Evolved bacterial resistance against fluoropyrimidines can lower chemotherapy impact in the Caenorhabditis elegans host
Figures
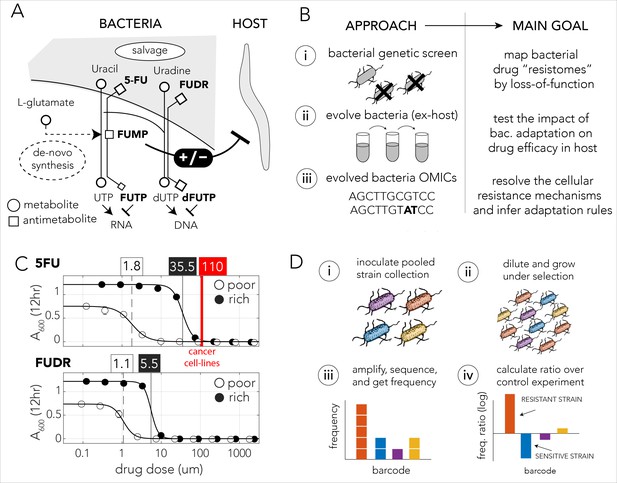
Bacteria effect on host drug toxicity and study design.
(A) Toxicity mechanisms of 5-FU and FUDR pro-drugs in bacteria and Caenorhabditis elegans. The two molecules, masqueraded as pyrimidines, are transported into bacteria through the nucleotide salvage pathway and are metabolized into active compounds that interfere with RNA and DNA synthesis. The intermediate derivative FUMP that is produced by the bacteria is highly toxic for the C. elegans host. Bacterial mutations effecting FUMP metabolism can increase or decrease drug toxicity in a C. elegans that is feeding on the bacteria and is exposed to the drug. (B) Overview of the study’s approach and aims of individual study stages. (C) Dose–response curves of 5-FU and FUDR in Escherichia coli and the calculated IC50 (half maximal inhibitory concentration). Both drugs are considerably more toxic for bacteria in nutrient-poor media. The mean IC50 of 5-FU from 806 human cancer (red line) is higher than the IC50 measured for E. coli. (D) Overall approach for pooled screening with the E. coli barcoded strain collection. The frequency of individual barcodes can be measured by deep sequencing of the barcode locus and can be used to infer changes in barcode representation in different conditions. The relative frequency of a barcode in screen and control experiments was used to infer if the gene knockout corresponding to the barcode increases drug resistance.
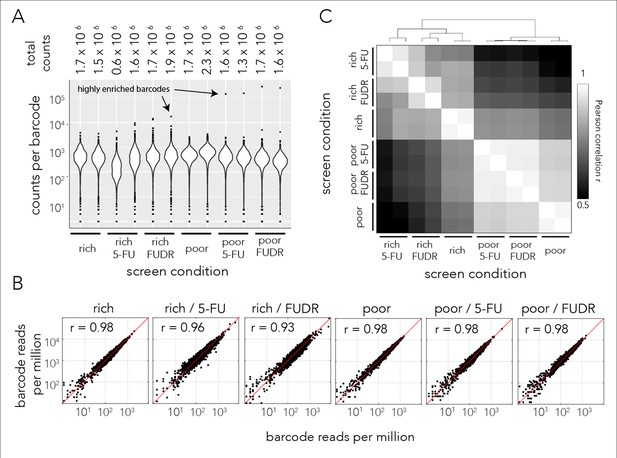
Sequencing statistics of pooled genetic screens.
(A) Barcode coverage in individual screen after de-multiplexing reads that were sequenced on the same flow-cell. We identified roughly 1.5 million barcodes in each screen condition (top). Each individual barcode was identified on average 500 times (bottom). (B) A comparison of individual barcode frequency of biological duplicates revealed they are highly correlated. The Pearson r correlation is shown for each replicate pair. (C) The Pearson correlation coefficients between barcode counts from all screens' conditions. The correlation matrix reveals a hierarchical correlation structure: biological replicates are highly correlated, followed by high correlation by media type.
