—also called Robertsonian fusion, this is a chromosomal rearrangement caused by the fusion of two acrocentric chromosomes into a single metacentric chromosome.
t-haplotypes—also called T-alleles, these are variants of a tightly-linked block of genes on the proximal end of chromosome 17 in mice that exhibit meotic drive.
https://doi.org/10.7554/eLife.05959.002The success of the house mouse as a genetic model organism is largely due to its unique natural history. In this article, we briefly introduce the evolutionary and natural history of house mice, as well as the origins of laboratory strains (Box 2). We highlight several examples in which wild house mice have provided important insights into mammalian biology and suggest future avenues for research.
The origins of laboratory mouse strains.
The trade of mice with distinct coat colors and behaviors has ancient origins in China, Japan, and Europe (reviewed in Keeler, 1931). Mice were used in experimental study as early as the 17th and 18th centuries (Morse, 1981). In the 19th century, Mendel is believed to have worked with mice before switching to pea plants after reportedly being admonished by his bishop for keeping organisms that had sex (Henig, 2000). When Mendel's laws were rediscovered in 1900, researchers saw the advantages of working with a mammal that could be housed in a small area, bred quickly, and that displayed many easily scored, variable traits.
Most of the inbred lines available today have their origins in the trade of fancy mice in the early 1900's (Morse, 1981). Abbie Lathrop, a retired schoolteacher in Granby, Massachusetts, USA, started breeding mice as pets, using mice purchased from other fanciers, including varieties like waltzing mice and creamy buffs. Among her best customers were scientists, including William E Castle and Clarence C Little at Harvard University, and Leo Loeb, who collaborated with Lathrop to investigate mammary tumors in her mouse strains (Morse, 1981). The need for truly genetically homogenous mice spurred these scientists and others to begin inbreeding their colonies. In 1929, CC Little founded the Roscoe B Jackson Memorial Laboratory, which now has the largest selection of inbred mice in the USA. Other major suppliers include Charles River Laboratories, Harlan Laboratories and Taconic Farms, Inc., among others.
https://doi.org/10.7554/eLife.05959.003The natural history of house mice
Phylogenetic history
House mice comprise three main subspecies of M. musculus with different global distributions: Mus musculus castaneus, Mus musculus domesticus and Mus musculus musculus (Figure 1). Their closest relatives are not human commensals (see ‘Glossary’) and include the mound building mouse, Mus spicilegus, and the Algerian mouse, Mus spretus (Chevret et al., 2005; Tucker et al., 2005). The ancestral range for M. musculus was likely in present-day India (Boursot et al., 1993). Genetic and genomic data indicate that the three subspecies of M. musculus started to diverge ∼350–500 thousand years ago (KYA) and that the split among the three subspecies occurred within a short period of time (Geraldes et al., 2008, 2011; Duvaux et al., 2011). Nonetheless, the available evidence suggests that M. m. castaneus and M. m. musculus are more closely related to each other than either is to M. m. domesticus (White et al., 2009; Keane et al., 2011). There is evidence of hybridization among the subspecies in zones of secondary contact (Tucker et al., 1992; Boursot et al., 1993; Duvaux et al., 2011). An additional subspecies, Mus musculus molossinus, is found in Japan and is derived from hybridization between M. m. castaneus and M. m. musculus (Yonekawa et al., 1988). Classical inbred strains of house mice are genetic mosaics of the three main subspecies, although they are primarily M. m. domesticus in origin (Yang et al., 2011; Didion and de Villena, 2013). Interestingly, contributions from the other two subspecies may be due to early crosses with M. m. molossinus, likely as a result of interactions between mouse fanciers in Japan and Western Europe (Yang et al., 2011). A fifth subspecies, Mus musculus gentilulus, has been described from collections in the southern Arabian Peninsula (Harrison, 1972; Harrison and Bates, 1991; Prager et al., 1998). Additional subspecies may yet be defined. In particular, populations in Afghanistan, Pakistan and Iran may be genetically distinct (Rajabi-Maham et al., 2012; Hardouin et al., 2015).
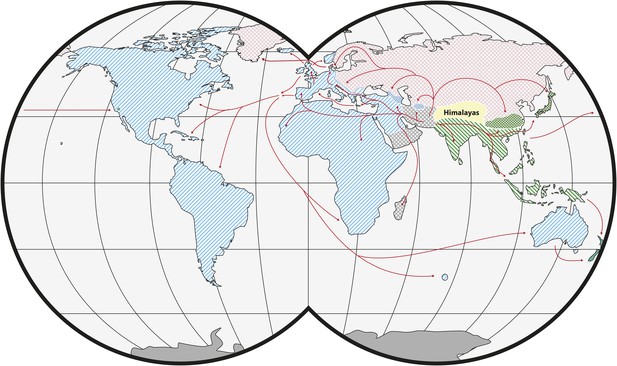
Worldwide distribution of Mus musculus subspecies (adapted from Didion and de Villena, 2013).
