Methionine restriction breaks obligatory coupling of cell proliferation and death by an oncogene Src in Drosophila
Figures

Src activation induces both cell proliferation and death, resulting in a mild tissue overgrowth.
(A) Src42A constitutively active (CA) expression induces cell proliferation, which was detected by phospho-histone 3 (pH3) staining. (B) Quantification of pH3 staining. The number of pH3+ cells was normalized by the area of GFP+ cells. Two-tailed unpaired t-test. (C) Src42A CA expression induces caspase activation, which was detected by cleaved DCP1 staining. (D) Quantification of percentage of cDCP1+ cells in GFP+ cells. Two-tailed unpaired t-test. (E) Quantification of the total volume of GFP+ cells (µm³). Two-tailed unpaired t-test. Scale bars, 100 µm.
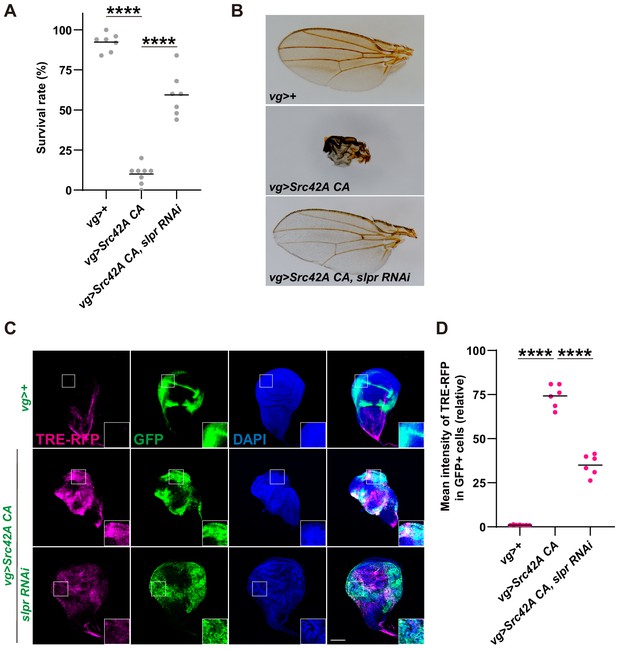
Inhibition of Slpr suppresses the phenotypes induced by Src activation.
(A) Src42A constitutively active (CA) expression in the wing disc induces organismal lethality, which is suppressed by knockdown of slpr. One-way ANOVA with Sidak’s post-test. (B) The small, disheveled wing phenotype of the rare escapers with Src42A CA is suppressed by knockdown of slpr. (C) Src42A CA-mediated JNK activation, which was detected by the TRE-RFP reporter, is suppressed by knockdown of slpr. (D) Quantification of TRE-RFP in C. One-way ANOVA with Sidak’s post-test. Scale bars, 100 µm.
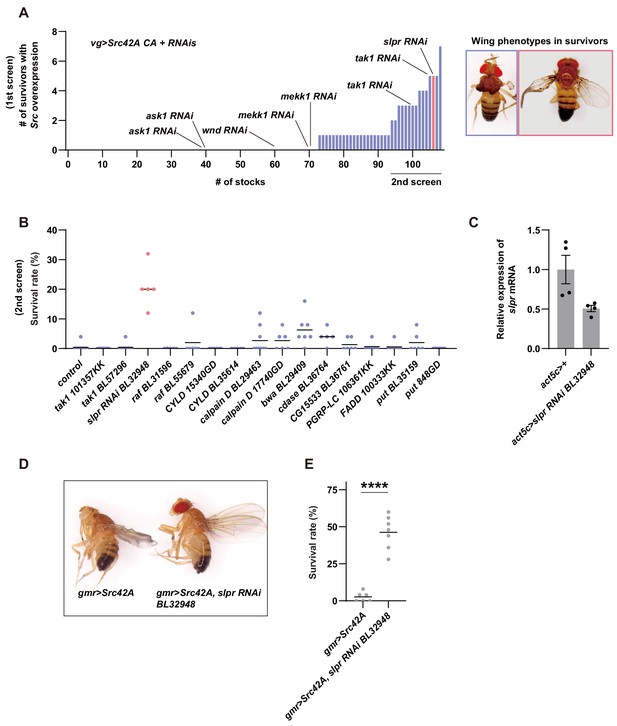
Inhibition of Slpr suppresses the phenotypes induced by Src activation.
(A) In the first screening, 107 RNAi stocks were used. The numbers of survivors were counted. The blue color indicates that survivors have disheveled wings while the magenta color indicates that survivors have the recovered wing phenotype. (B) In the second screening, the RNAi stocks that produced more than two survivors in the first screening were used. The blue color indicates that survivors have disheveled wings while the magenta color indicates that survivors have the recovered wing phenotype. (C) RT-qPCR of slpr using RNA from the larval whole body. slpr RNAi BL32948 lowered the transcriptional expression of slpr. (D) Src42A expression in the eye imaginal disc with gmr-Gal4 eliminates the adult eye, which can be suppressed by knockdown of slpr. Note that the disheveled wing phenotype induced by leaky expression of gmr-Gal4 in the wing disc is also suppressed by knockdown of slpr. (E) Src42A expression in the eye disc induces organismal lethality during development, which is suppressed by slpr knockdown. Two-tailed unpaired t-test.
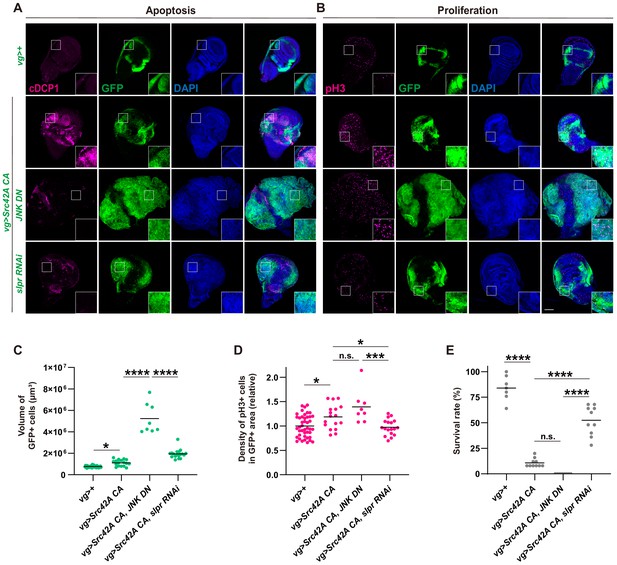
Slpr regulates both cell proliferation and cell death that are induced by Src activation.
