A positive feedback loop between Flower and PI(4,5)P2 at periactive zones controls bulk endocytosis in Drosophila
Figures
PI(4,5)P2 forms microdomains at periactive zones under conditions of intense stimulation.
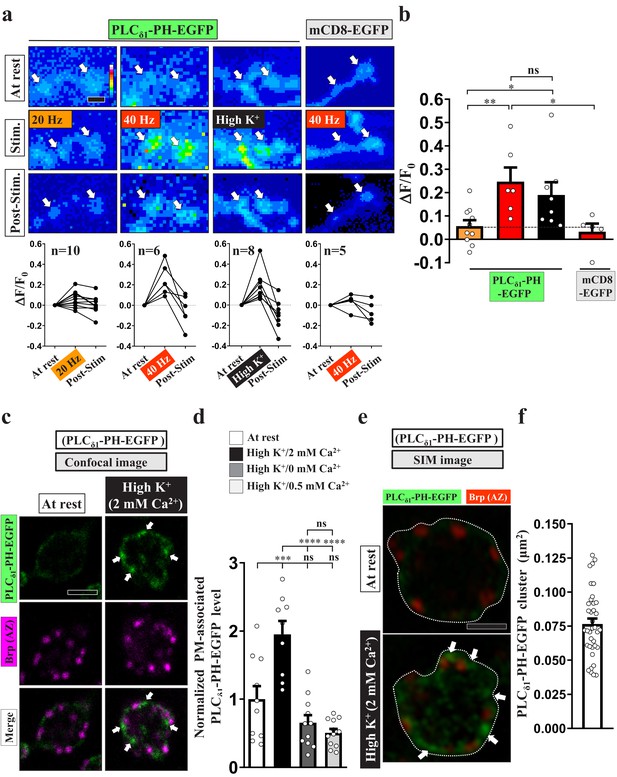
(a–b) Increased fluorescence of PLCδ1-PH-EGFP but not mCD8-GFP in NMJ boutons upon intense stimulation. (a) (Top) Live images of the boutons (arrows) expressing UAS-PLCδ1-PH-EGFP or UAS-mCD8-GFP. The larvae were reared at 25°C. Electrical (20 or 40 Hz) or chemical (90 mM K+) stimulation was conducted in a 2 mM-Ca2+ solution for 3 min (electrical) or 5 min (chemical) and then rested in 0 mM Ca2+ and 5 mM K+. Snapshot images taken before stimulation, at the third (electrical) and fifth (chemical) min of stimulation, and after stimulation. (Bottom) Traces of probe fluorescence for single boutons. The number of boutons imaged (N). (b) Quantification data for EGFP fluorescence change. The resting fluorescence level (F0). Fluorescence change evoked by stimulation (ΔF). (c–f) PLCδ1-PH-EGFP enrichment at periactive zones is dependent on Ca2+ upon intense stimulation. Single-plane confocal (c) or SIM (e) images of the boutons expressing PLCδ1-PH-EGFP. The larvae were reared at 25°C. The boutons subjected to high K+/2 mM Ca2+ (10 min stimulation of 90 mM K+/2 mM Ca2+), high K+/0 mM Ca2+ (10 min stimulation of 90 mM K+/0 mM Ca2+), or high K+/0.5 mM Ca2+ (1 min stimulation of 90 mM K+/0.5 mM Ca2+) treatments were fixed immediately and immunostained for PLCδ1-PH-EGFP (green) and Bruchpilot (Brp) [an active zone scaffold protein; magenta (c), red (e)]. The PLCδ1-PH-EGFP-enriched puncta (Arrows). (d) Quantification data for PLCδ1-PH-EGFP staining intensities, normalized to the value of the resting condition. (f) Average area of PLCδ1-PH-EGFP clusters in individual boutons, as measured by SIM. Individual data values are shown in graphs. p values: ns, not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. Mean ± SEM. Scale bar: 1 µm (e), 2 µm (a, c). Statistics: one-way ANOVA with Tukey’s post hoc test (b, d).
-
Figure 1—source data 1
Source data for Figure 1.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig1-data1-v2.xlsx

The change in PLCδ1-PH-EGFP fluorescence is responsible for an increase in the PI(4,5)P2 level.
(a–b) The protein expression level of PLCδ1-PH-EGFP was not changed upon high K+ stimulation. (a) Larval fillets expressing UAS-PLCδ1-PH-EGFP using nSyb-GAL4 were dissected, and the brain and ventral nerve cord were removed. The samples were subjected to the resting condition (10 min incubation of 5 mM K+/0 mM Ca2+) or high K+ stimulation (10 min stimulation of 90 mM K+/2 mM Ca2+), and then immediately lysed in 1x SDS sample buffer. The lysate of a single larva was loaded in each lane of the western blot, and α-GFP and α-actin were used to detect presynaptic levels of PLCδ1-PH-EGFP or to serve as loading control, respectively. (b) Quantification data for the ratio of α-GFP to α-actin blotting signal, normalized to the value of the resting condition. (c–f) Reducing Synj levels increased the levels of PI(4,5)P2 and thus association of PLCδ1-PH-EGFP on the plasma membrane, but it did not increase the protein expression level of PLCδ1-PH-EGFP in synaptic boutons. (c–d) Single-plane confocal images of NMJ boutons neuronally expressing UAS-PLCδ1-PH-EGFP using nSyb-GAL4 in the wild-type control or synj1/+ lines. The boutons were subjected to the resting condition (10 min incubation of 5 mM K+/0 mM Ca2+), followed by chemical fixation and α-GFP immunostaining. Line plot profiles of absolute fluorescence units (a.u.) of native fluorescence or α-GFP immunostaining signal of PLCδ1-PH-EGFP are shown at right. α-Hrp immunostaining was used to outline the presynaptic plasma membrane. (d) Quantification data for the plasma membrane-associated PLCδ1-PH-EGFP fluorescence or immunostaining intensities, normalized to the value of wild-type boutons. (e–f) Brain-removed larval fillets expressing PLCδ1-PH-EGFP were subjected to western blot using α-GFP and α-actin antibodies. Quantification data for the ratio of α-GFP to α-actin blotting signal, normalized to the value of the wild-type control fillets (f). (g–h) High K+ stimulation elevated plasma membrane localization but not protein expression level of PLCδ1-PH-EGFP. (g) Single-plane confocal images of NMJ boutons neuronally expressing UAS-PLCδ1-PH-EGFP using nSyb-GAL4. The boutons were subjected to the resting condition (10 min incubation of 5 mM K+/0 mM Ca2+) or high K+ stimulation (10 min stimulation of 90 mM K+/2 mM Ca2+), followed by chemical fixation and α-GFP immunostaining. Line plot profiles of absolute fluorescence units (a.u.) of native fluorescence or α-GFP immunostaining signal of PLCδ1-PH-EGFP are shown at right. α-Hrp immunostaining was used to outline the presynaptic plasma membrane. (h) Quantification data for the plasma membrane-associated PLCδ1-PH-EGFP fluorescence or immunostaining intensities, normalized to the value of the resting condition. Individual data values are shown in graphs. p values: ns, not significant; *p<0.05; **p<0.01. Mean ± SEM. Scale bar: 2 µm (c, g). Statistics: Student t-test.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig1-figsupp1-data1-v2.xlsx
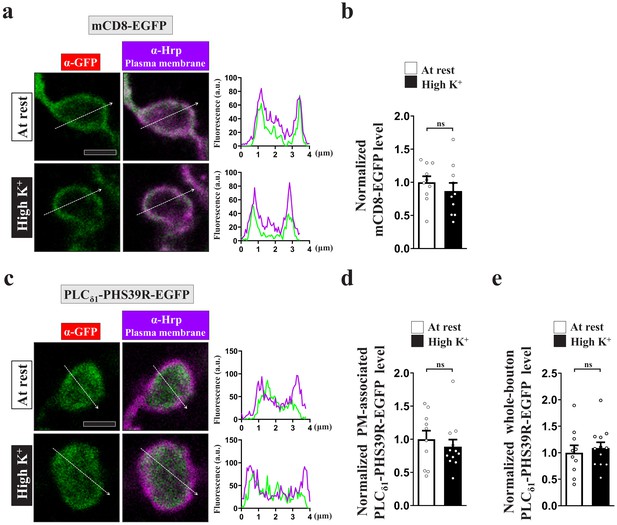
The distribution of mCD8-EGFP and PLCδ1-PHS39R-EGFP remains unchanged upon intense stimulation.