Ranges of M. musculus subspecies are indicated by hatching. Green: M. m. castaneus; blue: M. m. domesticus; red: M. m. musculus; grey: central populations and M. m. gentilulus. Note that house mice may not be found throughout the complete extent of hatched areas: for example, sub-arctic regions, the Sahara Desert, and the Amazon rainforest. Checkered areas indicate regions of hybridization. Red arrows indicate inferred routes of historical migrations and recent movements in association with humans. Reproduced from Springer and Didion J, de Villena FPM. 2013. Mammalian Genome 24:1–20.
© 2012 Springer-Verlag. All Rights Reserved. Figure 1 is reproduced with kind permission from Springer Science and Business Media.
Habitat and life style
Unique among other species in the genus Mus, house mice primarily live in close proximity to humans. Although feral populations exist, house mice are commonly found in residential, agricultural, and commercial structures. The rise of agrarian societies is very recent compared to the divergence among house mice subspecies, indicating that commensalism evolved independently in each of the three subspecies. House mice are omnivorous with varied diets including grains, seeds, and insects (Sage, 1981; Singleton and Krebs, 2007). They generally weigh less than 20 grams and serve as food for predatory snakes, birds and mammals. Laboratory mice tolerate regular handling by humans and live and breed in a small area. Their commensal life style, small size, and flexible diet have likely been key to their success as a model system.
Reproduction and social structure
The reproductive biology of mice is also favorable for laboratory breeding. Mice have short generation times; gestation lasts approximately 3 weeks and they are sexually mature at 6–8 weeks of age. Thus, generation times in the lab are between 9 and 11 weeks. In the wild, females may breed seasonally with one or two litters per year or they may breed year round if resources are available (Pocock et al., 2004; reviewed in Latham and Mason, 2004; Singleton and Krebs, 2007). In the lab, with unlimited food and good environmental conditions, females can breed year round. Litter sizes in the wild are large (∼4–9 pups), a trait that facilitates the efficient generation of inbred strains (see Sage, 1981; Singleton and Krebs, 2007). House mice in the wild have variable social structures, ranging from discrete ‘demes’ with a single male and one to several adult females and juveniles, to high-density aggregations in which adults are socially gregarious and have largely overlapping ranges (Singleton and Krebs, 2007). While males can be territorial, females are known to mate with multiple males (Dean et al., 2006; Firman and Simmons, 2008). Female house mice tend to live in extended family groups and can nest communally and nurse offspring communally (reviewed in König and Lindholm, 2012). Mice found in a limited geographic area are likely to be related, and the average inbreeding coefficient (see ‘Glossary’) of wild-caught mice in one study was ∼0.2 (Laurie et al., 2007). This natural level of inbreeding may help to purge recessive deleterious alleles and may have facilitated the creation of inbred strains in the laboratory.
Disease
Wild mice can carry a variety of diseases, including mouse hepatitis virus, mouse mammary tumor virus and mouse parvovirus. They can also be vectors for human diseases, such as leptospirosis, cryptosporidiosis, salmonellosis, and streptobacillosis (summarized in Singleton and Krebbs, 2007). However, unlike rats, they are not known to be major vectors of the plague or of hemorrhagic fevers like hantavirus. They are commonly parasitized by tapeworms, flukes, fleas and lice (Singleton and Krebbs, 2007). As such, wild mice are routinely quarantined before they enter mouse facilities.
Phenotypic variation
Wild house mice (Figure 2) exhibit considerable phenotypic variation, and it is likely that much more remains to be described. In association with humans, house mice have been spread around the world and have adapted to a wide range of environments (e.g., Jones et al., 2012). Mice can be found from sea level to over 4000 m in elevation, from the tropics to subarctic environments, and in both dry and wet environments. Studies of water balance in wild populations suggest that house mice can adapt to arid habitats (Mutze et al., 1991; Moro and Bradshaw, 1999).

Wild derived house mouse (Mus musculus domesticus) from Brazil.
Image credit: Taichi Suzuki.