(A) Apoptosis induced by Src42A constitutively active (CA) is suppressed by slpr RNAi and JNK DN. Note the aggressive overgrowth phenotype induced by combining Src with JNK DN. (B) JNK DN does not inhibit Src42A CA-mediated proliferation whereas slpr inhibition does. (C) Quantification of the total volume of GFP+ cells (µm³) in B. One-way ANOVA with Sidak’s post-test. (D) Quantification of phospho-histone 3 (pH3) staining in B. The number of pH3+ cells was normalized by the area of GFP+ cells. Src42A CA-induced proliferation is suppressed by knockdown of slpr but not by overexpression of JNK DN. One-way ANOVA with Sidak’s post-test. (E) Inhibition of JNK enhances organismal lethality induced by Src42A CA. One-way ANOVA with Sidak’s post-test. Scale bars, 100 µm.
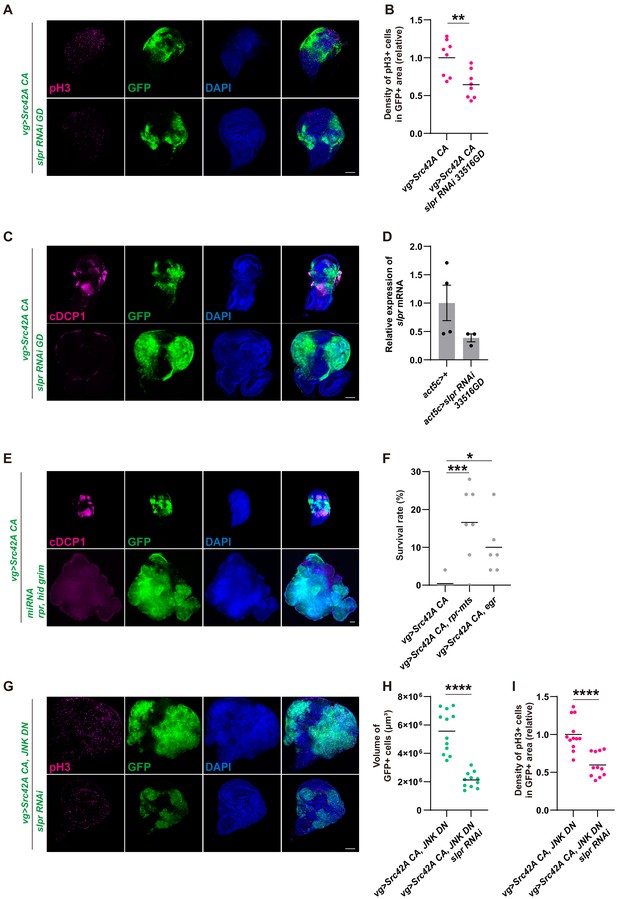
Inhibition of Slpr suppresses the phenotypes induced by Src activation.
(A) slpr RNAi 33516GD suppresses Src-induced cell proliferation, similarly to slpr RNAi BL32948. (B) Quantification of phospho-histone 3 (pH3) staining in A. The number of pH3+ cells was normalized by the area of GFP+ cells. Src42A constitutively active (CA)-induced proliferation is suppressed by slpr knockdown with the 33516GD line. Two-tailed unpaired t-test. (C) slpr RNAi 33516GD suppresses Src-induced cell death, similarly to slpr RNAi BL32948. (D) RT-qPCR of slpr using RNA from the larval whole body. slpr RNAi 33516GD lowered the transcriptional expression of slpr. (E) Apoptosis induced by Src42A CA is suppressed by miRNA for rpr, hid, grim. (F) Induction of cell death by rpr-mts or egr suppressed organismal lethality induced by Src42A CA. One-way ANOVA with Sidak’s post-test. (G) Overgrowth induced by Src activation and JNK inhibition is suppressed by slpr knockdown. (H) Quantification of the total volume of GFP+ cells (µm³) in G. Two-tailed unpaired t-test. (I) Quantification of pH3 staining in G. The number of pH3+ cells was normalized by the area of GFP+ cells. Cell proliferation induced by Src activation and JNK inhibition is suppressed by slpr knockdown. Two-tailed unpaired t-test. Scale bars, 100 µm.
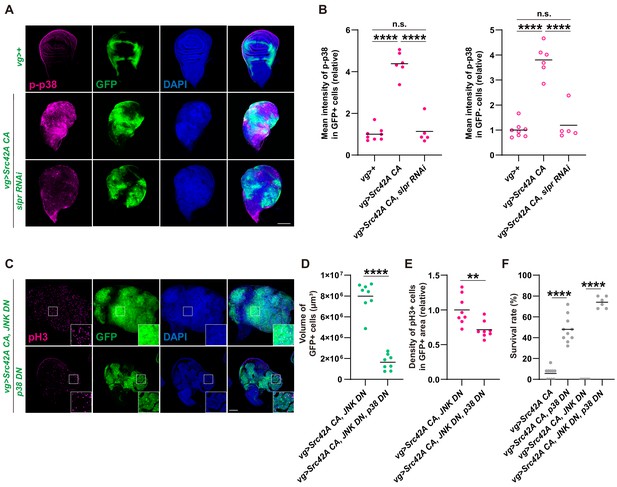
p38 mediates Src-induced cell proliferation.
(A) Src42A constitutively active (CA) expression induces phosphorylation of p38 both cell autonomously and non-cell autonomously, which is suppressed by slpr knockdown. (B) Quantification of phosphorylated p38 in A. One-way ANOVA with Sidak’s post-test. (C) Src42A CA-induced proliferation with/without JNK inhibition is suppressed by p38 DN. (D) Quantification of the total volume of GFP+ cells (µm³) in C. Two-tailed unpaired t-test. (E) Quantification of phospho-histone 3 (pH3) staining in C. The number of pH3+ cells was normalized by the area of GFP+ cells. Two-tailed unpaired t-test. (F) Inhibition of p38 suppresses organismal lethality induced by Src42A CA. One-way ANOVA with Sidak’s post-test. Scale bars, 100 µm.
