Ubiquitin-dependent regulation of a conserved DMRT protein controls sexually dimorphic synaptic connectivity and behavior
Figures
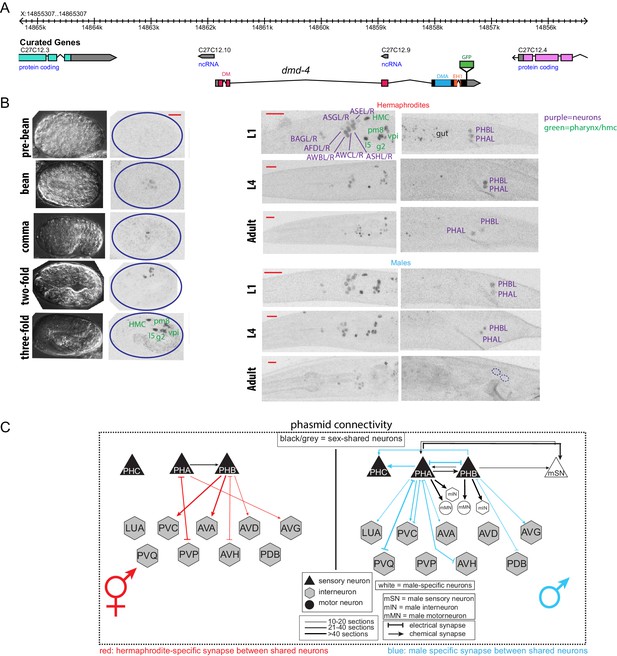
DMD-4 shows both sexually dimorphic and non-dimorphic expression.
(A) Schematic of the GFP-tagged dmd-4 locus in the dmd-4 (ot935) allele. The DM, DMA, and EH1 domains of DMD-4 are indicated in the colored boxes and labeled above the locus. (B) DMD-4::GFP is expressed in head sensory neurons and pharyngeal cells/the head mesodermal cell (hmc) in both sexes, and the phasmid sensory neurons PHA and PHB in both sexes until adulthood, when it is degraded in the male phasmids. Expression in the pharyngeal cells onsets during the ‘bean’ stage of embryogenesis. GFP is shown as color-inverted black and white. All scale bars (red horizontal bars) indicate 10 microns. Head images are maximum intensity projections of the entire worm; tail images are maximum intensity images of one half of the tail to clearly display phasmid nuclei. (C) Sexually dimorphic connectivity of the PHA and PHB neurons based on electron micrograph reconstruction (Cook et al., 2019; Jarrell et al., 2012).
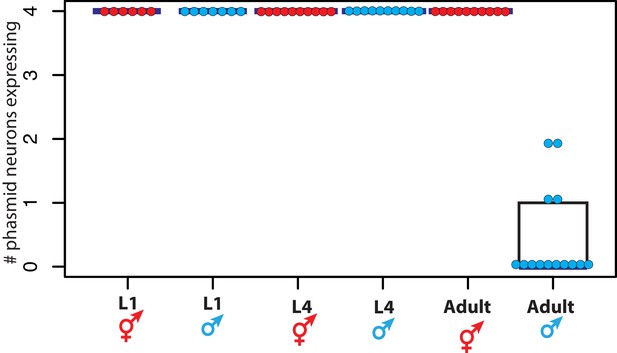
DMD-4::GFP is dimorphically expressed in the phasmid neurons.
Quantification of the number of phasmid neurons expressing DMD-4::GFP in L1/L4/adult animals of both sexes is shown (maximum is 4 cells for PHAL/R and PHBL/R), where each dot represents the number of neurons expressing in a single animal, blue horizontal bars indicate median, and black boxes indicate quartiles. Representative images are shown in Figure 1B.
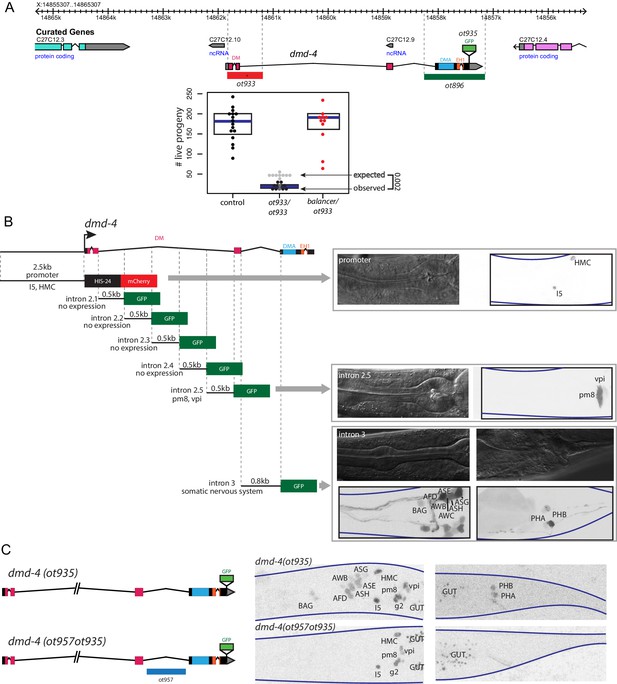
Generation of a viable cis-regulatory allele of dmd-4.