Phenotypic variation among wild house mice has been described along environmental gradients. For example, Lynch (1992) surveyed populations of house mice on the east coast of the United States and found that mice from more northern latitudes were bigger and built bigger nests than those from more southern populations. These differences persisted in the lab over several generations, demonstrating a genetic basis. Selection experiments also provide evidence that house mice have the ability to rapidly adapt to cold temperatures. Wild mice bred at 3°C for 10 generations were more fertile, with larger litters of larger body size, and reached sexual maturity earlier than mice bred at 23°C (Barnett and Dickson, 1984). Rapid phenotypic change has occurred on many islands, including shifts in size, morphology, and diet (e.g., Berry et al., 1978; Davis, 1983; Le Roux et al., 2002; Renaud et al., 2013). One example comes from mice on Gough Island, a small island in the Atlantic Ocean approximately halfway between South Africa and Argentina. House mice were likely introduced to this island by sealers or whalers in the 19th century and are the only terrestrial mammals living on it (Holdgate, 1965; Gray et al., 2014). Gough Island mice live independently of humans, are large compared to European house mice, and eat seabird chicks, including those of the critically endangered Tristan albatross (e.g., Rowe–Rowe and Crafford, 1992; Cuthbert and Hilton, 2004; Wanless et al., 2007). These mice are alarming from a conservation perspective, but they are intriguing from an evolutionary one. Variation in natural populations is fertile ground for exploring the genetic basis of complex traits. Such efforts could add to the decades of study of complex traits in inbred strains of mice (such as body size, Cheverud, 2005; Kenney-Hunt et al., 2006).
Genetic variation
Populations of wild house mice exhibit levels of genetic variation that are several-fold higher than those seen in human populations, consistent with the larger effective population size for house mice (Geraldes et al., 2008, 2011; Halligan et al., 2010, 2013). Patterns of genetic variation in natural populations have provided insight into the demographic and evolutionary history of wild mice (e.g., Bonhomme et al., 2007; Geraldes et al., 2008; Duvaux et al., 2011; Jones et al., 2012) and have helped us to understand how natural selection has shaped patterns of genetic variation (e.g., Halligan et al., 2010, 2013; Phifer-Rixey et al., 2012; for review, Teschke et al., 2012). For example, some beneficial variants appear to have come from introgression between subspecies or between closely related species (Song et al., 2011; Staubach et al., 2012; Liu et al., 2015).
Despite the substantial level of genetic variation seen in wild house mice, the commonly used classical inbred strains of laboratory mice derive from a limited set of founders and thus contain only a small subset of the genetic variation that is seen in nature (Salcedo et al., 2007; Yang et al., 2011). As a result, there are many genomic ‘blind spots’ in crosses involving classical inbred strains. Since the 1970's, new inbred lines have been established using wild mice (Guénet and Bonhomme, 2003). Levels of diversity are much higher among these wild-derived inbred strains than among classical inbred strains (e.g., Ideraabdullah et al., 2004; Yang et al., 2011). Several recent outcrossed populations have also been developed from classical and wild-derived inbred strains, including the Heterogeneous Stock mice (Valdar et al., 2006) and the Collaborative Cross mice (Churchill et al., 2004). Both of these outbred mapping populations contain more genetic variation than do mice derived from crossing classical inbred lines, and they also have more recombination, permitting traits of interest to be mapped with improved resolution (Yalcin et al., 2010). However, they are not without their limitations. First, much of the genetic variation is due to differences between subspecies. Second, since crosses between subspecies result in partial hybrid sterility, selection effectively eliminates genetic variation in some regions of the genome in these crosses. Third, mapping intervals are not typically at the resolution of individual genes. The development of additional inbred lines of wild-derived mice should help address these limitations and augment existing resources.
Insights from wild house mice
Here, we briefly highlight a few examples in which studies of wild house mice have provided insight into fundamental questions concerning mammalian biology. This is not a comprehensive review; research on wild house mice encompasses a diverse range of topics, only some of which are touched on here. For more information, we refer readers to ‘The mouse in biomedical research’ (Foster et al., 1981; Fox et al., 2007), ‘The genus Mus as a model for evolutionary studies’ (Britton-Davidian and Searle, 2005) and ‘Evolution of the house mouse’ (Macholán et al., 2012).