Single-plane confocal images of NMJ boutons neuronally expressing UAS-mCD8-EGFP (a) or UAS-PLCδ1-PHS39R-EGFP (c) using nSyb-GAL4. The boutons were subjected to the resting condition (10 min incubation of 5 mM K+/0 mM Ca2+) or high K+ stimulation (10 min stimulation of 90 mM K+/2 mM Ca2+), followed by chemical fixation and α-GFP immunostaining. Line plot profiles of absolute fluorescence units (a.u.) of α-GFP immunostaining signal of both probes are shown at right. α-Hrp immunostaining (magneta) was used to outline the presynaptic plasma membrane. mCD8-EGFP (green) was exclusively localized in the plasma membrane, whereas PLCδ1-PHS39R-EGFP (green) was mainly expressed in the cytosol. Quantification data for immunostaining intensities of the plasma membrane-associated mCD8-EGFP (b), the plasma membrane-associated PLCδ1-PHS39R-EGFP (d), or the whole-bouton PLCδ1-PHS39R-EGFP (e), normalized to the value of the resting condition. Individual data values are shown in graphs. p values: ns, not significant. Mean ± SEM. Scale bar: 2 µm (a, c). Statistics: Student t-test.
-
Figure 1—figure supplement 2—source data 1
Source data for Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig1-figsupp2-data1-v2.xlsx
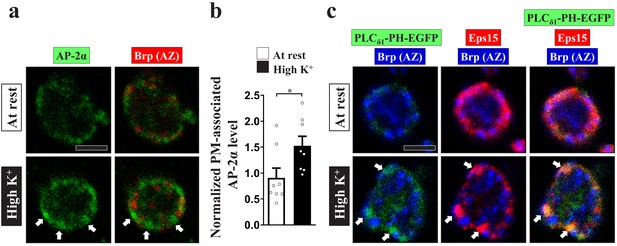
PI(4,5)P2 is increased at the periactive zone upon intense stimulation.
(a–b) AP-2 complex was actively recruited to the plasma membrane after the boutons were intensely stimulated. (a) Single-plane confocal images of NMJ boutons stained with α-AP-2α. The boutons were subjected to the resting condition (10 min incubation of 5 mM K+/0 mM Ca2+) or high K+ stimulation (10 min stimulation of 90 mM K+/2 mM Ca2+), followed by chemical fixation and α-AP-2α immunostaining (green). Active zones were labeled with Brp staining (red). An increase in AP-2α immunostaining signal was observed at the periactive zones (arrows). (b) Quantification data for the plasma membrane-associated α-AP-2α immunostaining intensities, normalized to the value of the resting condition. Individual data values are shown in graphs. p values: *p<0.05. Mean ± SEM. Scale bar: 2 µm. Statistics: Student t-test. (c) Upon high K+ stimulation, PLCδ1-PH-EGFP accumulated in PI(4,5)P2-enriched microdomains at the periactive zones. Single-plane confocal images of NMJ boutons neuronally expressing UAS-PLCδ1-PH-EGFP using nSyb-GAL4. The boutons were subjected to the resting condition (10 min incubation of 5 mM K+/0 mM Ca2+) or high K+ stimulation (10 min stimulation of 90 mM K+/2 mM Ca2+), followed by chemical fixation and immunostaining. Arrows indicate that Eps15 (red) was concentrated at the periactive zone and colocalized with PLCδ1-PH-EGFP (green) stained with α-GFP after intense stimulation. Brp (blue) labels active zones. Scale bar: 2 µm.
-
Figure 1—figure supplement 3—source data 1
Source data for Figure 1—figure supplement 3.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig1-figsupp3-data1-v2.xlsx
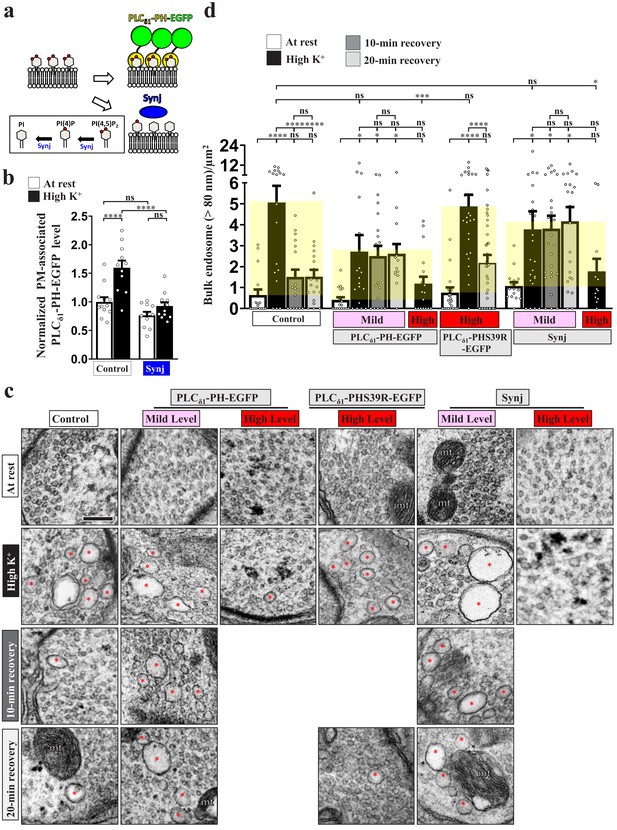
PI(4,5)P2 microdomains drive ADBE and SV reformation from bulk endosomes.
Reducing PI(4,5)P2 availability suppresses ADBE and subsequent SV reformation. (a) A schematic for PI(4,5)P2 suppression by PLCδ1-PH-EGFP or Synj expression. (b) Expression of Synj reduced presynaptic plasma membrane PI(4,5)P2 upon high K+ treatment. The boutons co-expressing UAS-PLCδ1-PH-EGFP with UAS-RFP (control) or UAS-synj using nSyb-GAL4 were reared at 25°C and subjected to resting condition (10 min incubation of 5 mM K+/0 mM Ca2+) or high K+ stimulation (10 min stimulation of 90 mM K+/2 mM Ca2+), followed by α-GFP immunostaining. Single-plane confocal images of the boutons are shown in Figure 2—figure supplement 1c. Quantification data for PLCδ1-PH-EGFP staining intensity are shown, normalized to the value of the resting condition of controls. (c) TEM images of the boutons of controls (nSyb-GAL4/+ at 29°C), mild PLCδ1-PH-EGFP expression (nSyb-GAL4/UAS-PLCδ1-PH-EGFP at 25°C), high PLCδ1-PH-EGFP expression (nSyb-GAL4/UAS-PLCδ1-PH-EGFP at 29°C), high PLCδ1-PHS39R-EGFP expression (nSyb-GAL4/UAS-PLCδ1-PHS39R-EGFP at 29°C), mild Synj expression (nSyb-GAL4/UAS-synj at 25°C), or high Synj expression (nSyb-GAL4/UAS-synj at 29°C). At rest (10 min incubation of 5 mM K+/0 mM Ca2+). High K+ (10 min stimulation of 90 mM K+/2 mM Ca2+). 10 min recovery (10 min stimulation of 90 mM K+/2 mM Ca2+, followed by 10 min incubation of 5 mM K+/0 mM Ca2+). 20 min recovery (10 min stimulation of 90 mM K+/2 mM Ca2+, followed by 20 min incubation of 5 mM K+/0 mM Ca2+). Bulk endosomes (>80 nm in diameter, red asterisks). Mitochondria (mt). Quantification data for total number of bulk endosomes per bouton area (d). Individual data values are shown in graphs. p values: ns, not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. Mean ± SEM. Scale bar: 500 nm. Statistics: one-way ANOVA with Tukey’s post hoc test.