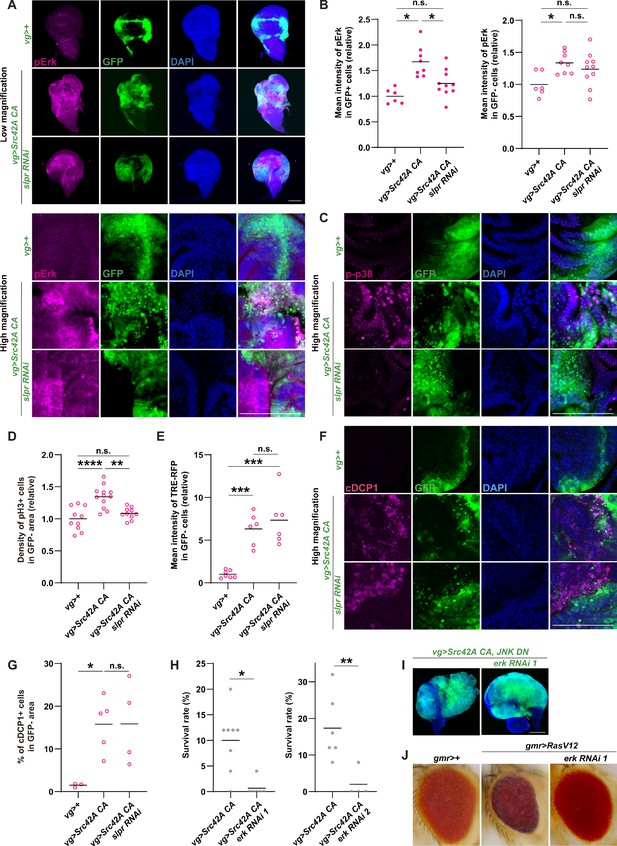
Erk is activated downstream of Src-Slpr signaling but does not mediate cell proliferation.
(A) Src42A constitutively active (CA) expression induces phosphorylation of Erk both cell autonomously and non-cell autonomously, which is suppressed by slpr knockdown. (B) Quantification of phosphorylated Erk in A. One-way ANOVA with Sidak’s post-test. (C) A high magnification picture demonstrating that Src42A CA expression induces phosphorylation of p38 both cell autonomously and non-cell autonomously, which is suppressed by slpr knockdown. (D) Src42 CA-induced non-cell autonomous proliferation is suppressed by slpr knockdown. One-way ANOVA with Sidak’s post-test. (E) Quantification of TRE-RFP in GFP-negative cells in Figure 2C. Src42A CA expression induces non-cell autonomous JNK activation, which is not suppressed by slpr knockdown. One-way ANOVA with Sidak’s post-test. (F) Src42A CA expression induces apoptosis both cell autonomously and non-cell autonomously. slpr knockdown suppresses Src42A CA-induced cell autonomous apoptosis but not non-cell autonomous one. (G) Quantification of percentage of cDCP1+ cells in GFP- cells in F. One-way ANOVA with Sidak’s post-test. (H) Knockdown of erk worsens organismal survival over the Src stress. Two-tailed unpaired t-test. (I) Inhibition of Erk does not inhibit cell proliferation induced by Src42A CA and JNK DN. (J) The erk RNAi stock suppresses the RasV12-induced rough eye phenotype, validating its knockdown. Scale bars, 100 µm.
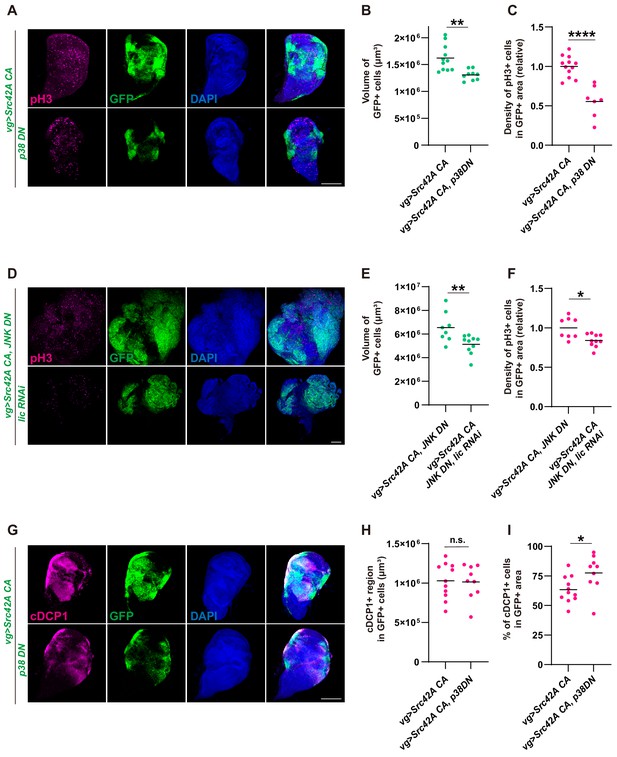
p38 inhibition does not inhibit Src-induced apoptosis.
(A) Src42A constitutively active (CA)-induced proliferation is suppressed by p38 inhibition. (B) Quantification of the total volume of GFP+ cells in A. p38 inhibition suppresses Src42A CA-induced proliferation. Two-tailed unpaired t-test. (C) Quantification of phospho-histone 3 (pH3) staining in A. The number of pH3+ cells was normalized by the area of GFP+ cells. Two-tailed unpaired t-test. (D) Cell proliferation induced by Src42A CA and JNK inhibition is suppressed by lic knockdown. (E) Quantification of the total volume of GFP+ cells in D. Two-tailed unpaired t-test. (F) Quantification of pH3 staining in D. The number of pH3+ cells was normalized by the area of GFP+ cells. Cell proliferation induced by Src42A CA and JNK inhibition is suppressed by lic knockdown. Two-tailed unpaired t-test. (G) Inhibition of p38 does not suppress DCP1 activation induced by Src42A CA. (H) Quantification of the volume of apoptotic cells in the Src42A CA expressing region. p38 inhibition does not affect the volume of cDCP1+ cells. Two-tailed unpaired t-test. (I) Percentage of cDCP1+ cells in GFP+ cells. Since p38 inhibition decreases proliferation but not the volume of cDCP1+ cells, the percentage of cDCP1+ cells in GFP+ cells increases by p38 inhibition. Two-tailed unpaired t-test. Scale bars, 100 µm.