(A) The ot933 probable null allele of dmd-4 results in embryonic lethality and severe feeding defects in mutants that survive to adulthood. Deletion alleles generated by CRISPR/Cas9 are shown in the context of the dmd-4 locus above, the penetrance of ot933 embryonic lethality was quantified below by comparing the number of expected ot933 homozygous progeny from a balanced heterozygote mother (25% of total brood) with observed ot933 homozygous progeny. The difference between the observed number of ot933 homozygous progeny and expected homozygous progeny gives a measure of the penetrance of embryonic lethality. We observe a mean of 52% embryonic lethality, and comparing observed to expected ot933 progeny counts is significant by Wilcoxon rank-sum test. Each dot represents the brood size of one hermaphrodite, with blue bars indicating median and black boxes indicating quartiles. We show both observed and expected numbers of ot933 progeny in the figure panel. (B) The enhancers for I5/hmc expression, pharyngeal muscle expression, and somatic nervous system expression are located in separable regions of the dmd-4 promoter and introns. All constructs we examined are schematized, those that showed expression (promoter, intron 2.5, and intron 3) are shown in adult hermaphrodites to the right. GFP and mCherry are shown as color-inverted black and white, boundaries of the heads and tails of animals are marked with navy lines in all panels. Expressing cells are labeled on the GFP/mCherry images. (C) Generation of the dmd-4 (ot957ot935) nervous system null allele. Deletion of the third intron is schematized in the context of the dmd-4 locus above, color-inverted black and white GFP images of dmd-4 (ot935[DMD-4::GFP]) and dmd-4 (ot957ot935) hermaphrodites are shown below.
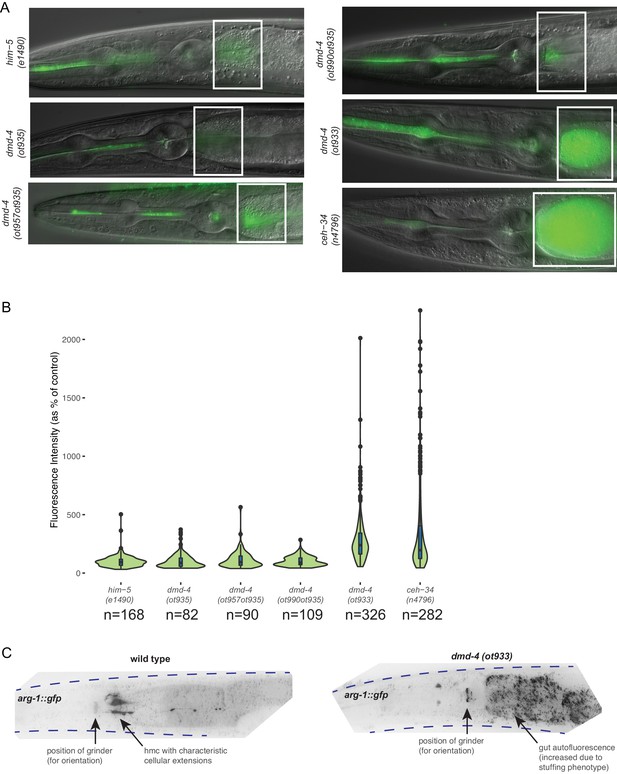
DMD-4 expression in the pharynx is required for proper anterior intestinal function.
(A) Constitutive loss of dmd-4 results in bacterial stuffing in the anterior intestine, as quantified by the accumulation of OP50-GFP. No anterior intestinal stuffing is observed in the him-5(e1490) control, dmd-4(ot935), dmd-4(ot957ot935), and dmd-4(ot990ot935). These four strains all maintain pharyngeal DMD-4 expression throughout the larval stages and into adulthood. The dmd-4(ot933) null mutant shows severe anterior intestinal stuffing similar to the ceh-34(n4796) strain, whose alimentary defect was used as a positive control (Hirose et al., 2010). Representative images of adult hermaphrodite animals are shown, white boxes indicate anterior intestine. (B) Quantification of OP50-GFP stuffing. Data are displayed as a violin plot, the internal box plots visualize the quartile distribution of data points, the center bar indicates the median, the blue boxes indicate quartiles, and the black bars indicate range. The black dots show outliers. (C) dmd-4(ot933) null mutants display defects in hmc development. The hmc is marked by a cytosolically localized arg-1::gfp transcriptional reporter (ccIs4443). In wild-type animals (n = 10), the hmc is localized posteriorly to the pharynx and always extends projections along the anterior intestine; in dmd-4 mutants (n = 10), no marker signal is visible in its normal localization, suggesting that the hmc does not develop properly. Representative color-inverted GFP images of adult hermaphrodite animals are shown; dotted lines show the outline of the animal. ‘GUT’ refers to autofluorescent gut granules (not GFP signal).