-
Figure 2—source data 1
Source data for Figure 2.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig2-data1-v2.xlsx
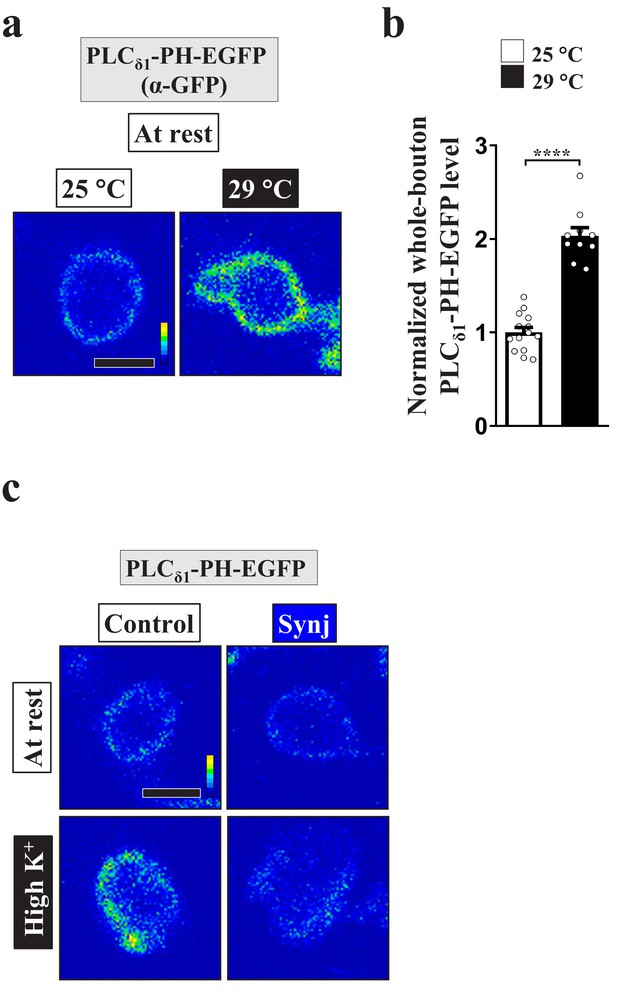
Blockade of the PI(4,5)P2 microdomains by expressing PLCδ1-PH-EGFP and Synj.
(a) Increasing rearing temperature from 25°C to 29°C elevated PLCδ1-PH-EGFP expression. Single-plane confocal images of NMJ boutons neuronally expressing UAS-PLCδ1-PH-EGFP using nSyb-GAL4. The boutons were situated at rest and immunostained with α-GFP. (b) Quantification data for the whole-bouton PLCδ1-PH-EGFP immunostaining intensities, normalized to the value for the 25°C condition. Individual data values are shown in graphs. p values: ****p<0.0001. Mean ± SEM. Scale bar: 2 µm. Statistics: Student t-test. (c) Synj expression suppressed PI(4,5)P2 microdomain formation. Single-plane confocal images of NMJ boutons co-expressing UAS-PLCδ1-PH-EGFP with UAS-RFP control or UAS-Synj at 25°C. The boutons were subjected to the resting condition (10 min incubation of 5 mM K+/0 mM Ca2+) or high K+ stimulation (10 min stimulation of 90 mM K+/2 mM Ca2+), followed by chemical fixation and α-GFP immunostaining. Scale bar: 2 µm.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig2-figsupp1-data1-v2.xlsx

Blockade of the PI(4,5)P2 microdomains abolishes SV reformation from the bulk endosome.
Quantification data for the ratio of total area of bulk endosomes to bouton area. Indicated genotypes were assessed. Representative TEM images are shown in Figure 2c. Individual data values are shown in graphs. p values: ns, not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. Mean ± SEM. Statistics: one-way ANOVA with Tukey’s post hoc test.
-
Figure 2—figure supplement 2—source data 1
Source data for Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig2-figsupp2-data1-v2.xlsx
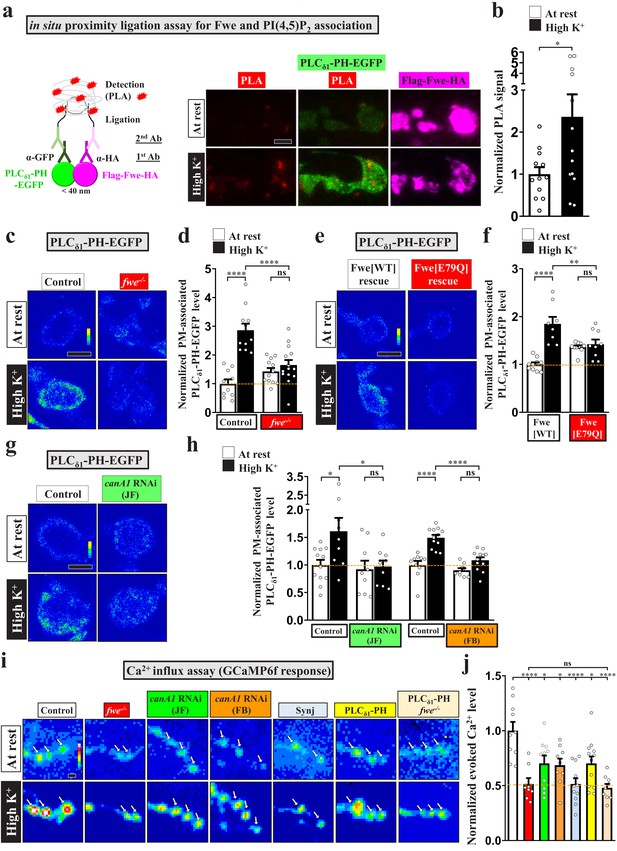
Fwe and PI(4,5)P2 form a positive feedback loop to establish PI(4,5)P2 microdomains.