Immunity: the Major Histocompatibility Complex
The Major Histocompatibility Complex (MHC) is a family of strikingly polymorphic genes that have a critically important role in the immune response of vertebrates. Proteins encoded by MHC genes bind peptides derived from pathogens and present them on the cell surface for recognition by T cells. MHC genes were first discovered in classical inbred strains of house mice (reviewed in Klein, 1981, 1986; Penn and Musolf, 2012). Wild house mice have also played a major role in advancing our understanding of the MHC. For example, the high level of variation seen at MHC genes was first discovered in populations of wild mice (Klein, 1970; Klein and Bailey, 1971) and has since been documented in many other vertebrates. This discovery spawned a considerable amount of research aimed at understanding the evolutionary forces that maintain such diversity. These studies have led to some intriguing findings regarding the possible functional significance of MHC diversity in wild mice and other organisms. One area of particular interest is mate choice. Data suggest that individual mice prefer mates with dissimilar MHC haplotypes, a form of disassortative mating (e.g., Penn and Potts, 1998; reviewed in Penn and Musolf, 2012). Such mating preferences could function to improve offspring resistance to pathogens, reduce inbreeding, or both. However, the relative importance of MHC in mate choice in wild populations remains unresolved; there are many other factors that have been shown to affect mate choice. Moreover, the specific mechanism by which individuals identify MHC-dissimilar individuals remains obscure.
Cancer: retroviruses and the Lake Casitas mice
In the late 1960's, virally induced cancers were known to occur in laboratory mice and in chickens, but had yet to be discovered in humans. Robert Huebner and Murray Gardner hypothesized that resistance to viral cancer was likely present in wild mouse populations but had been lost in laboratory strains through inbreeding (Gardner et al., 1991; O'Brien, 2003). Gardner embarked on a mission to sample wild populations of house mice. While he found disease-free mice at most sites, at a poultry farm near Lake Casitas in California, he found ∼85% of the mice carried murine leukemia virus (MuLV). Infected mice developed severe lymphoma and/or hind limb paralysis after sexual maturity (Gardner et al., 1991). Despite the severity of the disease and evidence of transmission, the population did not crash and many mice were apparently unaffected (Gardner et al., 1980, 1991; O'Brien, 2003). Gardener and his colleague, Stephen O'Brien, hypothesized that an anti-retroviral genetic element was segregating in the Lake Casitas mice, protecting some from infection. This proved to be the case; they found an allele at a single locus that conferred complete resistance in homozygotes and was inherited in a Mendelian fashion (Gardner et al., 1980). Researchers in Japan independently identified a common viral cancer, as well as endogenous retroviral sequences (see ‘Glossary’) in M. m. molossinus (Ikeda et al., 1981; Odaka et al., 1981). Ultimately, it was shown that resistance in the two populations localized to the same gene (O'Brien et al., 1983).
How did the resistance allele prevent infection? And why were two mouse populations on either side of the Pacific Ocean, thought to be different subspecies, polymorphic for the same protective allele? The resistance locus was itself a truncated endogenous retrovirus in which only the subunit responsible for the creation of the viral envelope remained intact (see ‘Glossary’). When a virus replicates in a cell, some of its envelope proteins bind to receptors on the cell, preventing additional viruses from entering. Thus, resistance to MuLV in the Lake Casitas and Japanese mice was conferred by cells that produce harmless endogenous viral envelope proteins, blocking the binding of the virulent MuLV viruses (Kozak et al., 1984; Ikeda et al., 1985; Dandekar et al., 1987; Ikeda and Sugimura, 1989). These populations of mice share the resistance allele because they have a shared evolutionary history. Japanese mice descend from hybridization between M. m. castaneus from Southern China and M. m. musculus from northern China. While the resistance gene and the virus were undetected in M. m. musculus, they were both found in M. m. castaneus (O'Brien, 2003). Although mice in North America were originally believed to be entirely of M. m. domesticus origin, the Lake Casitas mice are hybrids of M. m. domesticus and M. m. castaneus (Orth et al., 1998). The Lake Casitas farm was originally settled by immigrants from China in the 19th century, who likely brought M. m. castaneus with them (Gardner et al., 1991). The Lake Casitas mice have provided insight into the mechanisms of resistance to retroviruses, the dynamics of disease outbreaks, and the origins of sequences derived from retroviruses found throughout vertebrate genomes, including humans.
Structural rearrangements and non-Mendelian inheritance
Two kinds of structural rearrangements in the genomes of wild mice have provided insight into genome evolution and into chromosome transmission during meiosis: Robertsonian (whole-arm) chromosomal translocations (also called fusions) and t-haplotypes, which are associated with chromosomal inversions (see ‘Glossary’). The karyotype of wild mice typically consists of 2n = 40 acrocentric chromosomes. However, numerous populations exist within the subspecies M. m. domesticus in which the diploid number is as low as 2n = 22 as a result of Robertsonian fusions between two different acrocentric chromosomes, creating a metacentric chromosome (e.g., Gropp et al., 1972; Capanna et al., 1973). Otherwise indistinguishable from mice with the standard karyotype, some karyotypic races show some reproductive isolation as a consequence of the mis-pairing of chromosomal heterozygotes in hybrids and the production of aneuploid gametes or complete meiotic arrest (reviewed in Garagna et al., 2014). Interestingly, some Robertsonian heterozygotes show transmission distortion in meiosis, preferentially transmitting the acrocentric products (instead of the fused, metacentric chromosome) to the oocytes in female meiosis (Gropp and Winking, 1981). In these cases, the metacentric chromosomes are preferentially distributed to the first polar body.