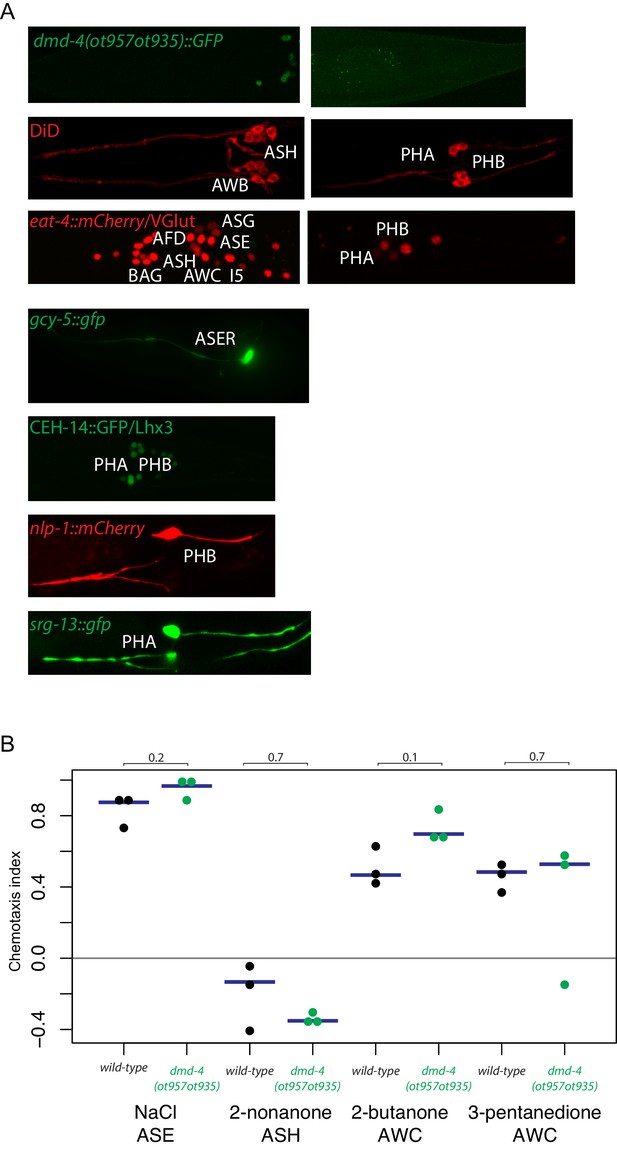
General neuron identity is unaffected in dmd-4 (ot957ot935) mutants.
(A) Markers for neuronal subtype differentiation of both head and tail neurons that express DMD-4 are shown in the dmd-4 (ot957ot935) mutant (where neuronal expression of DMD-4 is lost; shown at top). All markers continue to be expressed in dmd-4 mutant animals, suggesting normal generation and specification of all relevant neuron types. (B) Chemotaxis behavior generated by head sensory neurons is unaffected in dmd-4 (ot957ot935). Three experimental replicates were performed for all chemotaxis assays; each docant shows the chemotaxis index of one replicate, blue bars indicate median. P values are shown above black horizontal bars by Wilcoxon rank-sum test. The odorants used and relevant sensory neurons are shown below each experiment. Chemotaxis index calculation and assays were performed as previously described (see Materials and methods; Bargmann et al., 1993; Wes and Bargmann, 2001).
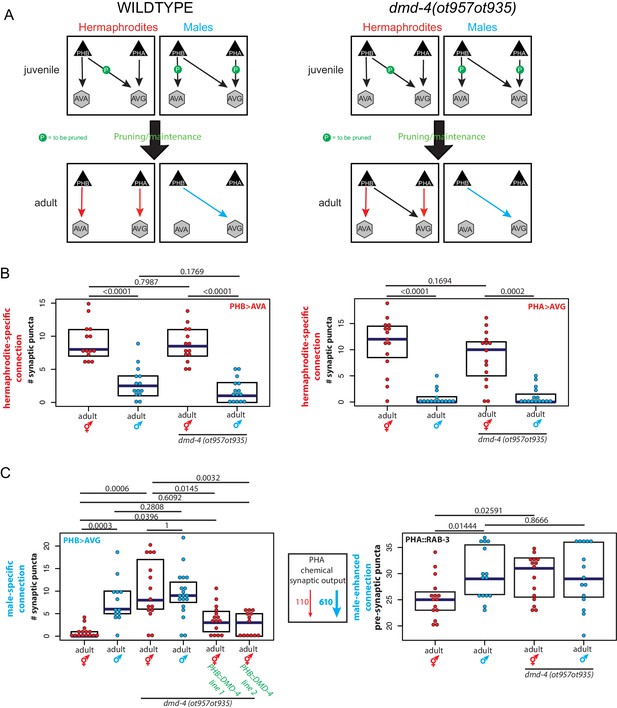
dmd-4 is required for sexually dimorphic synaptic connectivity.
(A) Sexually dimorphic connectivity of the phasmid neurons in wild-type animals and dmd-4 (ot957ot935) animals (quantified in B, representative images shown in Figure 3—figure supplement 2) is schematized. Adult sexual dimorphism is generated by synaptic pruning of juvenile ‘sex-hybrid’ connectivity (Oren-Suissa et al., 2016). (B) The hermaphrodite-specific connections between PHB>AVA and PHA>AVG are unaffected in dmd-4 (ot957ot935) mutants. Each dot represents one animal (red = hermaphrodite, cyan = male, in all figures), blue bars show median, black boxes represent quartiles. P values are shown above horizontal black bars by Wilcoxon rank-sum test with Bonferroni corrections for multiple testing where applicable (see Materials and methods). Representative images are shown in Figure 3—figure supplement 2. (C) The male-specific PHB>AVG connection is not pruned in dmd-4 (ot957ot935) mutant hermaphrodites, and this phenotype can be rescued by expressing dmd-4 cDNA specifically in the PHB neurons (‘PHB::DMD-4’). We quantified two independent transgenic lines for mutant rescue. dmd-4 (ot957ot935) mutant hermaphrodites also show an increased number of BFP puncta in the BFP::RAB-3 construct labeling pre-synaptic vesicles. The sexual dimorphism of PHA chemical synaptic output is shown on the left, based on the number of EM sections where PHA is presynaptic in both sexes, 110 sections in hermaphrodites and 610 sections in males, (Cook et al., 2019). We term this connectivity ‘male-enhanced’ as it is present in both sexes but stronger in the male. Representative images are shown in Figure 3—figure supplement 2.