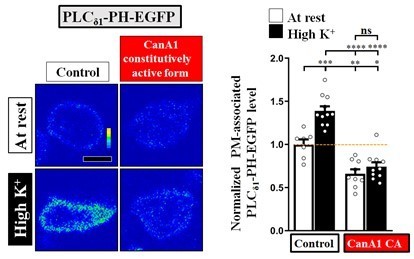
(a–b) Fwe and PLCδ1-PH-EGFP interact upon intense stimulation. (a) (Left) A schematic for PLA. (Right) Z-projected confocal images of fwe mutant boutons co-expressing Flag-Fwe-HA and PLCδ1-PH-EGFP (nSyb-GAL4/UAS-Flag-fwe-HA/UAS-PLCδ1-PH-EGFP in fweDB25/DB56 at 25°C). After resting condition or high K+ stimulation, the boutons were subjected to PLA. α-GFP and α-HA stained for PLCδ1-PH-EGFP (green) and Flag-Fwe-HA (magenta), respectively. PLA signals (red). (b) Quantification data for PLA signal intensities, normalized to the value of the resting condition. (c–f) Loss of Fwe or its Ca2+ channel activity perturbs PI(4,5)P2 microdomain formation. (c and e). (c) Single-plane confocal images of the boutons neuronally expressing UAS-PLCδ1-PH-EGFP in fweDB25/+ (control) or fweDB25/DB56 at 25°C. The boutons were subjected to resting condition or high K+ stimulation, followed by α-GFP immunostaining. Quantification data for PLCδ1-PH-EGFP staining intensity are shown, normalized to the value of the resting condition of controls (d). (e) Single-plane confocal images of the boutons neuronally expressing lexAop2-PLCδ1-PH-EGFP using vglut-lexA in wild-type Fwe rescue boutons (vglut-lexA/lexAop2-PLCδ1-PH-EGFP, nSyb(w)-GAL4/UAS-flag-fwe-RB-HA in fweDB25/fweDB56 at 25°C), or FweE79Q rescue boutons (vglut-lexA/lexAop2-PLCδ1-PH-EGFP, nSyb(w)-GAL4/UAS-flag-fweE79Q-RB-HA in fweDB25/fweDB5 at 25°C). When the weak nSyb(w)-GAL4 driver drove low expression of UAS-fwe transgenes, this binary system (vglut-lexA/lexAop2-PLCδ1-PH-EGFP) was used to produce detectable levels of PLCδ1-PH-EGFP in boutons. Quantification data for PLCδ1-PH-EGFP staining intensity are shown, normalized to the value of the resting condition of Fwe-rescued boutons (f). (g–h) canA1 RNAi knockdown impairs PI(4,5)P2 microdomain formation. Single-plane confocal images of the boutons co-expressing UAS-PLCδ1-PH-EGFP with UAS-RFP (control), UAS-canA1-RNAi (TRiP.JF01871), or UAS-canA1-RNAi (FB4) using nSyb-GAL4. The larvae were reared at 25°C. After resting condition or high K+ stimulation, the boutons were stained with α-GFP. (h) Quantification data for PLCδ1-PH-EGFP staining intensities, normalized to the value of the resting condition of controls. (i–j) Blockade of the PI(4,5)P2 microdomains attenuates Fwe Ca2+ conductance. (i) Snapshot Ca2+ images of the boutons (arrows) expressing lexAop2-GCaMP6f using vglut-lexA. The larvae of control (w1118), fwe mutant (fweDB25/DB56), canA1 RNAi (JF) (nSyb-GAL4/UAS-canA1-RNAi (TRiP.JF01871)), canA1 RNAi (FB) (nSyb-GAL4/UAS-canA1-RNAi (FB4)), Synj overexpression (nSyb-GAL4/UAS-synj), PLCδ1-PH-APEX2-HA expression (vglut-lexA/LexAop2-PLCδ1-PH-APEX2-HA), or fwe mutant expressing PLCδ1-PH-APEX2-HA (vglut-lexA/LexAop2-PLCδ1-PH-APEX2-HA in fweDB25/DB56) were reared at 25°C. Imaging was taken in the fifth minute for one minute after high K+ (2 mM Ca2+) stimulation. (j) Quantification data for evoked Ca2+ level, normalized to the value of controls. Evoked Ca2+ levels are shown as the increase in GCaMP6f fluorescence under high K+ stimulation. Individual data values are shown in graphs. p values: ns, not significant; *p<0.05; **p<0.01; ****p<0.0001. Mean ± SEM. Scale bar: 2 µm. Statistics: Student t-test (b). One-way ANOVA with Tukey’s post hoc test (d, f, h, j).
-
Figure 3—source data 1
Source data for Figure 3.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig3-data1-v2.xlsx
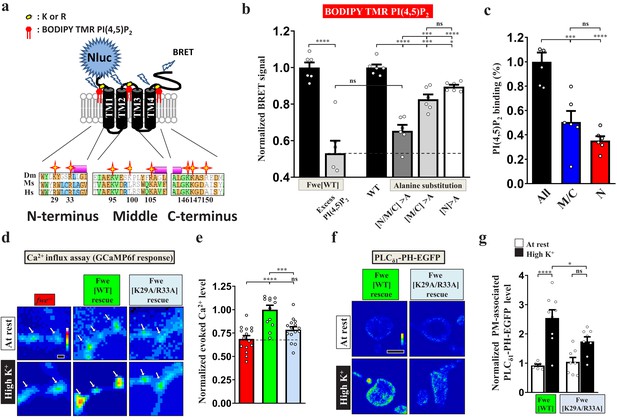
PI(4,5)P2 binds to Fwe and promotes its Ca2+ channelling activity.
(a–c) PI(4,5)P2 binds Fwe. (a) A schematic of the Fwe structure and a BRET assay. Stars highlight conserved lysine (K) and arginine (R) in juxta-transmembrane regions: N-terminus (N), middle-domain (M), and C-terminus (C). Drosophila melanogaster (Dm). Mus Musculus (Ms). Human Sapiens (Hs). N-terminal fusion of Nluc to Fwe allows detection of PI(4,5)P2 binding. A 1D4 epitope was used for protein purification. (b) Nluc-Fwe-1D4 in micelles excites BODIPY-TMR-conjugated PI(4,5)P2 to emit BRET signal, which is decreased by competitive cold PI(4,5)P2 (1 mM). Alanine substitution of positively-charged residues in all regions (N/M/C > A), both middle-domain and C-terminus (M/C > A), or N-terminus (N > A) reduced BRET signals. (c) Corresponding PI(4,5)P2 binding ability was calculated by subtracting the signal values of N/M/C > A, M/C > A or N > A from that for WT. Quantification data was normalized to the signal value of all mutations (N/M/C). (d) PI(4,5)P2 controls Fwe channel activity. Snapshot Ca2+ images of the boutons (arrows) expressing lexAop2-GCaMP6f using vglut-lexA. fwe mutant (fweDB25/DB56). HA-Fwe[WT]-APEX2 rescue (nSyb-GAL4/UAS-HA-Fwe[WT]-APEX2 in fweDB25/DB56). HA-Fwe[K29A/R33A]-APEX2 rescue (nSyb-GAL4/UAS-HA-Fwe[K29A/R33A]-APEX2 in fweDB25/DB56). The larvae were reared at 25°C. Images were taken in the fifth minute for one minute after high K+ stimulation. (e) Quantification data for evoked Ca2+ level, normalized to the value of HA-Fwe-APEX2 rescue larvae. (f) Single-plane confocal images of the boutons expressing UAS-PLCδ1-PH-EGFP. The larvae were reared at 25°C. After resting conditions or high K+ stimulation, the boutons were stained with α-GFP. (g) Quantification data for the PLCδ1-PH-EGFP staining intensity, normalized to the value of the resting condition of HA-Fwe-APEX2 rescue larvae. Individual data values are shown in graphs. p values: ns, not significant; *p<0.05; ***p<0.001; ****p<0.0001. Mean ± SEM. Scale bar: 2 µm (d, f). Statistics: Student t-test (b). One-way ANOVA with Tukey’s post hoc test (b, c, e, g).