T-haplotypes are meiotic-drive systems (see ‘Glossary’) found at low frequency in natural populations (Ardlie and Silver, 1996). T-alleles are associated with inversions on chromosome 17, and they show a strong transmission distortion in meiosis, with heterozygotes transmitting the t-bearing allele to over 90% of gametes (Lyon, 2003). The responder locus (see ‘Glossary’) was first identified and molecularly characterized 15 years ago (Herrmann et al., 1999), but transmission distortion was first documented in 1927 (reviewed in Willison and Lyon, 2000; Herrmann and Bauer, 2012). Decades of research have made t-haplotypes one of the best-studied meiotic-drive systems in any organism, providing a model for understanding non-Mendelian inheritance.
Adaptation: rodenticide resistance
One consequence of a commensal lifestyle is an ongoing arms race with humans intent on extirpating mice and reducing food loss and the spread of disease. Warfarin-based rodenticides were introduced in the 1950's and have been periodically replaced by other rodenticides that have anticoagulant properties owing to the development of resistance among rats and mice (e.g., reviewed in Pelz et al., 2005; Ishizuka et al., 2008; Buckle, 2011). The study of rodenticide resistance in wild populations has provided insights into the molecular biology and biochemistry of anticoagulants and mechanisms of resistance to those agents. Importantly, the spread of rodenticide resistance has also led to insights into the process of adaptation (e.g., Rost et al., 2004, 2009; Pelz et al., 2005). For example, Song et al. (2011) found that an allele of the mouse gene Vkorc1, which underlies resistance to warfarin in M. m. domesticus, resulted from adaptive introgressive hybridization (see ‘Glossary’) with the Algerian mouse (M. spretus). This study is one of many recent studies that highlight that adaptive introgression is an often overlooked source of new variation in animals (Hedrick, 2013).
Speciation and hybrid male sterility
Laboratory crosses between some subspecies of house mice produce hybrid males that are sterile or have reduced fertility (e.g., Forejt and Ivanyi, 1974; Britton-Davidian et al., 2005). In nature, house mouse subspecies hybridize where they meet in secondary contact. The best-studied hybrid zone is between M. m. domesticus and M. m. musculus in Central Europe (e.g., Vanlerberghe et al., 1986; Dod and Jermlin, 1993, 2005; Payseur et al., 2004; Macholán et al., 2007; Teeter et al., 2008; Wang et al., 2011; Janoušek et al., 2012). This hybrid zone is young; colonization of this area is relatively recent (∼3000 YA; for example, Cucchi et al., 2005; reviewed in Baird and Macholán, 2012). Studies of this hybrid zone, and of laboratory crosses between these subspecies, have helped us to understand speciation genetics, particularly the genetic basis of hybrid male sterility (e.g., Mihola et al., 2009; White et al., 2011; Turner et al., 2014). For example, evidence from natural populations and from laboratory crosses has revealed an important role of the X chromosome in reproductive incompatibilities (Tucker et al., 1992; Oka et al., 2004; Storchová et al., 2004; Dod et al., 2005; Good et al., 2008; Oka and Shiroishi, 2012), a pattern that appears to be quite general across animals (Coyne and Orr, 2004). Studies of wild house mice have also found that hybrid male sterility has a complex basis, involving many genes (Oka et al., 2007; Good et al., 2008; White et al., 2011, 2012; Turner and Harr, 2014). Laboratory crosses between M. m. domesticus and M. m. musculus also led to the positional cloning of Prdm9, the only gene known to contribute to hybrid sterility in vertebrates (Mihola et al., 2009). This gene has a role in recombination rate variation in both mice and humans (e.g., Baudat et al., 2010; Berg et al., 2010; Parvanov et al., 2010). The identification of other genes underlying hybrid male sterility remains a challenge, but the combination of mapping studies in the lab and studies of regions showing limited introgression in nature might identify good candidates for future study (Janoušek et al., 2012; Phifer-Rixey et al., 2014; Turner and Harr, 2014).