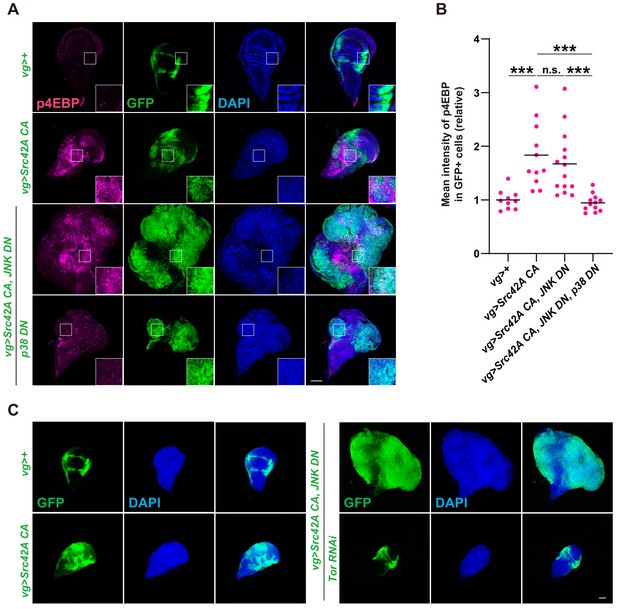
Tor signaling functions downstream of the Src-p38 pathway.
(A) Src42A constitutively active (CA) induces phosphorylation of 4EBP, a readout of Tor activation. Src42A CA-mediated phosphorylation of 4EBP was suppressed by p38 inhibition but not by JNK inhibition. (B) Quantification of phosphorylated 4EBP in A. One-way ANOVA with Sidak’s post-test. (C) Knockdown of Tor suppresses Src42A CA-induced cell proliferation. Scale bars, 100 µm.
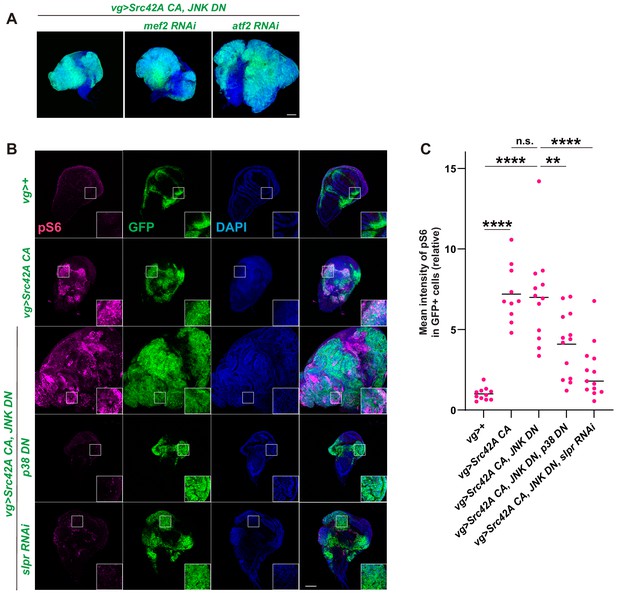
Inhibition of Mef2 or Atf2 does not suppress Src42A constitutively active (CA)-induced cell proliferation.
(A) Knockdown of mef2 or atf2, well-known downstream factors of p38, does not suppress Src42A CA-induced proliferation. (B) Src42A CA induces phosphorylation of S6, another readout of Tor activation. Src42A CA-mediated phosphorylation of S6 was suppressed by p38 inhibition or slpr knockdown but not by JNK inhibition. (C) Quantification of phosphorylated S6 in B. One-way ANOVA with Sidak’s post-test. Scale bars, 100 µm.
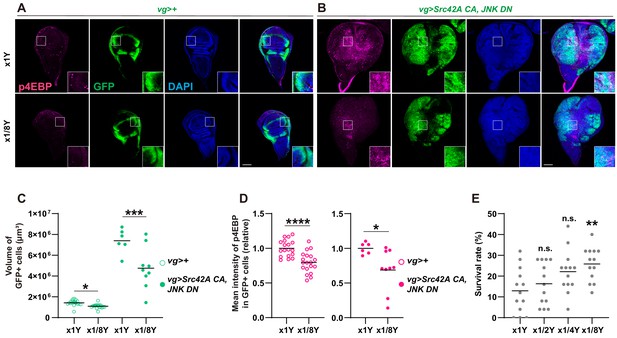
Yeast dilution affects Src-induced Tor signaling, tissue growth, and organismal lethality.
(A-B) Dietary restriction of yeast suppresses both Src42A constitutively active (CA)-induced proliferation and phosphorylation of 4EBP. (C) Quantification of the total volume of GFP+ cells (µm³) in A-B. Mann-Whitney test. (D) Quantification of phosphorylated 4EBP in A-B. Two-tailed unpaired t-test. (E) Dietary restriction of yeast reduces organismal lethality caused by Src42A CA expression in the wing disc in a dose-dependent manner. One-way ANOVA with Sidak’s post-test.
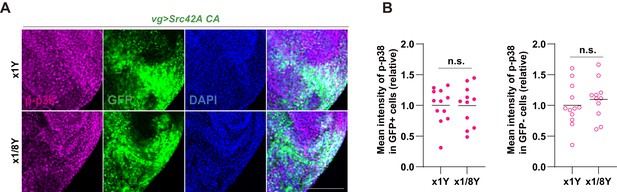
Yeast dilution does not affect Src-induced p38 phosphorylation.
(A) Yeast dilution does not suppress phosphorylation of p38 induced by Src42A constitutively active (CA). (B) Quantification of phosphorylated p38 in A. Both cell autonomous and non-cell autonomous p38 activation by Src42A CA are not suppressed by dietary restriction of yeast. Two-tailed unpaired t-test. Scale bars, 50 µm.

Methionine regulates Src-induced Tor signaling, tissue growth, and organismal lethality.
(A) Addition of essential amino acids enhances organismal lethality caused by Src42A constitutively active (CA) expression in the wing disc, whereas addition of non-essential amino acids does not. Kruskal-Wallis test with Dunn’s post-test. (B) Only methionine subtraction from the diet improves organismal survival over the Src42A CA stress. One-way ANOVA with Sidak’s post-test. (C) Addition of methionine reduces organismal survival over the Src42A CA stress in a dose-dependent manner. One-way ANOVA with Sidak’s post-test. (D) An amount of methionine in the hemolymph was measured by LC-MS/MS. Expression of Src42A CA in the wing disc decreases the circulating methionine in the hemolymph. Two-tailed unpaired t-test. (E-F) Dietary methionine activates both cell proliferation and phosphorylation of 4EBP that are induced by Src42A CA and JNK DN. (G) Quantification of the total volume of GFP+ cells (µm³) in E-F. Mann-Whitney test. (H) Quantification of phosphorylated 4EBP in E-F. Two-tailed unpaired t-test. (I) SamS knockdown suppresses phosphorylation of 4EBP and overgrowth induced by Src42A CA and JNK DN. (J) Quantification of the total volume of GFP+ cells (µm³) in I. One-way ANOVA with Sidak’s post-test. (K) Quantification of phosphorylated 4EBP in I. One-way ANOVA with Sidak’s post-test. Scale bars, 100 µm.