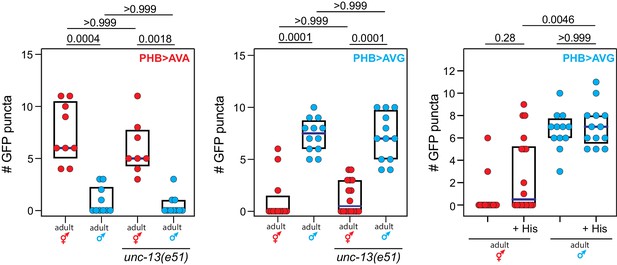
Phasmid synaptic pruning is not activity-dependent.
Reducing phasmid circuit activity either by mutation of the unc-13 regulator of synaptic vesicle exocytosis, or by chemogenetic, HisCl-mediated silencing of the PHB neuron, does not affect normal sex-specific synaptic pruning. Each dot represents one animal (red = hermaphrodite, cyan = male), blue bars show median, black boxes represent quartiles. P values are shown above horizontal black bars by Kruskal-Wallis with Dunn’s multiple comparisons test.
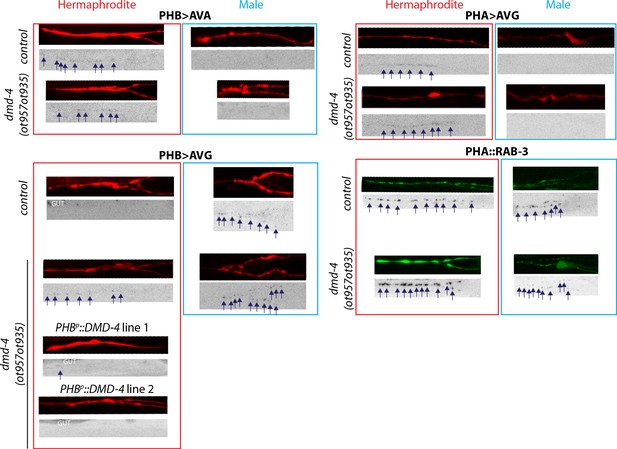
dmd-4 is required for dimorphic synaptic connectivity.
Representative images of data quantified in Figure 3B,C. Axon traces are shown above (RFP for GRASP and GFP for the PHA::RAB-3 strain), puncta are shown below in color-inverted black and white images. Intestinal auto-fluorescence is labeled ‘GUT’. Synaptic puncta are labeled with navy arrows.
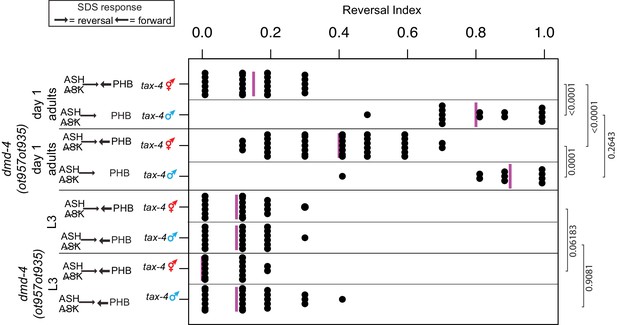
dmd-4 affects sexually dimorphic behavior.
Noxious chemosensory avoidance is mediated by the PHB neuron (Hilliard et al., 2002), and sexually dimorphic in adults (Oren-Suissa et al., 2016). Left panel shows predicted synaptic input into avoidance behavior by relevant amphid and phasmid neurons. All experiments were done using tax-4 (p678) mutants to disable amphid input and uncover phasmid function, as previously described (Hilliard et al., 2002). dmd-4 (ot957ot935) adult hermaphrodites show reduced chemosensory avoidance behavior, but this is not evident in larval animals. Each dot represents the average reversal index of one animal over 10 experimental trials, median shown with vertical magenta bar. P values shown to the right by Wilcoxon rank-sum test with Bonferroni corrections for multiple testing (where applicable; see Materials and methods).
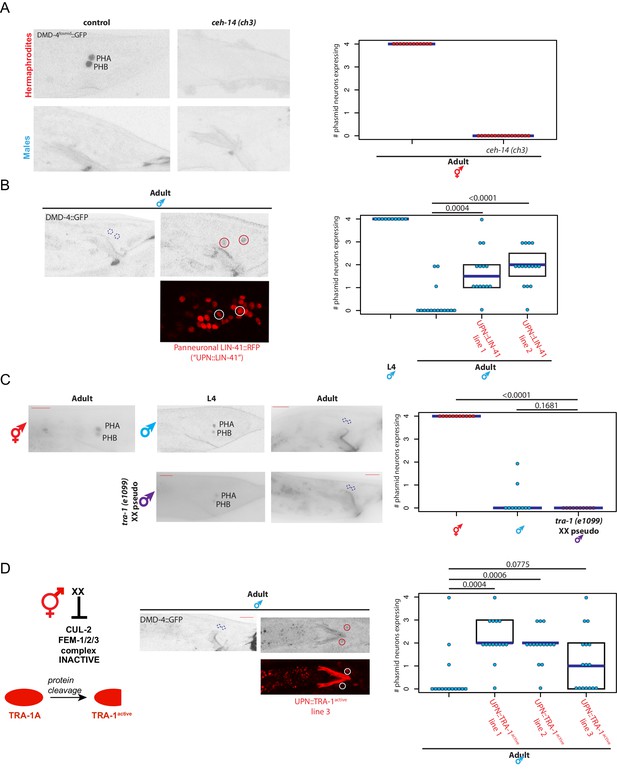
dmd-4 is regulated by spatial, temporal, and sex-specific cues in the phasmid neurons.