Conclusions: the next frontiers
As outlined in Box 3, much remains to be learned about wild mice, even in the areas of research highlighted above. Genetic variation, phenotypic variation, variation in disease and parasite load remain uncharacterized in many wild populations throughout the world. The genetic bases for adaptive phenotypes are largely unknown. Contact zones between subspecies outside of Europe are mostly unexplored. Wild mice are an untapped reservoir of genetic variation. The generation of new wild-derived inbred strains would add a valuable component to existing mouse resources by adding new genetic variants. Moreover, since haplotype blocks are much shorter in wild mice than in classical inbred strains (Laurie et al., 2007), association mapping (see ‘Glossary’) with wild mice could help link individual genes to phenotypic differences. New more efficient genome-editing methods (Jinek et al., 2012) will also enable the direct testing of the effects of wild alleles on phenotypes. Thus, there is great potential for wild mice to continue to fuel new advances in the biosciences.
Outstanding questions about the natural history of house mice.
Although house mice have been studied for more than a century, there are still important questions about the basic biology of wild house mice that remain largely unanswered. Answers to these questions would further strengthen the mouse as a model for research.
What is the nature and extent of variation in morphology, physiology, reproduction, and development among wild house mice that have adapted to live in different environments? Although house mice are known to occur in a wide variety of environments, we still know relatively little about their physiological ecology. For example, how do some house mice survive extreme cold, high elevations, or extremely arid regions? Further study of such populations would likely provide additional mouse models for important phenotypes.
What are the determinants of social structure in mice? Mice sometimes live in small demes and sometimes live in larger aggregations, but much remains to be learned about the causes of these differences.
Which pathogens and parasites are present in house mice from different areas? Infectious agents can be a powerful evolutionary force, yet we know little about natural infections of mice from different places. Pathogens are likely to vary between temperate and tropical areas, but this remains largely uninvestigated. Similarly, mice from different areas may have evolved resistance to different pathogens, but this too is mostly unstudied.
What determines the limits to the distribution of house mice? They are amazingly successful at colonizing new areas, but they are not found everywhere. How important is competition with native rodents in determining the distribution of house mice?
Which genes underlie adaptation? Our understanding of the genetic basis of adaptive differences is in its infancy. There have been some genome scans for selection, but few instances in which specific genes have been linked to specific phenotypes.
What is the structure of haplotype blocks in natural wild mouse populations? Understanding haplotype structure is important for characterizing and understanding the evolution of recombination and will also lay the groundwork for association studies using wild mice.
References
-
Low frequency of mouse t haplotypes in wild populations is not explained by modifiers of meiotic driveGenetics 144:1787–1797.
-
BookWhat can the Mus musculus musculus/M. m. domesticus hybrid zone tell us about speciation?In: Macholán M, Baird SJE, Munclinger P, Piálek J, editors. Evolution of the house mouse. Cambridge: Cambridge University Press. pp. 334–372.
-
Sub‐antarctic house mice, colonization, survival and selectionJournal of Zoology 184:127–141.https://doi.org/10.1111/j.1469-7998.1978.tb03270.x
-
The evolution of house miceAnnual Review of Ecology and Systematics 24:119–152.https://doi.org/10.1146/annurev.es.24.110193.001003
-
Postzygotic isolation between the two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybridsBiological Journal of the Linnean Society 84:379–393.https://doi.org/10.1111/j.1095-8312.2005.00441.x
-
The genus Mus as a model for evolutionary studiesBiological Journal of the Linnean Society 84:321–674.https://doi.org/10.1111/j.1095-8312.2005.00490.x
-
A chromosomal polymorphism in an alpine population of Mus musculus. LBolletino Di Zoologia 40:379–383.https://doi.org/10.1080/11250007309429254
-
The heredity of albinismProceedings of the National Academy of Sciences of USA 38:603–622.
-
BookGenetics of growth in the mouseIn: Eisen EJ, editors. Mouse in animal genetics and breeding research. London: Imperial College Press. pp. 113–130.
-
Molecular phylogeny of the genus Mus (Rodentia: Murinae) based on mitochondrial and nuclear dataBiological Journal of the Linnean Society 84:417–427.https://doi.org/10.1111/j.1095-8312.2005.00444.x
-
Molecular characterization of the Akvr-1 restriction gene: a defective endogenous retrovirus-borne gene identical to Fv-4rJournal of Virology 61:308–314.