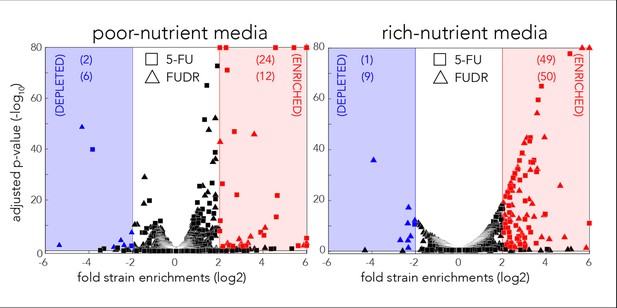
Volcano plots for the identification of screen hits.
Hits were determined by fold-change and false-discovery-rate adjusted p-value that was calculated with DEBRA and DESeq2 tools.
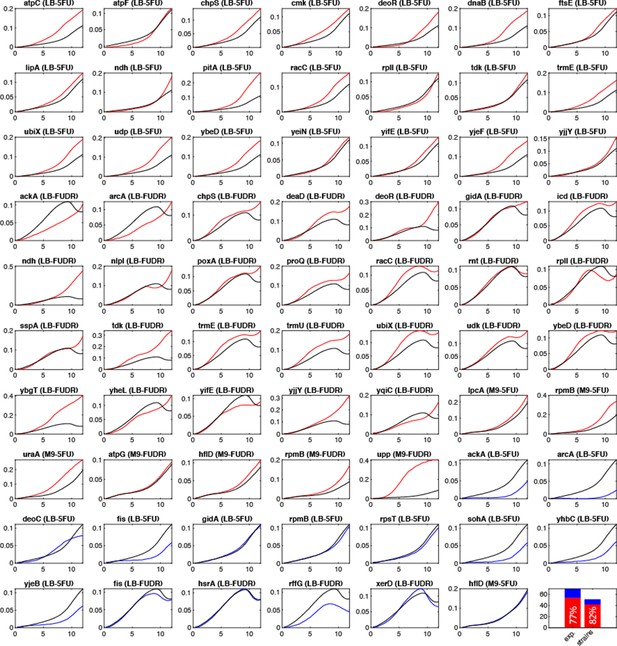
Drug resistance of individual knockout strains.
We monitored the growth of the wild-type strain and 50 individual knockout strains in 69 different growth conditions (each strain was tested in the pooled genetic screen condition it was found in). The graphs show the mean growth curve for replicate cultures (duplicates for knockout strains and 10 replicates for the wild-type strain). To allow comparison with the wild-type strains, we only tested hit strains that are not slow growing strains in the tested media (without drug) and are not annotated as auxotrophs in the EcoCyc database. A knockout strain was considered a hit if its absorbance in the last time point was higher than that of the wild-type strain (red curves). A strain was counted as a non-validated strain if its absorbance in the last time point was equal or lower than the wild type. The lower right bar graph shows the percentage of validated hits (by the 69 growth conditions or by the 50 tested strains).
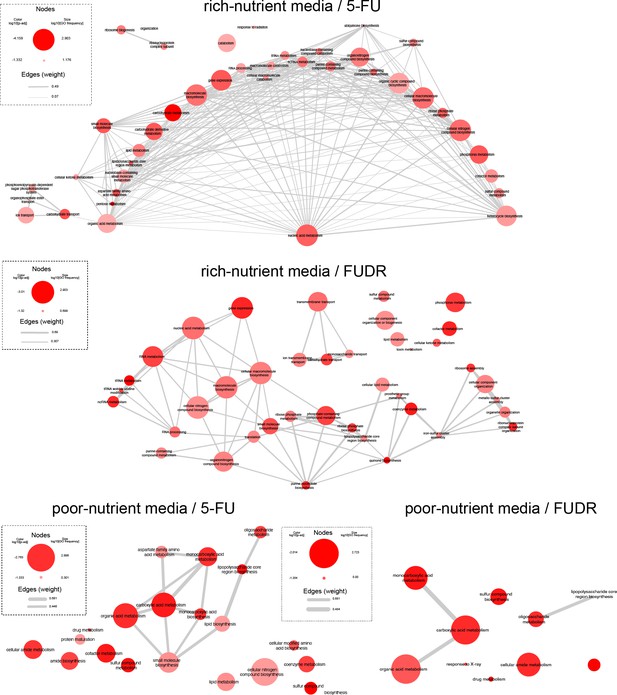
Network of related GO categories that are enriched by the genetic screen.
Node size indicates the frequency of the particular GO within the Escherichia coli database (on a log scale). Colors represent the p-adj for that category according to GAGE analysis (on a log scale). Edge width correlates with the similarity of the GO pairs. Plot was generated using REVIGO and plotted with Cytoscape (https://cytoscape.org/).