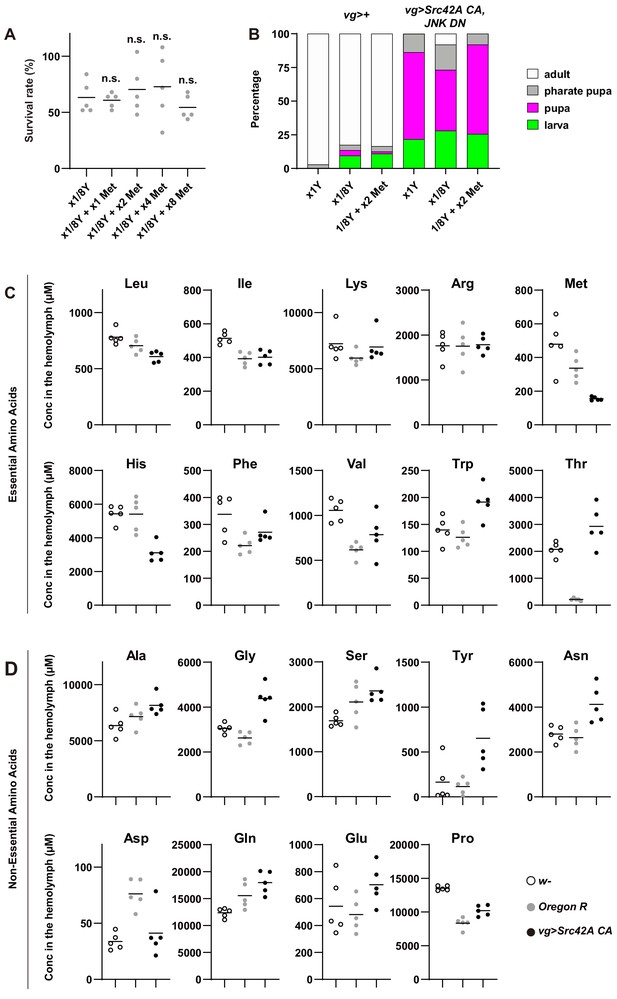
Involvement of amino acids in Src-induced oncogenic stress.
(A) Methionine addition does not affect survival of control flies without tumor burden. One-way ANOVA with Sidak’s post-test. (B) The graph shows which stage animals could reach. Dietary restriction of yeast suppresses organismal lethality induced by Src42A constitutively active (CA) overexpression with JNK inhibition. Methionine supplementation suppresses the dietary restriction-mediated effect on organismal lethality. (C) The concentrations of EAAs in the hemolymph were measured by CE-MS. (D) The concentrations of NEAAs in the hemolymph were measured by CE-MS.
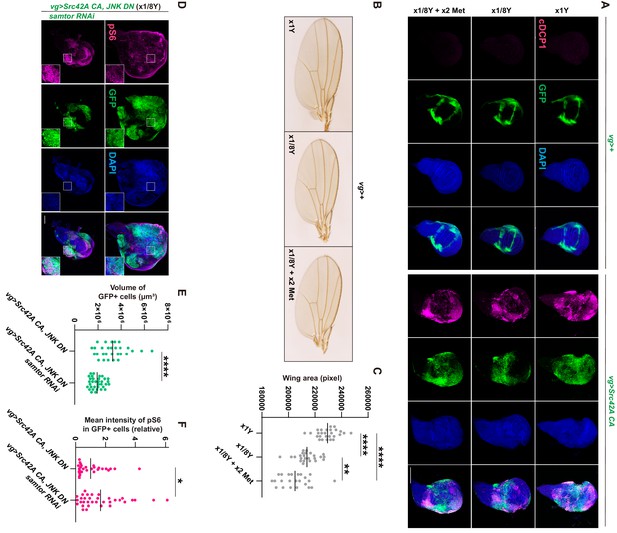
Involvement of methionine in Src-induced Tor activation.
(A) Neither dietary restriction of yeast nor methionine supplementation affects Src42A constitutively active (CA)-induced apoptosis. (B-C) Dietary restriction of yeast reduces the wing tissue development, which is estimated by the size of wings. Methionine supplementation does not rescue the tissue growth defects caused by yeast restriction. One-way ANOVA with Sidak’s post-test. (D) Samtor knockdown enhances Tor activation induced by Src42A CA and JNK DN in a diluted yeast condition, where Tor activation is suppressed. On the other hand, the overgrowth phenotype induced by Src42A CA and JNK DN was suppressed. (E) Quantification of the total volume of GFP+ cells (µm³) in D. Two-tailed unpaired t-test. (F) Quantification of phosphorylated S6 in D. Two-tailed unpaired t-test. Scale bars, 100 µm.
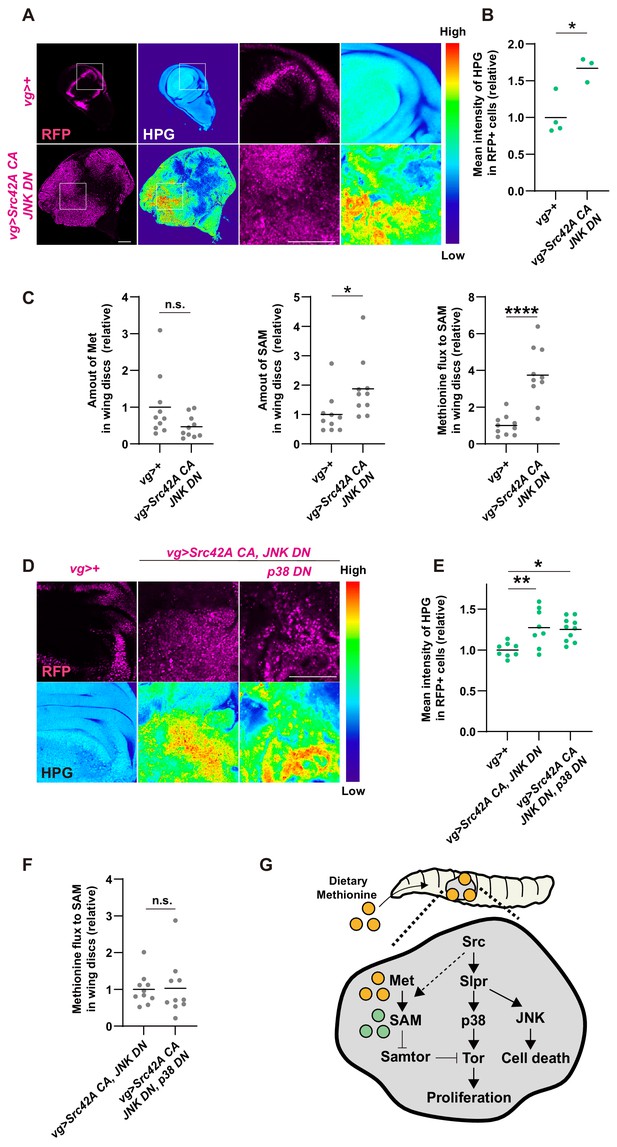
Cross talk between Src signaling and methionine-Tor signaling.