(A) DMD-4 expression is lost in hermaphrodite animals when the ceh-14 terminal selector of phasmid neuron fate is mutated. Male expression is unaffected. Color-inverted black and white GFP images are shown to the left in all panels, quantification of phasmid neurons expressing DMD-4 in wild-type and mutant animals is shown to the right in all panels. Horizontal blue line indicates median. (B) Constitutive, panneuronal expression of the LIN-41 heterochronic gene maintains DMD-4 in the adult male phasmids. Color-inverted black and white GFP images of adult males are shown in addition to the panneuronal, RFP-tagged LIN-41 overexpression (‘UPN::LIN-41’). L4 male quantification of DMD-4 expression in the phasmids (from Figure 1B) is shown again for reference comparison to control adults (siblings of transgenic animals) and LIN-41 overexpressing males. We quantified two independent transgenic lines. P values are shown above horizontal black bars by Wilcoxon rank-sum test with Bonferroni corrections for multiple testing where applicable (see Materials and methods) in all panels. (C) DMD-4 is degraded in the phasmid neurons of tra-1 (e1099) XX pseudomales in adulthood, but not in the L4 stage. (D) Constitutive expression of proteolytically cleaved TRA-1active feminizes adult male phasmid neurons and maintains DMD-4 expression. The hermaphrodite sex determination pathway represses the CUL-2/FEM-1/2/3 ubiquitin ligase pathway, resulting in TRA-1 stabilization and proteolytic cleavage, which is schematized to the left. Color-inverted black and white GFP images of adult males are shown in addition to the panneuronal, RFP-tagged TRA-1active in the center (‘UPN::TRA-1active::RFP’). We examined three independent transgenic lines; two had a significant effect on DMD-4 expression in the male phasmid neurons and one did not.
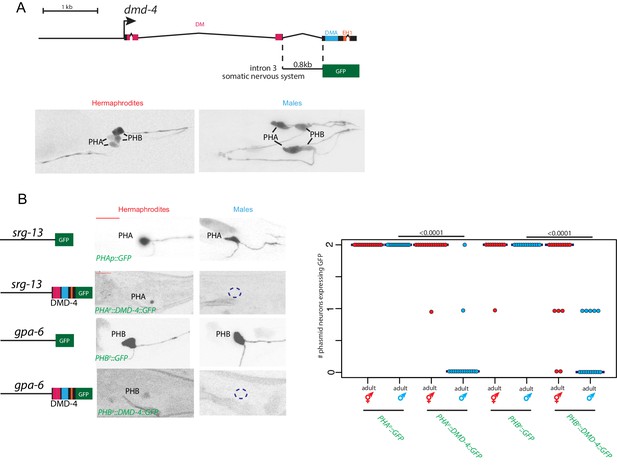
Sexually dimorphic degradation of DMD-4.
(A) Dimorphic expression is not regulated on the level of transcription. The enhancer driving DMD-4 expression in the somatic nervous system is schematized above (same as shown in Figure 2B), color-inverted black and white GFP images in adult hermaphrodite and male are shown below. (B) Driving dmd-4 cDNA under heterologous promoters in the phasmid neurons recapitulates the endogenous expression pattern. Heterologous promoters driving gfp with and without the dmd-4 cDNA are shown to the left in adult hermaphrodites and males, quantification is shown to the right. Scale bar (horizontal red bar) denotes 10 microns. For quantification, each dot represents the number of phasmid neurons expressing GFP in one animal (maximum is two neurons for PHAL/R or PHBL/R), red = hermaphrodite and cyan = male. Horizontal blue bars indicate median. P values are shown above horizontal black bars by Wilcoxon rank-sum test.
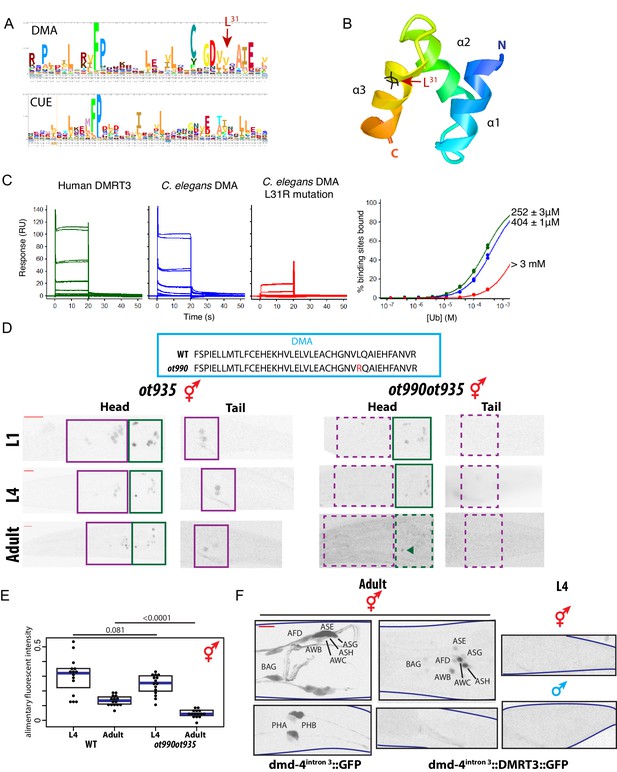
The conserved DMA domain is a novel, ubiquitin binding degron.