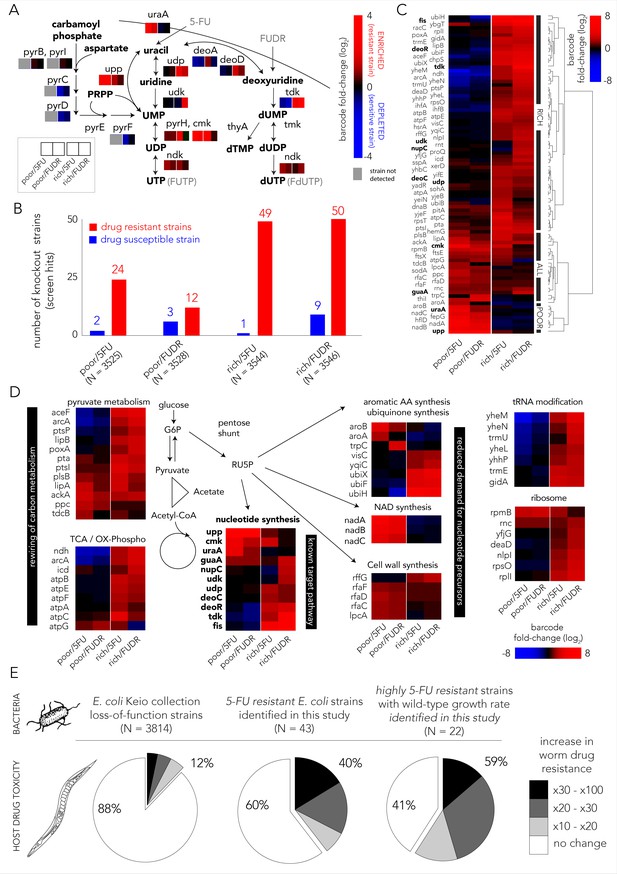
Multiple gene knockouts increase drug resistance in bacteria and are expected to lower drug toxicity in the Caenorhabditis elegans host.
(A) Screen results projected on the network of nucleotide synthesis. Compatible with the known toxicity mechanism, we observed that mutations in the salvage and pro-drug activation pathways increase drug resistance (red) while loss-of-function mutations in the de-novo synthesis pathway increase drug sensitivity (blue). (B) The number of strains found to be differentially represented in our screens. Hits were determined by fold-change and false-discovery-rate adjusted p-value that was calculated with DEBRA and DESeq2 tools. The number of hits in nutrient-rich media is considerably higher than the number of hits in nutrient-poor media. (C) Unsupervised hierarchical clustering of hits identified in all screen conditions. The clustering uncovers a media-dependent Resistome pattern. The majority of hits are nutrient-rich media specific. Only a minority of hits are nutrient-poor media specific. The genes from nucleotide synthesis network are marked bold. (D) The functional characterization of identified hits reveals a plausible metabolic connection through RU5P, a metabolite from the pentose phosphate pathway that is a precursor for nucleotide synthesis. Hits were assigned to broad functional categories according to their annotated function in the KEGG and EcoCyc databases. (E) Gene-knockouts that increase bacterial resistance will likely reduce drug toxicity in the C. elegans host. The three pie charts show the proportion of bacterial strains that reduce drug toxicity in C. elegans for different subsets of bacterial strains: all tested strains from the Keio library collection (left) previously tested in the C. elegans screen (Scott et al., 2017), all 5-FU resistant bacterial strains we identified in our screens and that are present in the C. elegans screen (middle), and a subset of highly 5-FU resistant strains we identified (fold enrichment > 6) that also have an un-impaired growth rate in either nutrient-rich or nutrient-poor media (right).