-
Figure 4—source data 1
Source data for Figure 4.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig4-data1-v2.xlsx
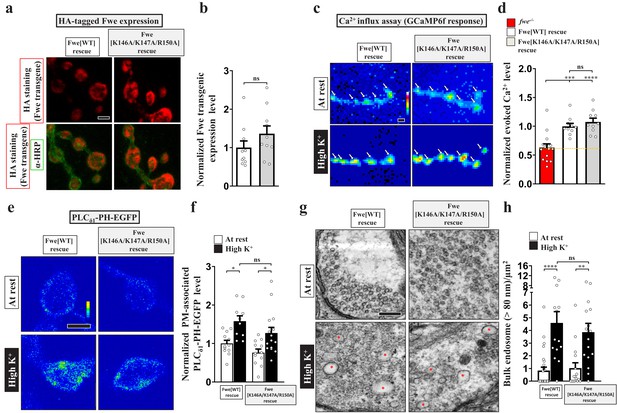
The C-terminal residues (K146/K147/R150) of Fwe are not involved in regulating Ca2+ channel activity.
(a–b) Alanine substitution of the C-terminal residues K146/K147/R150 does not influence the expression or SV localization of Fwe. (a) Z-projected confocal images of NMJ boutons derived from the larvae of Flag-Fwe[WT]-HA rescue (nSyb-GAL4/UAS-Flag-Fwe-HA in fweDB25/DB56) or Flag-Fwe[K146A/K147A/R150A]-HA rescue (nSyb-GAL4/UAS-Flag-Fwe[K146A/K147A/R150A]-HA in fweDB25/DB56) lines. The larvae were reared at 25°C. α-HA (red) and α-HRP (green) staining was used to detect Fwe transgenic proteins and neuronal membranes, respectively. α-HRP staining was also used as the staining control. (b) Quantification data for the staining intensity ratio of α-HA to α-HRP, normalized to the value of the Flag-Fwe[WT]-HA rescue line. (c–h) Alanine substitution of the C-terminal residues K146/K147/R150 does not influence Ca2+ channel activity and the ability to induce PI(4,5)P2 microdomains and ADBE. (c) Snapshot Ca2+ images of the NMJ boutons. fwe-/- (fweDB25/DB56). The boutons were subjected to 5 min stimulation of 90 mM K+/2 mM Ca2+. The images were taken in the fifth minute. Evoked Ca2+ levels are represented as the increase in GCaMP6f fluorescence in response to high K+ stimulation. (d) Quantification data for evoked Ca2+ level, normalized with the value of the Flag-Fwe[WT]-HA rescue line. (e) Single-plane confocal images of the NMJ boutons. The boutons were subjected to the resting condition (10 min incubation of 5 mM K+/0 mM Ca2+) or high K+ stimulation (10 min stimulation of 90 mM K+/2 mM Ca2+), followed by chemical fixation and α-GFP immunostaining. (f) Quantification data for the PLCδ1-PH-EGFP staining intensity, normalized to the value of the resting condition of the Flag-Fwe[WT]-HA rescue line. (g) TEM images of NMJ boutons. TEM processing was performed under the following conditions: at rest (10 min incubation of 5 mM K+/0 mM Ca2+); or high K+ (10 min stimulation of 90 mM K+/2 mM Ca2+). Bulk endosomes (red asterisks). (h) Quantification data of total numbers of bulk endosomes per bouton area. Individual data values are shown in the graphs. p values: ns, not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. Mean ± SEM. Scale bar: 2 µm (a, c, e), 500 nm (g). Statistics: Student t-test (b). One-way ANOVA with Tukey’s post hoc test (d, f, h).
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig4-figsupp1-data1-v2.xlsx

Expression of Fwe[R29A/K33A] in boutons.
(a–b) The Fwe[K29A/R33A] mutant variant properly localizes to the presynaptic compartment. (a) Z-projected confocal images of NMJ boutons derived from the larvae of the HA-Fwe[WT]-APEX2 rescue (nSyb-GAL4/UAS-HA-Fwe[WT]-APEX2 in fweDB25/DB56) or HA-Fwe[K29A/R33A]-APEX2 rescue (nSyb-GAL4/UAS- HA-Fwe[K29A/R33A]-APEX2 in fweDB25/DB56) lines. The larave were reared at 25°C. α-HA (red) and α-HRP (green) staining was used to detect Fwe transgenic proteins and neuronal membranes, respectively. α-HRP staining was also used as the staining control. (b) Quantification data for the staining intensity ratio of α-HA to α-HRP, normalized to the value of the Flag-Fwe[WT]-HA rescue line. Individual data values are shown in the graphs. p values: ns, not significant. Scale bar: 2 µm. Statistics: Student t-test.
-
Figure 4—figure supplement 2—source data 1
Source data for Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig4-figsupp2-data1-v2.xlsx
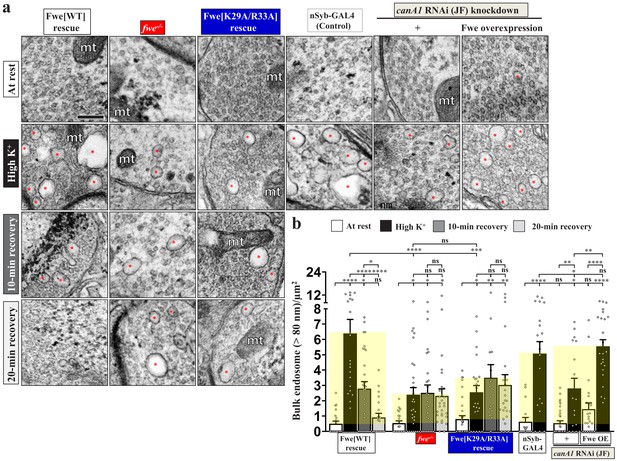
Perturbations of the positive feedback loop involving Fwe and PI(4,5)P2 suppress ADBE and SV reformation from bulk endosomes.
(a) TEM images of the boutons of HA-Fwe-APEX2 rescue (nSyb-GAL4/UAS-HA-Fwe-APEX2 in fweDB25/DB56 at 25°C), fwe mutant (fweDB25/DB56 at 25°C), HA-Fwe[K29A/R33A]-APEX2 rescue (nSyb-GAL4/UAS-HA-Fwe[K29A/R33A]-APEX2 in fweDB25/DB56 at 25°C), control (nSyb-GAL4 at 25°C), canA1 RNAi (JF) knockdown (nSyb-GAL4/UAS-canA1-RNAi (TRiP.JF01871) at 29°C), canA1 RNAi (JF) knockdown plus Fwe overexpression (nSyb-GAL4/UAS-canA1-RNAi (TRiP.JF01871)/UAS-HA-Fwe-APEX2 at 29°C). TEM processing was performed after the following treatments: at rest (10 min incubation of 5 mM K+/0 mM Ca2+); high K+ stimulation (10 min stimulation of 90 mM K+/2 mM Ca2+); 10 min recovery (10 min stimulation of 90 mM K+/2 mM Ca2+, followed by 10 min incubation of 5 mM K+/0 mM Ca2+); or 20 min recovery (10 min stimulation of 90 mM K+/2 mM Ca2+, followed by 20 min incubation of 5 mM K+/0 mM Ca2+). Bulk endosomes (>80 nm in diameter, red asterisks). Mitochondria (mt). (b) Quantification data of total numbers of bulk endosomes per bouton area. Individual data values are shown in graphs. p values: ns, not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. Mean ± SEM. Scale bar: 500 nm. Statistics: one-way ANOVA with Tukey’s post hoc test.