(A) Weighted sequence alignment of DMA domains and CUE domains across phyla. The similarities behind these two sequence profiles is demonstrated by pairwise alignments of profile hidden Markov models using HHsearch (Soding, 2005) and is noted in the PF03474 Pfam record of the DMA domain. (B) Homology model of the DMD-4 DMA domain based on the known structure of a ubiquitin-binding CUE domain (Kang et al., 2003). The three alpha-helices are labeled; helices 1 and 3 form a binding surface for contact with ubiquitin. The leucine 31 side chain mutated in panel D is highlighted in black; note the position on the ubiquitin-binding surface. (C) SPR binding analysis of the C. elegans dmd-4 and human Dmrt3 DMA domains shows that both bind to monoubiquitin, and this binding is lost in the L31R mutation (see panel A). (D) CRISPR/Cas9 mutation of leucine 31 (see panel B) to arginine, which we predicted to impair DMA binding to ubiquitin, promotes DMD-4::GFP degradation. ot935 is the gfp-tagged dmd-4 locus and the ot990 allele carries the leucine 31 mutation (to Arg) as indicated. Color-inverted black and white GFP images of L1/L4/adult hermaphrodite heads and tails are shown below, purple boxes indicate nervous system expression, green boxes indicate pharyngeal expression. Dashed boxes indicate missing expression. I5 sometimes maintains DMD-4::GFP expression in adults (green arrowhead). Scale bars (red horizontal bars) denote 10 microns. (E) DMD-4::GFP fluorescence intensity in gfp tagged animals (ot935) and ot935 animals in which the ot990 mutation is incorporated (shown in panel D) is quantified in a defined region of the posterior pharynx (alimentary tissues excluding I5) in L4 and adult wildtype and mutant hermaphrodites. Each dot represents fluorescence intensity in one animal (normalized to background fluorescence), blue bars indicate medians, black boxes indicate quartiles. P values are shown above horizontal black bars by Wilcoxon rank-sum test. (F) Expressing human DMRT3 under the dmd-4intron3 enhancer ("dmd-4intron3::DMRT3::GFP") recapitulates DMD-4 expression in the head neurons, but in the phasmid neurons DMRT3 is degraded constitutively and is absent in both sexes in both larval and adult stages (compared to expression of the enhancer alone, 'dmd-4intron3::GFP'). Color-inverted black and white GFP images of adult hermaphrodite heads and tails and L4 hermaphrodite and male tails are shown, red scale bar indicates 10 microns.
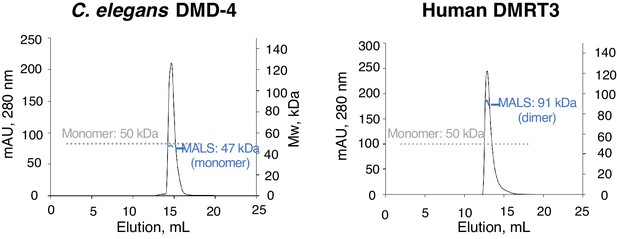
Analysis of DMA domain dimerization.
SEC-MALS experiments for C. elegans DMD-4 and human DMRT DMA domains. Plots show A280 for each MBP-fusion protein plotted against elution volume. The grey dotted line shows the monomer molecular weight based on sequence, and apparent molecular weights determined from light scattering analysis is shown in blue at the peak position.
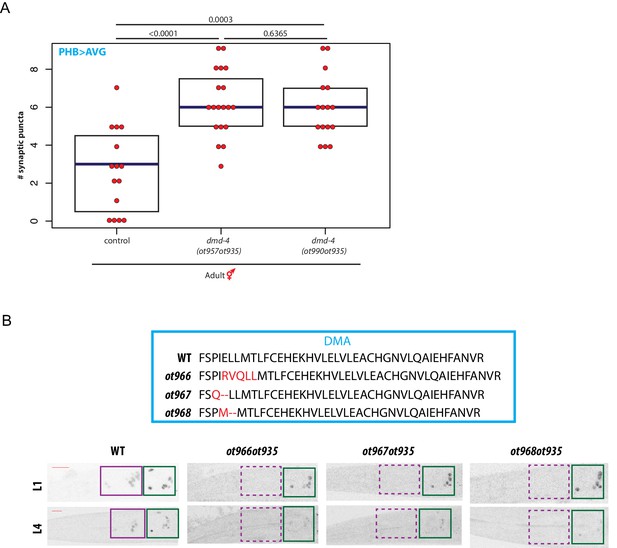
Disrupting the DMA domain promotes DMD-4::GFP degradation.
(A) Loss of DMD-4 expression in the nervous system in the L31R dmd-4(ot990) mutant disrupts pruning of the male-specific PHB>AVG connection in adult hermaphrodites. GRASP data from L31R mutants are shown alongside wild-type and dmd-4(ot957ot935) null mutants. Each dot represents one adult hermaphrodite animal, blue bars show median, black boxes represent quartiles. P values are shown above horizontal black bars by Wilcoxon rank-sum test (B) CRISPR/Cas9 mutation of the first alpha helix in the DMA domain promotes DMD-4::GFP degradation. The sequences of the ot966, ot967, and ot968 mutations are given relative to wild-type above. Color-inverted black and white GFP images of L1 and L4 hermaphrodite heads and tails are shown below, purple boxes indicate nervous system expression, green boxes indicate pharyngeal expression. Dashed boxes indicate missing expression. Scale bars (red horizontal bars) denote 10 microns.