(A) An in vitro culture of the wing disc with a methionine analog homopropargylglycine (HPG) demonstrates that the tumor disc induced by Src42A constitutively active (CA) and JNK DN uptakes more methionine than the control disc. (B) Quantification of the HPG intensity in A. Two-tailed unpaired t-test. (C) The amounts of methionine and SAM in the wing discs were measured by LC-MS. The tumor disc induced by Src42A CA and JNK DN contains a higher amount of SAM, whereas the amount of methionine is not significantly different. Methionine flux was calculated as a ratio of SAM and methionine. Two-tailed unpaired t-test. (D) The increase of methionine incorporation in the Src tumor is not mediated by p38. (E) Quantification of the HPG intensity in D. One-way ANOVA with Sidak’s post-test. (F) p38 inhibition does not suppress the upregulated methionine flux by Src42A CA and JNK DN. Two-tailed unpaired t-test. (G) A schematic of the Src42A CA-mediated coupling of cell proliferation and death. JNK activates cell death, while p38 activates cell proliferation, which is regulated by methionine-mediated Tor signaling. Scale bars, 100 µm.
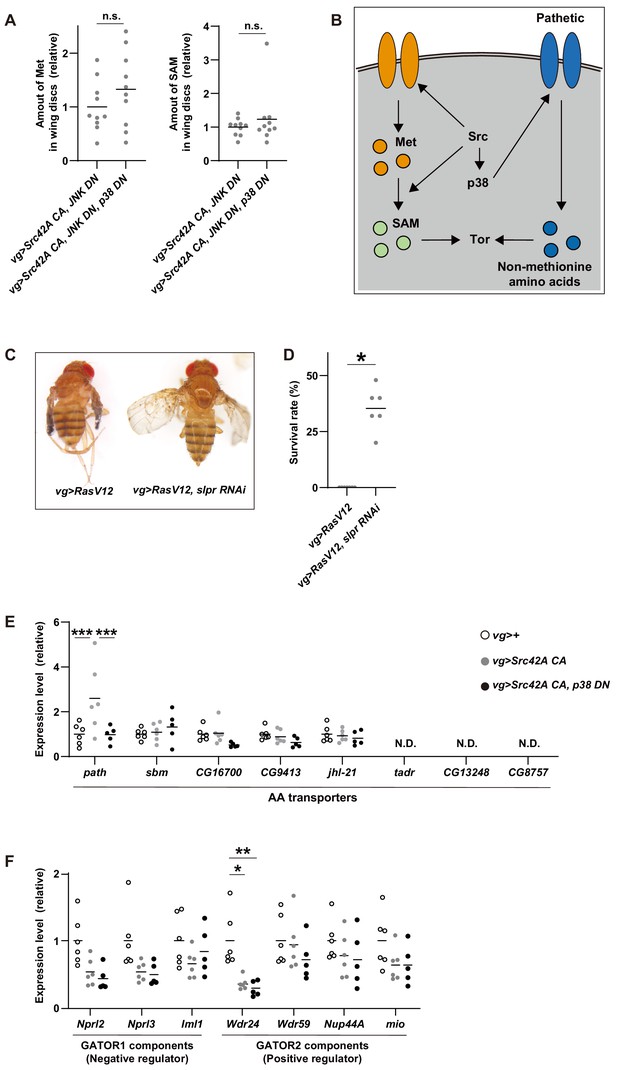
Cross-talk between Src signaling and Tor signaling.
(A) The data show the amounts of methionine and SAM in wing discs, which were used to calculate methionine flux in Figure 8F. p38 inhibition does not affect the amounts of methionine and SAM in tumorous discs induced by Src42A constitutively active (CA) and JNK DN. Two-tailed unpaired t-test. (B) A schematic of Src42A CA-mediated Tor regulation. Src activates Tor pathway via both p38-dependent and -independent pathways. Src promotes methionine incorporation and methionine metabolism flux independently of p38, which could activate Tor. On the other hand, Src increases expression of an amino acid transporter, path, in a p38-dependent manner, which could also activate Tor activity through uptake of non-methionine amino acids. (C) RasV12 expression in the wing imaginal disc induces small wings. Knockdown of slpr reverses the wing phenotype induced by RasV12.(D) RasV12 expression in the wing discs induces organismal lethality during development. Knockdown of slpr suppresses organismal lethality induced by RasV12 expression in the wing discs. Two-tailed unpaired t-test. (E) RT-qPCR of amino acid transporters using RNA from wing discs. One-way ANOVA with Sidak’s post-test. (F) RT-qPCR of GATOR1/2 components using RNA from wing discs. One-way ANOVA with Sidak’s post-test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene Drosophila melanogaster | ras | Flybase | FLYB: FBgn0003205 | NA |
| Gene Drosophila melanogaster | src42A | Flybase | FLYB: FBgn0264959 | NA |
| Gene Drosophila melanogaster | vg | Flybase | FLYB: FBgn0003975 | NA |
| Gene Drosophila melanogaster | slpr | Flybase | FLYB: FBgn0030018 | NA |
| Gene Drosophila melanogaster | rpr | Flybase | FLYB: FBgn0011706 | NA |
| Gene Drosophila melanogaster | grim | Flybase | FLYB: FBgn0015946 | NA |
| Gene Drosophila melanogaster | hid | Flybase | FLYB: FBgn0003997 | NA |
| Gene Drosophila melanogaster | bsk | Flybase | FLYB: FBgn0000229 | NA |
| Gene Drosophila melanogaster | lic | Flybase | FLYB: FBgn0261524 | NA |
| Gene Drosophila melanogaster | tor | Flybase | FLYB: FBgn0021796 | NA |
| Gene Drosophila melanogaster | rl | Flybase | FLYB: FBgn0003256 | NA |
| Gene Drosophila melanogaster | p38b | Flybase | FLYB: FBgn0024846 | NA |
| Gene Drosophila melanogaster | mef2 | Flybase | FLYB: FBgn0003256 | NA |
| Gene Drosophila melanogaster | atf2 | Flybase | FLYB: FBgn0265193 | NA |
| Gene Drosophila melanogaster | SamS | Flybase | FLYB: FBgn0005278 | NA |
| Gene Drosophila melanogaster | samtor | Flybase | FLYB: FBgn0035035 | NA |
| Gene Drosophila melanogaster | CG13248 | Flybase | FLYB: FBgn0036984 | NA |
| Gene Drosophila melanogaster | tadr | Flybase | FLYB: FBgn0036984 | NA |
| Gene Drosophila melanogaster | CG9413 | Flybase | FLYB: FBgn0030574 | NA |
| Gene Drosophila melanogaster | jhl-21 | Flybase | FLYB: FBgn0028425 | NA |
| Gene Drosophila melanogaster | sbm | Flybase | FLYB: FBgn0030574 | NA |
| Gene Drosophila melanogaster | CG8757 | Flybase | FLYB: FBgn0036380 | NA |
| Gene Drosophila melanogaster | CG16700 | Flybase | FLYB: FBgn0030816 | NA |
| Gene Drosophila melanogaster | path | Flybase | FLYB: FBgn0036007 | NA |
| Gene Drosophila melanogaster | nprl3 | Flybase | FLYB: FBgn0036397 | NA |