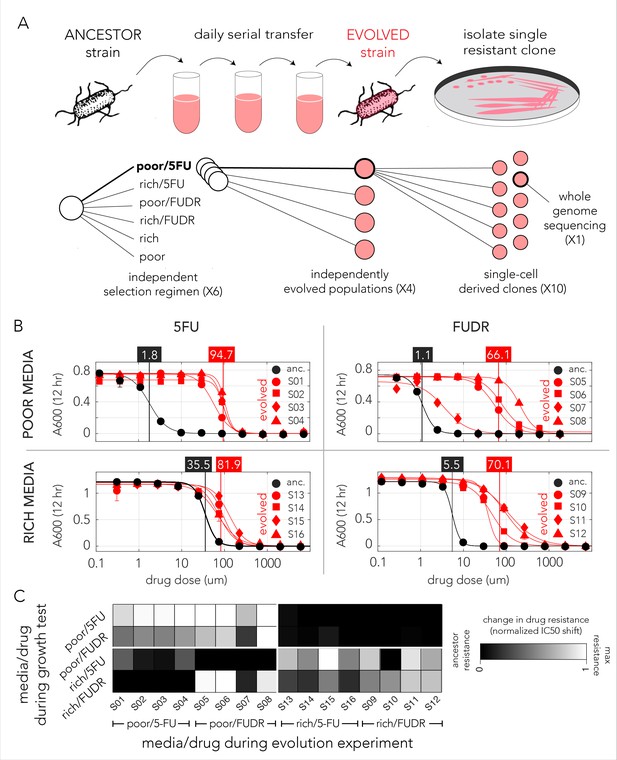
Evolved bacteria are drug resistant and display a media-type dependent cross-resistance pattern.
(A) Overall approach for the lab evolution experiment. We evolved 16 resistant bacterial populations with serial transfer evolution protocol. We evolved four individual replicates on each of the drug and media combinations. We isolated single colonies from each independently evolved population and tested for drug resistance. The lower panel shows the number of replicates used for each step. (B) Drug resistance increased in all evolved strains. The curves show the inferred drug sensitivity of clones from four independently evolving populations (red) in comparison to the ancestral strain (black). Increased resistance is reflected by the shift in the IC50 to a higher concentration in evolved clones. (C) Drug cross-resistance in evolved strains reveals a media-dependent pattern. The heatmap shows the relative change in IC50, normalized to the range of IC50 in other strains (from the ancestor strain IC50 to the maximum measured IC50 in strains that evolved on the tested condition).
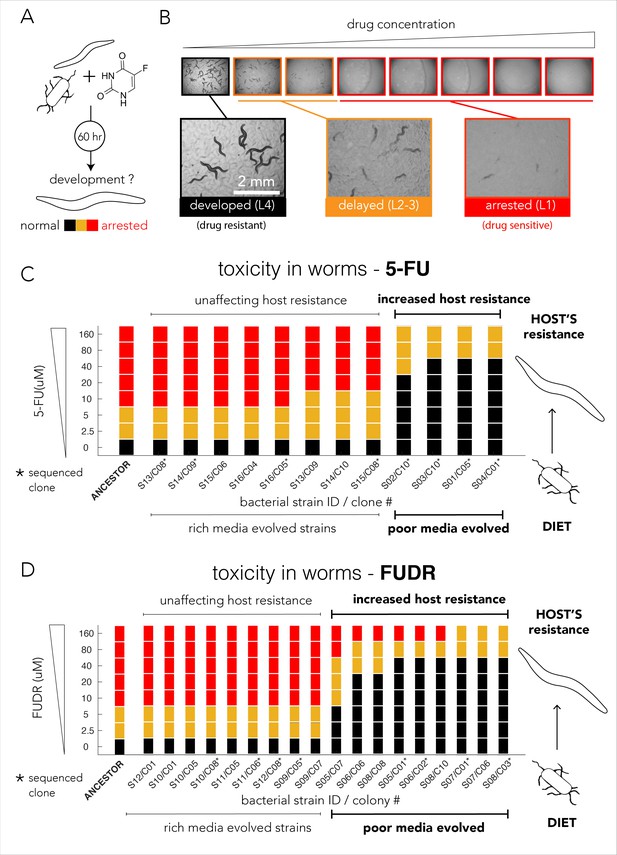
Evolved bacterial resistance lowers drug efficacy in the Caenorhabditis elegans host.