-
Figure 5—source data 1
Source data for Figure 5.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig5-data1-v2.xlsx

Blockade of the positive feedback loop between Fwe and PI(4,5)P2 abolishes SV reformation from the bulk endosome.
Quantification data for the ratio of total area of bulk endosomes to bouton area of the indicated genotypes. Individual data values are shown in the graphs. p values: ns, not significant; *p<0.05; **p<0.001; ****p<0.0001. Mean ± SEM. Statistics: One-way ANOVA with Tukey’s post hoc test.
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig5-figsupp1-data1-v2.xlsx
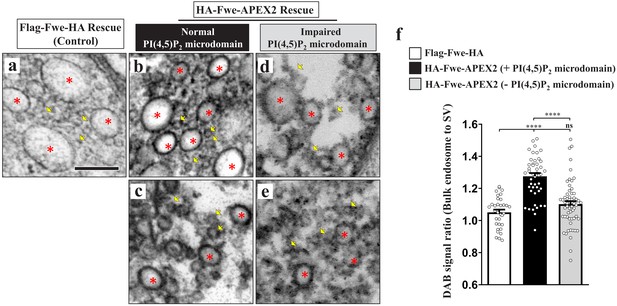
PI(4,5)P2 facilitates recycling of Fwe to bulk endosomes.
(a–f) PI(4,5)P2 retrieves Fwe to bulk endosomes. (a–e) TEM images of the boutons of Flag-Fwe-HA rescue (nSyb-GAL4/UAS-Flag-Fwe-HA in fweDB25/DB56 at 25°C, a), HA-Fwe-APEX2 rescue (nSyb-GAL4/UAS-HA-Fwe-APEX2 in fweDB25/DB56 at 25°C, b–c), or HA-Fwe-APEX2 rescue coexpressing Synj (nSyb-GAL4/UAS-HA-Fwe-APEX2/UAS-synj in fweDB25/DB56 at 25°C, d–e). After high K+ stimulation, the boutons were subjected to DAB labeling. (a) The Flag-Fwe-HA rescue boutons presented DAB-negative bulk endosomes (red asterisks) and SVs (yellow arrows). (b and c) In the HA-Fwe-APEX2 rescue boutons, DAB signals on bulk endosomes (red asterisks) were higher than those on the SVs (yellow arrows). The views in b and c are from different boutons. (d and e) Under the condition of Synj overexpression, perturbation of PI(4,5)P2 microdomain formation predominantly reduced DAB levels on the bulk endosomes (red asterisks). Views in d and e are from different boutons. (f) Quantification data for the DAB staining intensity ratio of bulk endosomes to surrounding SVs. The number of bulk endosomes, NMJ boutons, and larvae counted (Flag-Fwe-HA rescue control): Bulk endosomes (n = 33) derived from 5 NMJ boutons of two different larvae; HA-Fwe-APEX2 rescue: Bulk endosomes (n = 47) derived from 6 NMJ boutons of two different larvae; HA-Fwe-APEX2 rescue expressing Synj: Bulk endosomes (n = 59) derived from 9 NMJ boutons of two different larvae. Individual values were shown in graphs. p values: ns, not significant; ****p<0.0001. Mean ± SEM. Scale bar: 200 nm (a–e). Statistics: one-way ANOVA with Tukey’s post hoc test.
-
Figure 6—source data 1
Source data for Figure 6.
- https://cdn.elifesciences.org/articles/60125/elife-60125-fig6-data1-v2.xlsx
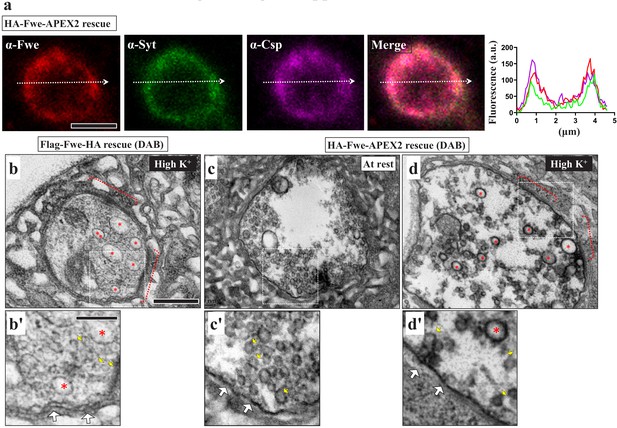
HA-Fwe-APEX2 behaves like endogenous Fwe.
(a) HA-Fwe-APEX2 localizes in the SVs. Single-plane confocal images of NMJ boutons of the HA-Fwe-APEX2 rescue line (nSyb-GAL4/UAS-HA-Fwe[WT]-APEX2 in fweDB25/DB56). The boutons were stained for Fwe (red) and the SV markers, Syt (green), and Csp (magenta). Line plot profiles of absolute fluorescence units (a.u.) of immunostaining signals are shown at right. (b–d) TEM images of the boutons of Flag-Fwe-HA rescue (nSyb-GAL4/UAS-Flag-Fwe-HA in fweDB25/DB56 at 25°C), (b) or HA-Fwe-APEX2 rescue (nSyb-GAL4/UAS-HA-Fwe-APEX2) in fweDB25/DB56 at 25°C, (c and d) lines. The boutons were subjected to the resting condition (c) or high K+ stimulation (b and d), followed by DAB labeling. High-magnification images are shown at bottom. In the Flag-Fwe-HA rescue boutons, SVs (yellow arrows), bulk endosomes (red asterisks), and plasma membrane (white arrows) lack specific DAB signals. In the HA-Fwe-APEX2 rescue boutons at rest, both SVs (yellow arrows) and plasma membrane (white arrows) were significantly labelled with DAB. Upon high stimulation, high levels of DAB signals were observed on the bulk endosomes of HA-Fwe-APEX2 rescue boutons. Active zones (red dashed lines). Scale bar: 2 µm (a), 500 nm (b, c, d), 200 nm (b’, c’, d’).
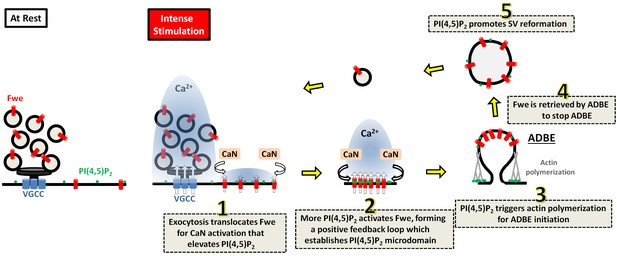
A proposed model for the role of Fwe-dependent PI(4,5)P2 microdomains in coordinating ADBE and SV reformation from bulk endosomes.
Details are described in the Discussion section.