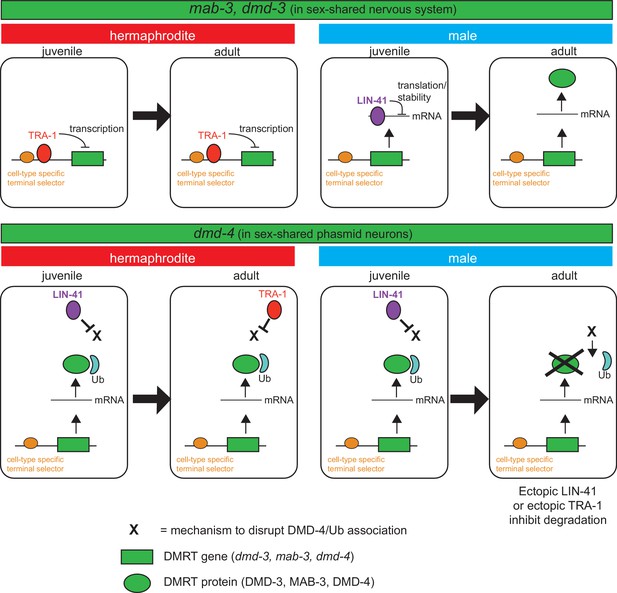
Spatiotemporal specificity and regulation of DMD-4 expression differs from the regulation of other C. elegans DMRT genes.
Similar pathways are used to control the spatial, temporal and sexual specificity of C. elegans DMRT genes, but these pathways are utilized in a fundamentally distinct manner for different DMRT proteins, ranging from transcriptional to posttranslation regulation. See text for more discussion.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Caenorhabditis elegans) | him-5 (e1490) V; dmd-4 (ot935) X | This work | OH15814 ‘DMD-4::GFP’ | Figure 1 |
| Genetic reagent (C. elegans) | dmd-4 (ot896) X | This work | OH15302 | Figure 2 |
| Genetic reagent (C. elegans) | dmd-4(ot933)/tmC24 X | This work | OH16260 | Figure 2 |
| Genetic reagent (C. elegans) | ccIs4251; stIs10539 | CGC | SD1633 | Figure 2 ccIs4251 [(pSAK2) myo-3p::GFP::LacZ::NLS + (pSAK4) myo-3p::mitochondrial GFP + dpy-20(+)] I; stIs10539 [dmd-4p::HIS-24::mCherry + unc-119(+)] |
| Genetic reagent (C. elegans) | otEx6774; him-5 (e1490) | This work | OH14506 | Figure 2 dmd-4int3::GFP, pRF4 |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH15908 | Figure 2 |
| Genetic reagent (C. elegans) | otIs612; him-5 (e1490) | Oren-Suissa et al., 2016 | OH13575 ‘PHB > AVA’ | Figure 3 MVC12 (flp-18p::nlg-1::gfp11)15 ng/ul, MVC6 (gpa-6p::nlg-1::gfp1- 10)15 ng/ul, MVC11 (flp- 18 p::mcherry)10 ng/ul, MVC15 (nlp- 1::mcherry)10 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otIs612; him-5 (e1490); dmd-4 (ot957ot935) X | This work | OH16014 ‘PHB > AVA’ | Figure 3 MVC12 (flp-18p::nlg-1::gfp11) 15 ng/ul, MVC6 (gpa-6p::nlg-1::gfp1- 10)15 ng/ul, MVC11 (flp- 18 p::mcherry)10 ng/ul, MVC15 (nlp- 1::mcherry)10 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otEx6347; him-5 (e1490) V | Oren-Suissa et al., 2016 | OH13696 ‘PHA > AVG’ | Figure 3 srg-13::BirA::nrx-1 25 ng/ul, inx-18p::AP::nlg-1 25 ng/ul, inx-18p::wcherry 10 ng/ul, unc-122::streptavidin::2xsfGFP 25 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otEx6347; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16067 ‘PHA > AVG’ | Figure 3 srg-13::BirA::nrx-1 25 ng/ul, inx-18p::AP::nlg-1 25 ng/ul, inx-18p::wcherry 10 ng/ul, unc-122::streptavidin::2xsfGFP 25 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otIs614; him-5 (e1490) | Oren-Suissa et al., 2016 | OH13577 ‘PHB > AVG’ | inx-18p::NLG-1::GFP1130 ng/ul, MVC6 gpa-6::NLG-1:::GFP1-10 30 ng/ul, MVC15 nlp-1::mcherry 5 ng/ul, inx-18::wcherry 10 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otIs614; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16017 ‘PHB > AVG’ | Figure 3 inx-18p::NLG-1::GFP11 30 ng/ul, MVC6 gpa-6::NLG-1:::GFP1-10 30 ng/ul, MVC15 nlp-1::mcherry 5 ng/ul, inx-18::wcherry 10 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otEx7431; otIs614; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16217 ‘PHBp::DMD-4 line 1’ | Figure 3 gpa-6p::DMD-4::GFP, ttx-3p::GFP |
| Genetic reagent (C. elegans) | otEx7433; otIs614; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16208 ‘PHBp::DMD-4 line 2’ | Figure 3 gpa-6p::DMD-4::GFP, ttx-3p::GFP |
| Genetic reagent (C. elegans) | otIs702; him-5 (e1490) V | This work | OH15565 ‘PHA::RAB-3’ | Figure 3 srg-13p::RAB-3::BFP, srg-13p::gfp, ttx-3::mCherry |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot935ot957) X | This work | OH16030 ‘PHA::RAB-3’ | Figure 3 srg-13p::RAB-3::BFP, srg-13p::gfp, ttx-3::mCherry |
| Genetic reagent (C. elegans) | tax-4 (p678); him-5 (e1490) | Oren-Suissa et al., 2016 | OH15339 | Figure 4 |
| Genetic reagent (C. elegans) | tax-4 (p678); him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16115 | Figure 4 |