-
Counterselection on sex chromosomes in the Mus musculus European hybrid zoneJournal of Evolutionary Biology 6:529–546.https://doi.org/10.1046/j.1420-9101.1993.6040529.x
-
Testing for selection on the androgen-binding protein in the Danish mouse hybrid zoneBiological Journal of the Linnean Society 84:447–459.https://doi.org/10.1111/j.1095-8312.2005.00446.x
-
The frequency of multiple paternity predicts variation in testes size among island populations of house miceJournal of Evolutionary Biology 21:1524–1533.https://doi.org/10.1111/j.1420-9101.2008.01612.x
-
Akvr-1, a dominant murine leukemia virus restriction gene, is polymorphic in leukemia-prone wild miceProceedings of the National Academy of Sciences of USA 77:531–535.https://doi.org/10.1073/pnas.77.1.531
-
Demographic history of a recent invasion of house mice on the isolated Island of GoughMolecular Ecology 23:1923–1939.https://doi.org/10.1111/mec.12715
-
Biology of the house mouse141–181, Biology of the house mouse, London, Academic Press, Symposia of the Zoological Society of London 47.
-
BookThe mouse t-haplotype: a selfish chromosome – genetics, molecular mechanism, and evolutionIn: Macholán M, Baird SJE, Munclinger P, Piálek J, editors. Evolution of the house mouse. Cambridge: Cambridge University Press. pp. 297–314.
-
The fauna of the Tristan da Cunha IslandsPhilosophical Transactions of The Royal Society B Biological Sciences 249:361–402.https://doi.org/10.1098/rstb.1965.0015
-
Genetic and haplotype diversity among wild-derived mouse inbred strainsGenome Research 14:1880–1887.https://doi.org/10.1101/gr.2519704
-
Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistanceJournal of Virology 55:768–777.
-
Mapping of the Fv-4 mouse gene controlling resistance to murine leukemia virusesInternational Journal of Cancer 28:237–240.https://doi.org/10.1002/ijc.2910280218
-
Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference propertiesJournal of Virology 63:5405–5412.
-
Pesticide resistance in wild mammals–mechanisms of anticoagulant resistance in wild rodentsJournal of Toxicological Sciences 33:283–291.https://doi.org/10.2131/jts.33.283
-
BookThe Laboratory Mouse: Its Origins, Heredity and CultureCambridge, Massachusetts: Harvard University Press.
-
Quantitative trait loci for body size components in miceMammalian Genome 17:526–537.https://doi.org/10.1007/s00335-005-0160-6
-
BookThe histocompatibility-2 (H-2) complexIn: Foster HL, Small JD, Fox JG, editors. The mouse in biomedical research. New York: Academic Press. pp. 120–158.
-
BookNatural History of the Major Histocompatability ComplexNew York: John Wiley & Sons.
-
Histocompatibility differences in wild mice; further evidence for the existence of deme structure in natural populations of the house mouseGenetics 68:287–297.
-
BookThe complex social environment of female house mice (Mus domesticus)In: Macholán M, Baird SJE, Munclinger P, Piálek J, editors. Evolution of the house mouse. Cambridge: Cambridge University Press. pp. 114–134.
-
A unique sequence related to the ecotropic murine leukemia virus is associated with the Fv-4 resistance geneProceedings of the National Academy of Sciences of USA 81:834–837.https://doi.org/10.1073/pnas.81.3.834
-
From house mouse to mouse house, the behavioural biology of free-living Mus musculus and its implications in the laboratoryApplied Animal Behaviour Science 86:261–289.https://doi.org/10.1016/j.applanim.2004.02.006
-
Linkage disequilibrium in wild micePLOS Genetics 3:e144.https://doi.org/10.1371/journal.pgen.0030144
-
Interspecific introgressive origin of genomic diversity in the house mouseProceedings of the National Academy of Sciences of USA 112:196–201.https://doi.org/10.1073/pnas.1406298111
-
Clinal variation in cold adaptation in Mus domesticus: verification of predictions from laboratory populationsThe American Naturalist 139:1219–1236.https://doi.org/10.1086/285383
-
Transmission ratio distortion in miceAnnual Review of Genetics 37:393–408.https://doi.org/10.1146/annurev.genet.37.110801.143030
-
BookEvolution of the house mouseMacholán M, Baird SJE, Munclinger P, Piálek J, editors. Cambridge: Cambridge University Press.
-
Water and sodium requirements of field populations of house mice, Mus domesticus and short tailed mice (Leggadina lakedownensis) on Thevenard Island, in the arid Pilbara region of Western AustraliaJournal of Comparative Physiology B, Biochemical, Systemic, and Environmental Physiology 169:419–428.https://doi.org/10.1007/s003600050238
-
BookThe laboratory mouse—A historical perspectiveIn: Foster HL, Small JD, Fox JG, editors. The mouse in biomedical research. New York: Academic Press. pp. 1–16.