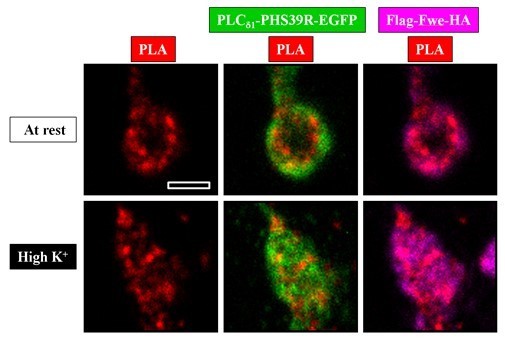
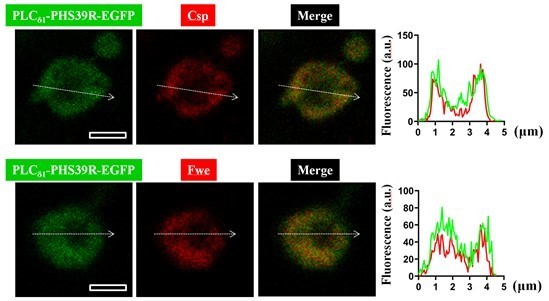
A close association between PLCδ1-PHS39R-EGFP and Flag-Fwe-HA on the SVs generates PLA signals at rest or after high K+ stimulation.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | fweDB25 | Yao et al., 2009 | fwe mutant allele used in Figures 3–6, Figure 4—figure supplements 1–2, Figure 5—figure supplement 1, and Figure 6—figure supplement 1 | |
| Genetic reagent (D. melanogaster) | fweDB56 | Yao et al., 2009 | fwe mutant allele used in Figures 3–6, Figure 4—figure supplements 1–2, Figure 5—figure supplement 1, and Figure 6—figure supplement 1 | |
| Genetic reagent (D. melanogaster) | UAS-Flag-Fwe-HA | Yao et al., 2017 | Wild-type version of fwe transgene used in Figure 3a–b, Figure 6a, Figure 4—figure supplement 1, and Figure 6—figure supplement 1b | |
| Genetic reagent (D. melanogaster) | UAS-Flag-Fwe[E79Q]-HA | Yao et al., 2017 | Channel deficient version of fwe transgene used in Figure 3e–f | |
| Genetic reagent (D. melanogaster) | UAS-canA1-RNAi (FB4) | Dijkers and O'Farrell, 2007 | RNAi line of canA1 used in Figure 3g–j | |
| Genetic reagent (D. melanogaster) | UAS-canA1-RNAi(TRiP.JF01871) | Bloomington Drosophila Stock Center Dijkers and O'Farrell, 2007 | RRID:BDSC_25850 | RNAi line of canA1 used in Figure 3g–h, 5 |
| Genetic reagent (D. melanogaster) | UAS-synj | Khuong et al., 2013 | Wild-type version of synj transgene used in Figure 2b–d, Figure 3i–j, Figure 6d–f, and Figure 2—figure supplement 1c. | |
| Genetic reagent (D. melanogaster) | UAS-PLCδ1-PH-EGFP | Bloomington Drosophila Stock Center Verstreken et al., 2009 | RRID:BDSC_39693 | PI(4,5)P2 reporter transgene used in Figures 1–4, Figure 1—figure supplement 1, Figure 1—figure supplement 3c, Figure 2—figure supplements 1–2, Figure 4—figure supplement 1e–f. |
| Genetic reagent (D. melanogaster) | UAS-PLCδ1-PHS39R-EGFP | Bloomington Drosophila Stock Center Verstreken et al., 2009 | RRID:BDSC_39694 | PI(4,5)P2-binding mutant transgene used in Figure 2c–d, Figure 1—figure supplement 2c–e |
| Genetic reagent (D. melanogaster) | nSyb-GAL4 | Bloomington Drosophila Stock Center Pauli et al., 2008 | RRID:BDSC_51635 | Neuronal GAL4 driver used in all figures |
| Genetic reagent (D. melanogaster) | vglut-lexA | Bloomington Drosophila Stock Center | RRID:BDSC_60314 | Motor neuron lexA driver used in Figure 3e–f, Figure 3i–j, Figure 4d–e, and Figure 4—figure supplement 1c–d |
| Genetic reagent (D. melanogaster) | 13XLexAop2-IVS-GCaMP6f | Bloomington Drosophila Stock Center | RRID:BDSC_44277 | Ca2+ indicator transgene used in Figure 3i–j, Figure 4d–e, and Figure 4—figure supplement 1c–d |
| Genetic reagent (D. melanogaster) | synj1 | Bloomington Drosophila Stock Center Verstreken et al., 2003 | RRID:BDSC_24883 | synj mutant allele used in Figure 1—figure supplement 1c–d |
| Genetic reagent (D. melanogaster) | UAS-mCD8-EGFP | Kyoto Stock Center | RRID:DGRC_108068 | mCD8-EGFP transgene used in Figure 1—figure supplement 2a–b |
| Genetic reagent (D. melanogaster) | UAS-HA-Fwe[WT]-APEX2 | This paper | APEX2 fusion version of wild-type fwe transgene used in Figure 4d–g, Figure 5, Figure 6b–f, Figure 4—figure supplement 2, and Figure 6—figure supplement 1c–d | |
| Genetic reagent (D. melanogaster) | UAS-HA-Fwe[K29A/R33A]-APEX2 | This paper | APEX2 fusion version of fwe[K29A/R33A] transgene used in Figure 4d–g, Figure 5, Figure 4—figure supplement 2 | |
| Genetic reagent (D. melanogaster) | UAS-Flag-Fwe[K29A/R33A/K95A/K100A/R105A/K146A/K147A/R150A]-HA | This paper | fwe[K29A/R33A/K95A/K100A/R105A/K146A/K147A/R150A] transgene was not expressed stably | |
| Genetic reagent (D. melanogaster) | UAS-Flag-Fwe[K146A/K147A/R150A]-HA | This paper | fwe[K146A/K147A/R150A] transgene used in Figure 4—figure supplement 1 | |
| Genetic reagent (D. melanogaster) | UAS-Flag-Fwe[K95A/K100A/R105A]-HA | This paper | fwe[K95A/K100A/R105A] transgene was not expressed stably | |
| Genetic reagent (D. melanogaster) | LexAop2-PLCδ1-PH-APEX2-HA | This paper | APEX2 fusion version of PLCδ1-PH transgene used in Figure 3i–j | |
| Genetic reagent (D. melanogaster) | LexAop2-PLCδ1-PH-EGFP | This paper | lexAop2 version of PLCδ1-PH-EGFP transgene used in Figure 3e–f | |
| Antibody | α-GFP (Chicken polyclonal) | Invitrogen | Cat #A10262, RRID:AB_2534023 | IF: 1:500 WB: 1:5000 |
| Antibody | α-HA (Mouse monoclonal) | Sigma | Cat # H3663, RRID:AB_262051 | IF: 1:400 PLA: 1:200 |
| Antibody | α-Bruchpilot (Mouse monoclonal) | DSHB Wagh et al., 2006 | Cat# nc82, RRID:AB_2314866 | IF: 1:100 |
| Antibody | α-AP-2α (Rabbit polyclonal) | González-Gaitán and Jäckle, 1997 | IF: 1:3000 | |
| Antibody | α-Eps15 (Guinea pig polyclonal) | Koh et al., 2007 | IF: 1:3000 | |
| Antibody | α-Fwe B isoform (Guinea pig polyclonal) | Yao et al., 2017 | IF: 1:400 | |
| Antibody | Cy3 AffiniPure Rabbit Anti-Horseradish Peroxidase | Jackson ImmunoResearch Labs | Cat# 323-165-021, RRID:AB_2340262 | IF: 1:250 |
| Antibody | Alexa Fluor 488 AffiniPure Rabbit Anti-Horseradish Peroxidase | Jackson ImmunoResearch Labs | Cat# 323-545-021, RRID:AB_2340264 | IF: 1:250 |
| Antibody | α-GFP (Rabbit polyclonal) | Thermo Fisher Scientific | Cat# A-6455, RRID:AB_221570 | PLA: 1:500 |