| Gene Drosophila melanogaster | nprl2 | Flybase | FLYB: FBgn0030800 | NA |
| Gene Drosophila melanogaster | iml1 | Flybase | FLYB: FBgn0035227 | NA |
| Gene Drosophila melanogaster | wdr24 | Flybase | FLYB: FBgn0027518 | NA |
| Gene Drosophila melanogaster | wdr59 | Flybase | FLYB: FBgn0032339 | NA |
| Gene Drosophila melanogaster | nup44A | Flybase | FLYB: FBgn0033247 | NA |
| Gene Drosophila melanogaster | mio | Flybase | FLYB: FBgn0031399 | NA |
| Gene Drosophila melanogaster | RpL32 | Flybase | FLYB: FBgn0002626 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-RasV12 | Iswar Hariharan lab | UAS-RasV12 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-Src42A CA | Bloomington Drosophila Stock Center | BDSC: 6410 RRID:BDSC_6410 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-Src42A | Tian Xu lab | UAS-Src42A | NA |
| Genetic reagent (Drosophila melanogaster) | vg-Gal4 | Bloomington Drosophila Stock Center | BDSC: 6819 RRID:BDSC_6819 | NA |
| Genetic reagent (Drosophila melanogaster) | gmr-Gal4 | Iswar Hariharan lab | gmr-Gal4 | NA |
| Genetic reagent (Drosophila melanogaster) | TRE-RFP | Bloomington Drosophila Stock Center | BDSC: 59011 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-slpr RNAi | Bloomington Drosophila Stock Center | BDSC: 32948 RRID:BDSC_32948 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-slpr RNAi | Vienna Drosophila Resource Center | VDRC ID: 33516 RRID:FlyBase_FBst0460140 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-GFP | Iswar Hariharan lab | UAS-GFP | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-his2B RFP | Iswar Hariharan lab | UAS-his2B RFP | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-miRNA RGH | Siegrist et al., 2010 | PMID:20346676 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-JNK DN | Iswar Hariharan lab | UAS-JNK DN | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-p38 DN | Bloomington Drosophila Stock Center | BDSC: 59005 RRID:BDSC_59005 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-lic RNAi | Bloomington Drosophila Stock Center | BDSC: 31643 RRID:BDSC_31643 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-Tor RNAi | Bloomington Drosophila Stock Center | BDSC: 34639 RRID:BDSC_34639 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-erk RNAi 1 | Vienna Drosophila Resource Center | VDRC ID: 35641 RRID:FlyBase_FBst0461260 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-erk RNAi 2 | Vienna Drosophila Resource Center | VDRC ID: 109573 RRID:FlyBase_FBst0481239 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-mef2 RNAi | Vienna Drosophila Resource Center | VDRC ID: 15550 RRID:FlyBase_FBst0451917 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-atf2 RNAi | Bloomington Drosophila Stock Center | BDSC: 60124 RRID:BDSC_60124 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-SamS RNAi 1 | Vienna Drosophila Resource Center | VDRC ID: 7167 RRID:FlyBase_FBst0470579 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-SamS RNAi 2 | Vienna Drosophila Resource Center | VDRC ID: 103143 RRID:FlyBase_FBst0475005 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-samtor RNAi | Bloomington Drosophila Stock Center | BDSC: 54010 RRID:BDSC_54010 | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-eiger | Iswar Hariharan lab | UAS-eiger | NA |
| Genetic reagent (Drosophila melanogaster) | UAS-rpr mts | Herman Steller lab | PMID:20837774 | NA |
| Genetic reagent (Drosophila melanogaster) | w¹¹¹⁸ | Erina Kuranaga lab | w¹¹¹⁸ | NA |
| Genetic reagent (Drosophila melanogaster) | Oregon R | Bloomington Drosophila Stock Center | BDSC: 4269 RRID:BDSC_4269 | NA |
| Antibody | Rabbit polyclonal phospho H3 antibody | Merck | Cat# 06–570 RRID:AB_310177 | Immunostaining (1:200) |
| Antibody | Rabbit polyclonal cleaved Drosophila Dcp-1 antibody | Cell Signaling | Cat# 9578 RRID:AB_2721060 | Immunostaining (1:100) |
| Antibody | Rabbit monoclonal phospho-p38 MAPK antibody | Cell Signaling | Cat# 4631 RRID:AB_331765 | Immunostaining (1:100) |
| Antibody | Mouse monoclonal phospho-Erk MAPK antibody | Merck | Cat# M8159 RRID:AB_477245 | Immunostaining (1:100) |
| Antibody | Rabbit monoclonal phospho-4EBP1 antibody | Cell Signaling | Cat# 2855 RRID:AB_560835 | Immunostaining (1:100) |
| Antibody | Rabbit polyclonal phospho-S6 antibody | Kim et al., 2017 | PMID:28829944 | Immunostaining (1:300) |
| Antibody | Alexa mouse Fluor 568 secondary antibody | Thermo Fisher | Cat# A-11004 RRID:AB_253407 | Immunostaining (1:300) |