-
The Bussey Institute and the early days of mammalian geneticsImmunogenetics 21:109–116.https://doi.org/10.1007/BF00364862
-
BookBuilding a better mouse: One hundred years of genetics and biologyIn: Fox JG, Barthold SW, Davisson MT, Newcomer CE, Quimby FW, Smith SL, editors. The mouse in biomedical research second edition. San Diego, CA: Academic Press. pp. 1–11.
-
Murine retroviral restriction genes Fv-4 and Akvr-1 are alleles of a single locusJournal of Virology 47:649–651.
-
BookTears of the Cheetah and Other Tales from the Genetic FrontierNew York: St. Martins Press.
-
Fv-4: gene controlling resistance to NB-tropic Friend murine leukemia virus. Distribution in wild mice, introduction into genetic background of BALB/c mice, and mapping of chromosomesJournal of the National Cancer Institute 67:1123–1127.
-
BookThe role of the X chromosome in house mouse speciationIn: Macholán M, Baird SJE, Munclinger P, Piálek J, editors. Evolution of the house mouse. Cambridge: Cambridge University Press. pp. 431–454.
-
BookThe evolution of MHC diversity in house miceIn: Macholán M, Baird SJE, Munclinger P, Piálek J, editors. Evolution of the house mouse. Cambridge: Cambridge University Press. pp. 221–252.
-
MHC-disassortative mating preferences reversed by cross-fosteringProceedings of the Royal Society of London B 265:1299–1306.https://doi.org/10.1098/rspb.1998.0433
-
Adaptative evoluton and effective population size in wild house miceMolecular Biology and Evolution 29:2949–2955.
-
Genetic variation and phylogeography of central asian and other house mice, including a major new mitochondrial lineage in YemenGenetics 150:835–861.
-
The south-eastern house mouse Mus musculus castaneus (Rodentia, Muridae) is a polytypic subspeciesBiological Journal of the Linnean Society 107:295–306.https://doi.org/10.1111/j.1095-8312.2012.01957.x
-
Invasive house mice facing a changing environment on the Sub-Antarctic Guillou Island (Kerguelen Archipelago)Journal of Evolutionary Biology 26:612–624.https://doi.org/10.1111/jeb.12079
-
Density, body size, and reproduction of feral house mice on Gough IslandSouth African Journal of Zoology 27:1–5.
-
BookOrigins and history of mouse inbred strains: Contributions of clarence cook littleIn: Morse HC, editors. Origins of inbred mice. New York: Academic Press. pp. 33–44.
-
BookWild miceIn: Foster HL, Small JD, Fox JG, editors. The mouse in biomedical research. New York: Academic Press. pp. 40–90.
-
Nucleotide variation in wild and inbred miceGenetics 177:2277–2291.https://doi.org/10.1534/genetics.107.079988
-
BookThe secret world of wild miceIn: Fox JG, Barthold SW, Davisson MT, Newcomer CE, Quimby FW, Smith SL, editors. The mouse in biomedical research second edition. San Diego: Academic Press. pp. 25–51.
-
Genetic analysis of X-linked hybrid sterility in the house mouseMammalian Genome 15:515–524.https://doi.org/10.1007/s00335-004-2386-0
-
BookTracing recent adaptations in natural populations of the house mouseIn: Macholán M, Baird SJE, Munclinger P, Piálek J, editors. Evolution of the house mouse. Cambridge: Cambridge University Press. pp. 315–333.
-
Genomic networks of hybrid sterilityPLOS Genetics 10:e1004162.https://doi.org/10.1371/journal.pgen.1004162
-
Can predation by invasive mice drive seabird extinctions?Biology Letters 3:241–244.https://doi.org/10.1098/rsbl.2007.0120
-
Fine-scale phylogenetic discordance across the house mouse genomePLOS Genetics 5:e1000729.
-
A UK-centric history of studies on the mouse t-complexThe International Journal of Developmental Biology 44:57–63.
-
Subspecific origin and haplotype diversity in the laboratory mouseNature Genetics 43:648–655.https://doi.org/10.1038/ng.847
-
Hybrid origin of Japanese mice ‘Mus musculus molossinus’: evidence from restriction analysis of mitochondrial DNAMolecular Biology and Evolution 5:63–78.
Article and author information
Author details
Funding
National Institutes of Health (NIH) (RO1 GM074245)
- Michael W Nachman
The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
This research was funded by an NIH grant (R01 GM074245) to MWN. We thank JP Didion and F Pardo-Manuel de Villena for permission to use a figure, and TA Suzuki for permission to use an image.
Publication history
- Received:
- Accepted:
- Version of Record published:
Copyright
© 2015, Phifer-Rixey and Nachman
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Ecology
- Evolutionary Biology
Essays on the wild lives of model organisms, from Arabidopsis to the zebrafish.