| Genetic reagent (C. elegans) | wgIs418; him-5 (e1490) V | This work | OH16268 | Figure 5 DMD-4fosmid::gfp |
| Genetic reagent (C. elegans) | wgIs418; ceh-14 (ch3) X | This work | OH14503 | Figure 5 DMD-4fosmid::gfp |
| Genetic reagent (C. elegans) | otEx7429; him-5 (e1490) V; dmd-4 (ot935) X | This work | OH16159 ‘UPN::LIN-41 line 1’ | Figure 5 UPN::LIN-41A, rab-3p::tagRFP, pRF4 |
| Genetic reagent (C. elegans) | otEx7451; him-5 (e1490) V; dmd-4 (ot935) X | This work | OH16235 ‘UPN::LIN-41 line 2’ | Figure 5 UPN::LIN-41A, rab-3p::tagRFP, pRF4 |
| Genetic reagent (C. elegans) | tra-1 (e1099) III/hT2 [bli-4(e937) umnIs36] (I;III); dmd-4 (ot935) X | This work | OH16194 | Figure 5 |
| Genetic reagent (C. elegans) | otEx7352; him-5 (e1490) V; dmd-4 (ot935) X | This work | OH15952 ‘UPN::TRA-1active line 1’ | Figure 5 UPN::tagRFP::TRA-1active, pRF4 |
| Genetic reagent (C. elegans) | otEx7353; him-5 (e1490) V; dmd-4 (ot935) X | This work | OH16158 ‘UPN::TRA-1active line 2’ | Figure 5 UPN::tagRFP::TRA-1active, pRF4 |
| Genetic reagent (C. elegans) | otEx7354; him-5 (e1490) V; dmd-4 (ot935) X | This work | OH15993 ‘UPN::TRA-1active line 3’ | Figure 5 UPN::tagRFP::TRA-1active, pRF4 |
| Genetic reagent (C. elegans) | otIs627; him-8 (e1489) | This work | OH14022 ‘PHAp::GFP’ | Figure 6 srg-13p::GFP |
| Genetic reagent (C. elegans) | otEx7435; him-5 (e1490) V ; dmd-4 (ot957ot935) X | This work | OH16177 ‘PHAp::DMD-4::GFP’ | Figure 6 srg-13p::DMD-4::GFP, ttx-3p::GFP |
| Genetic reagent (C. elegans) | otEx6456; him-5 (e1490) V | This work | OH13892 ‘PHBp::GFP’ | Figure 6 gpa-6p::GFP |
| Genetic reagent (C. elegans) | otEx7433; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16175 ‘PHBp::DMD-4::GFP’ | Figure 6 gpa-6p::DMD-4::GFP, ttx-3p::GFP |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot990ot935) X | This work | OH16125 | Figure 7 |
| Genetic reagent (C. elegans) | nIs175 [ceh-28p::4xNLS::GFP + lin-15AB(+)] IV; ceh-34(n4796) V | CGC | MT15695 | Figure 2—figure supplement 1 |
| Genetic reagent (C. elegans) | otIs518; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16054 | Figure 2—figure supplement 2 |
| Genetic reagent (C. elegans) | ntIs1; dmd-4 (ot957ot935) X | This work | OH16225 | Figure 2—figure supplement 2 |
| Genetic reagent (C. elegans) | wgIs73; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16018 | Figure 2—figure supplement 2 |
| Genetic reagent (C. elegans) | otIs612; unc-13(e51) I;him-5 (e1490) V | This work | MOS14 | Figure 3—figure supplement 1 |
| Genetic reagent (C. elegans) | otEx6913; unc-13(e51) I; him-5 (e1490) V | This work | MOS234 | Figure 3—figure supplement 1 inx-18p::NLG-1::GFP11, MVC6 gpa-6::NLG-1:::GFP1-10, MVC15 inx-18::wcherry, pRF4 |
| Genetic reagent (C. elegans) | etyEx2; otIs614; him-5 (e1490) V | This work | MOS243 | Figure 3—figure supplement 1 gpa-6::HisCl1, myo-2::GFP |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot966ot0935) X | This work | OH16013 | Figure 7—figure supplement 2 |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot967ot935) X | This work | OH16015 | Figure 7—figure supplement 2 |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot968ot935) X | This work | OH16016 | Figure 7—figure supplement 2 |
| Genetic reagent (C. elegans) | otIs614; him-5 (e1490) V; dmd-4 (ot990ot935) X | This work | OH16771 | Figure 7—figure supplement 2 |
| Genetic reagent (C. elegans) | ccIs4443 IV; him-5 (e1490) V | This work | OH16545 | Figure 2—figure supplement 1 arg-1::GFP + dpy-20(+) |
| Genetic reagent (C. elegans) | ccIs4443 IV; him-5 (e1490) V; dmd-4 (ot933) X | This work | OH16770 | Figure 2—figure supplement 1 |