(A) Experimental approach for testing the impact of bacteria on drug toxicity in C. elegans. The tested bacterial strain was incubated with L1 animals and drug was added in different concentrations. Plates were imaged after 48–72 hr and development stage of the animals was scored into three categories: normally developed (L4 stage), developmentally delayed (L2-3 stages), or completely arrested (L1 stage). Phenotype scoring was performed blindly. (B) Representative images showing the development of C. elegans growing on different FUDR concentrations when the ancestor strain was used as the bacterial diet. (C) Bacteria that evolved on nutrient-poor media and 5-FU reduced 5-FU toxicity C. elegans. The color bars indicate the C. elegans developmental phenotypes at different 5-FU concentrations and bacterial strains are ordered by their impact on C. elegans development. A bacterial strain was characterized as reducing drug efficacy if C. elegans development was not completely arrested at the same 5-FU concentration as the animals fed with ancestor strain (20 µM). The set of strains that impact the drug efficacy in the host perfectly coincides with their evolutionary history (only nutrient-poor media evolved strains impact host drug efficacy). (D) Bacteria that evolved on nutrient-poor media and FUDR reduced FUDR toxicity C. elegans. Similarly, to 5-FU (shown in C), only bacterial strains that evolved in poor-nutrient media impact the host.
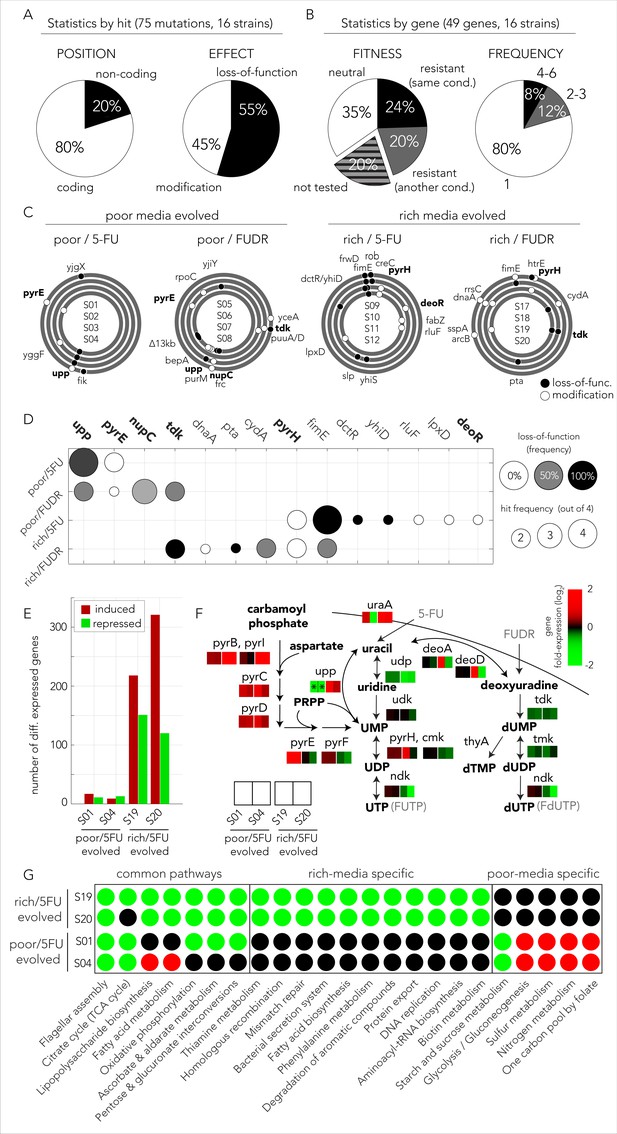
The evolved mechanisms underlying bacterial drug resistance are media-type dependent.
(A) Characterization of mutation positions and effects on the nearest open reading frame. Mutations that clearly disrupt the reading frame are annotated as loss-of-function mutations (frameshift mutation, large deletion, or transposon insertion). Other mutations are annotated as modifying mutations (point mutations, in-frame indels, and promoter mutations). (B) Characterization of mutations by their effect on genes. Mutations are annotated as leading to resistance if a gene-knockout strain is drug resistant. (C) The position of all mutations in the genomes of the sequenced evolved clones. The concentric circa plots show the genomes of four clones taken from four individually evolved populations. Circles represent the annotated strains from inner to outer order. Black circles mark loss-of-function mutations and white circles mark gene modifying mutations (similar to A). The genes from nucleotide synthesis network are marked bold. (D) Summary table of putative driver mutations across all sequenced strains. A mutation was characterized as a driver mutation if a loss-of-function of the corresponding gene was observed to significantly increase or decrease drug resistance. (E) Number of differentially expressed genes in four 5-FU resistant strains growing without any drug. The nutrient-rich media evolved strains showed a significantly higher number of differentially expressed genes relative to the strains that evolved on nutrient-poor media. (F) Changes in gene expression in evolved strains projected on the network of nucleotide synthesis. Asterisks signs mark loss-of-function mutations detected during whole genome sequencing. (G) Differentially expressed KEGG pathways in 5-FU-evolved strains. The circles mark individual pathways that were identified as significantly induced (red), or repressed (green), by the GAGE tool.