| Antibody | Alexa rabbit Fluor 568 secondary antibody | Thermo Fisher | A-11036 RRID:AB_10563566 | Immunostaining (1:300) |
| Sequence-based reagent | slpr (primer) | This paper | NA | F: 5'-CTACAAGGGCTTCGATCCGTTG-3 R: 5'-GTTTGCCAGCAGCTCTTCATCAG-3 |
| Sequence-based reagent | slpr (primer) | This paper | NA | F: 5'-CAATCATCTGCAGCAGAAGACGC-3' R: 5'-CATCGGAGAATTTGGAATAGGTGC-3' |
| Sequence-based reagent | SamS (primer) | Obata and Miura, 2015 | PMID:32938923 | F: 5'-GCCAACGGCGTTCATATC-3' R: 5'-GGCATATCCAAACATGATACCC-3' |
| Sequence-based reagent | CG13248 (primer) | FlyPrimerBank | PP18106 | F: 5'-AAACCGATGCCTCAACACCTT-3' R: 5'-CAGTCAGCACGTAGATGCCA-3' |
| Sequence-based reagent | tadr (primer) | FlyPrimerBank | PP20579 | F: 5'-CAGCCCGCTGTAAAACTAGC-3' R: 5'-GGCCAGAGCATCTAGCCAG-3' |
| Sequence-based reagent | CG9413 (primer) | FlyPrimerBank | PP29104 | F: 5'-TGGGGTGGCTTTAATTGTTGG-3' R: 5'-CAGTGCGAACCAGTAAACCG-3' |
| Sequence-based reagent | jhl-21 (primer) | Newton et al., 2020 | PMID:32938923 | F: 5'-TCAAGCGGAAGCTAACACTCA-3' R: 5'-TTCGGTGTAAATAAAGACTCCCG-3' |
| Sequence-based reagent | sbm (primer) | FlyPrimerBank | PP3597 | F: 5'-AATGTGCCAACAAAAACAACGA-3' R: 5'-GTCCCTGATGAGTCGGTCTC-3' |
| Sequence-based reagent | CG8757 (primer) | Newton et al., 2020 | PMID:32938923 | F: 5'-AGAAACGATTGGATCGGGCA-3' R: 5'-ATCTGCCATCTTTTGGACCGA-3' |
| Sequence-based reagent | CG16700 (primer) | FlyPrimerBank | PP25676 | F: 5'-CCTACAAGCTATCTGGAGACCA-3' R: 5'-GAGACCTCCGTTCTTGAAGGC-3' |
| Sequence-based reagent | path (primer) | Newton et al., 2020 | PMID:32938923 | F: 5'-TGTTTGATTTGCGCGGCATT-3' R: 5'-TTCGACCCGCTGTCCACTAT-3' |
| Sequence-based reagent | nprl3 (primer) | FlyPrimerBank | PP28256 | F: 5'-GTTAAACCACAGCTATGCAACCA-3' R: 5'-CAGAGTGGGATGACTGACAAAG-3' |
| Sequence-based reagent | nprl2 (primer) | FlyPrimerBank | PP27923 | F: 5'-TTCAACGCTGCATTCTCACC-3' R: 5'-ATTCCGTGCGTACTTCTGCTG-3' |
| Sequence-based reagent | iml1 (primer) | FlyPrimerBank | PP8389 | F: 5'-CGTGGCTGCAACAAATCCTAC-3' R: 5'-GCCCGATTCTATGCTTATCACA-3' |
| Sequence-based reagent | wdr24 (primer) | FlyPrimerBank | PP3395 | F: 5'-GCCCTGGCCCTGAATAAGG-3' R: 5'-TGAAGCCATTGCTGTTTATGGAG-3' |
| Sequence-based reagent | wdr59 (primer) | FlyPrimerBank | PP2977 | F: 5'-GCACCCGAACAAACGTACATC-3' R: 5'-CCGAGTAATCAACCGACATGG-3' |
| Sequence-based reagent | nup44A (primer) | FlyPrimerBank | PP28620 | F: 5'-GAGGAGGTGATTGGCGAAAAG-3'' R: 5'-GCGAGTCTACAAGGGTGGTG-3' |
| Sequence-based reagent | mio (primer) | NA | PMID:26024590 | F: 5'-AGCGAGACGAGCTAAACAATTC-3' R: 5'-GTGTAAGAGGCAAGCAAAGGTT-3' |
| Sequence-based reagent | RpL32 (primer) | This paper | NA | F: 5'-CCAGCATACAGGCCCAAGATCGTG-3' R: 5'-TCTTGAATCCGGTGGGCAGCATG-3' |
| Commercial assay or kit | Methionine analog homopropargylglycine (HPG)- based on Click-iT HPG Alexa Fluor 488 Protein Synthesis Assay kit Single Cell 3’ Library and Gel Bead Kit v2 | Invitrogen | C10428 | NA |
| Commercial assay or kit | Maxwell RSC simply RNA Tissue Kit | Promega | AS1340 | NA |
| Commercial assay or kit | ReverTra Ace qPCR RT Kit | Toyobo | FSQ-101 | NA |
| Commercial assay or kit | BCA protein assay kit | Thermo Fisher | 23225 | NA |
| Software, algorithm | ImageJ | NA | https://imagej.nih.gov/ij/ RRID:SCR_003070 | NA |
| Software, algorithm | IMARIS 9.5.1 | Oxford Instrument | https://imaris.oxinst.com/packages RRID:SCR_007370 | NA |
| Other | DAPI | Sigma | D9542 | 1:1000 |
Additional files
-
Supplementary file 1
Stock information and results in the first screening.
The table shows stock IDs and results of survivor numbers and the wing phenotype.
- https://cdn.elifesciences.org/articles/59809/elife-59809-supp1-v1.xlsx
-
Supplementary file 2
Food composition.
The table shows components of the fly foods that were used for dietary restriction, amino acid subtraction, and methionine addition experiments.
- https://cdn.elifesciences.org/articles/59809/elife-59809-supp2-v1.xlsx
-
Supplementary file 3
All the statistics data.
The table shows all the plotted data and information of statistic analyses performed in this manuscript.
- https://cdn.elifesciences.org/articles/59809/elife-59809-supp3-v1.xlsx
-
Supplementary file 4
Primer information.
The table shows sequences of the primers used in this study.
- https://cdn.elifesciences.org/articles/59809/elife-59809-supp4-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59809/elife-59809-transrepform-v1.docx