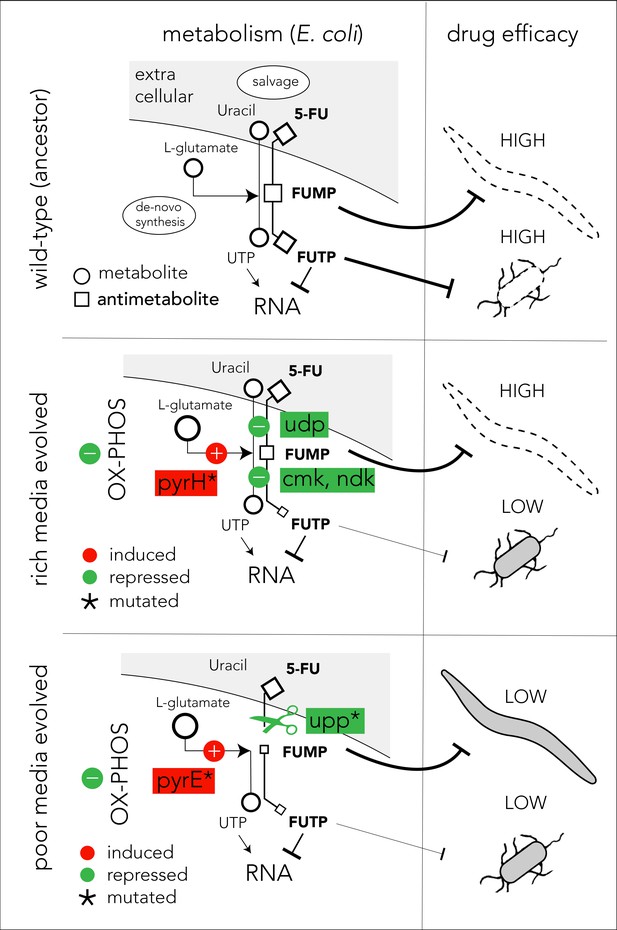
Proposed model of evolutionary adaptation to 5-FU.
The nutrient availability during the period of 5-FU selection leads to alternative adaptation mechanisms in bacteria and culminate in different outcomes in the Caenorhabditis elegans host. The top panel shows the drug toxicity mechanism in C. elegans feeding on the ancestor bacterial strain. Although evolutionary adaptation always selected for resistant bacteria, different mechanisms of adaptation lead to different impacts on the host (middle and lower). The genes that were repeatedly mutated in evolved strains are marked with an asterisk sign.
Additional files
-
Supplementary file 1
Table of barcode frequencies and enrichment across genetic screens.
- https://cdn.elifesciences.org/articles/59831/elife-59831-supp1-v2.xlsx
-
Supplementary file 2
Table of gene-set enrichment across genetic screens.
- https://cdn.elifesciences.org/articles/59831/elife-59831-supp2-v2.xlsx
-
Supplementary file 3
Table of growth of 43 strains with single gene knockout.
- https://cdn.elifesciences.org/articles/59831/elife-59831-supp3-v2.xlsx
-
Supplementary file 4
Table of drug resistance and growth of evolved strains.
- https://cdn.elifesciences.org/articles/59831/elife-59831-supp4-v2.xlsx
-
Supplementary file 5
Table of mutations in evolved strains.
- https://cdn.elifesciences.org/articles/59831/elife-59831-supp5-v2.xlsx
-
Supplementary file 6
Table of differential gene expression in 5-FU evolved strains.
- https://cdn.elifesciences.org/articles/59831/elife-59831-supp6-v2.xlsx
-
Supplementary file 7
Table of gene-set enrichment in 5-FU evolved strains.
- https://cdn.elifesciences.org/articles/59831/elife-59831-supp7-v2.xlsx
-
Supplementary file 8
Table of Caenorhabditis elegans developmental phenotypes.
- https://cdn.elifesciences.org/articles/59831/elife-59831-supp8-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59831/elife-59831-transrepform-v2.docx