| Antibody | α-actin (Mouse monoclonal) | Sigma | Cat# A7732, RRID:AB_2221571 | WB:1:20000 |
| Antibody | α−1D4 (Mouse monoclonal) | Yao et al., 2009 | WB: 1:2000 | |
| Strain, strain background (Escherichia coli) | DH5α | |||
| Strain, strain background (Escherichia coli) | yeast strain BJ5457 | Yao et al., 2009 | ||
| Peptide, recombinant protein | Nluc-Fwe-1D4 fusion protein | This paper | Used in Figure 4b–c | |
| Peptide, recombinant protein | Nluc-Fwe[K29A/R33A]−1D4 | This paper | Used in Figure 4b–c | |
| Peptide, recombinant protein | Nluc-Fwe[K95A/K100A/R105A/K146A/K147/R150A]−1D4 | This paper | Used in Figure 4b–c | |
| Peptide, recombinant protein | Nluc-Fwe[K29A/R33A/K95A/K100A/R105A/K146A/K147A/R150A]−1D4 | This paper | Used in Figure 4b–c | |
| Chemical compound, drug | n-Dodecyl-β-D-Maltopyranoside | Anatrace | Cat# D310 | Used in Figure 4b–c |
| Chemical compound, drug | BODIPY-TMR Phosphatidylinositol 4,5-bisphosphate | Echelon Bioscience | Cat#C-45M16A | Used in Figure 4b–c |
| Chemical compound, drug | Furimazine | Promega | Cat# N1110 | Used in Figure 4b–c |
| Chemical compound, drug | Brain Phosphatidylinositol 4,5-bisphosphate | Avanti Polar Lipids | AV-840046P | Used in Figure 4b–c |
| Commercial assay or kit | Duolink In Situ Red Starter Kit Mouse/Rabbit | Sigma | Cat# DUO92101 | Used in Figure 3a–b |
| Commercial assay or kit | NEBuilder HiFi DNA Assembly Master Mix | NEB | Cat# E2621 | |
| Recombinant DNA reagent | YeMP | Yao et al., 2009 | ||
| Recombinant DNA reagent | pcDNA3 APEX2-NES | Addgene | RRID:Addgene_49386 | |
| Recombinant DNA reagent | pJFRC19-13XLexAop2-IVS-myr:GFP | Addgene | RRID:Addgene_26224 | |
| Recombinant DNA reagent | pNL1.1[Nluc] | Promega | Cat# #N1001 | |
| Recombinant DNA reagent | pUAST-HA-Fwe[WT]-APEX2 | This paper | DNA construct of APEX2 fusion version of wild-type fwe transgene used in Figure 4d–g, Figure 5, Figure 6b–f, Figure 4—figure supplement 2, and Figure 6—figure supplement 1c–d | |
| Recombinant DNA reagent | pUAST-HA-Fwe[K29A/R33A]-APEX2 | This paper | DNA construct of APEX2 fusion version of fwe[K29A/R33A] transgene used in Figure 4d–g, Figure 5, Figure 4—figure supplement 2 | |
| Recombinant DNA reagent | pUAST-Flag-Fwe-HA | Yao et al., 2017 | ||
| Recombinant DNA reagent | pUAST-Flag-Fwe[K29/R33/K95/K100/R105/K146A/K147A/R150A]-HA | This paper | DNA construct of fwe[K29A/R33A/K95A/K100A/R105A/K146A/K147A/R150A] transgene | |
| Recombinant DNA reagent | pUAST-Flag-Fwe[K146A/K147A/R150A]-HA | This paper | DNA construct of fwe[K146A/K147A/R150A] transgene used in Figure 4—figure supplement 1 | |
| Recombinant DNA reagent | pUAST-Flag-Fwe[K95/K100/R105A]-HA | This paper | DNA construct of fwe[K95A/K100A/R105A] transgene | |
| Recombinant DNA reagent | plexAop2-PLCδ1-PH-APEX2-HA | This paper | DNA construct of APEX2 fusion version of PLCδ1-PH transgene used in Figure 3i–j | |
| Recombinant DNA reagent | plexAop2-PLCδ1-PH-EGFP | This paper | DNA construct of lexAop2 version of PLCδ1-PH-EGFP transgene used in Figure 3e–f | |
| Recombinant DNA reagent | YeMP-Nluc-Fwe-1D4 | This paper | DNA construct of expression of Nluc-Fwe-1D4 recombinant protein used in Figure 4b–c | |
| Recombinant DNA reagent | YeMP-Nluc-Fwe[K29A/R33A]−1D4 | This paper | DNA construct of expression of Nluc-Fwe[K29A/R33A]−1D4 recombinant protein used in Figure 4b–c | |
| Recombinant DNA reagent | YeMP-Nluc-Fwe[K95A/K100A/R105A/K146A/K147/R150A]−1D4 | This paper | DNA construct of expression of Nluc-Fwe[K95A/K100A/R105A/K146A/K147/R150A]−1D4 recombinant protein used in Figure 4b–c | |
| Recombinant DNA reagent | YeMP-Nluc-Fwe[K29A/R33A/K95A/K100A/R105A/K146A/K147A/R150A]−1D4 | This paper | DNA construct of expression of Nluc-Fwe[K29A/R33A/K95A/K100A/R105A/K146A/K147A/R150A]−1D4-1D4 recombinant protein used in Figure 4b–c | |
| Sequence-based reagent | HA-Fwe[WT]-APEX2 forward-1 | This paper | 5'- tcgttaacagatcttGCGGCCGCATGTACCCATACGATGTTCCAGATTACG-3' | |
| Sequence-based reagent | HA-Fwe[WT]-APEX2 Reverse-1 | This paper | 5'- tgaacctcctccgccTGTGGGGCGCCAGACATC-3' | |
| Sequence-based reagent | HA-Fwe[WT]-APEX2 forward-2 | This paper | 5'- gccccacaggcggaggaggttcaggaggcggaggttcgGGTACCGGCGGAAAGAGTTACCCGAC-3' | |
| Sequence-based reagent | HA-Fwe[WT]-APEX2 Reverse-2 | This paper | 5'- tccttcacaaagatccTCTAGAGGCGTCGGCGAATCCCAG −3' | |
| Sequence-based reagent | HA-Fwe[K29A/R33A]-APEX2-HA forward | This paper | 5'-CAGCCGTGGTATCTCGCATATGGAAGTGCATTGCTGGGCATTG-3' | |
| Sequence-based reagent | Flag-Fwe[K95/K100/R105A]-HA forward | This paper | 5'-GGAATCTGCGCCCTTGTACTTCGCTGCCGGGCTCTACATTGCCATGGCCATTCCGCCCATTATT-3' | |
| Sequence-based reagent | Flag-Fwe[K146A/K147A/R150A]-HA forward | This paper | 5'-CAGAGGACATGGCCGCCGCTGCCACATCGCCCACACAGATGGCCGGCAGTCAGGCGGGCGG-3' | |
| Sequence-based reagent | PLCδ1-PH-APEX2-HA forward-1 | This paper | 5'-tcttatcctttacttcagGCGGCCGCATGGACTCGGGCCGGGAC-3' | |
| Sequence-based reagent | PLCδ1-PH-APEX2-HA forward-2 | This paper | 5'- GCTGTACAAGggcggaggaggttcaggaggcggaggttcgGGCGGAAAGAGTTACCCGAC-3' | |
| Sequence-based reagent | PLCδ1-PH-APEX2-HA reverse-1 | This paper | 5'- cctccgccggtaccAAGATCTTCCGGGCATAGCTGTCG-3' | |
| Sequence-based reagent | PLCδ1-PH-APEX2-HA reverse-2 | This paper | 5'- ggttccttcacaaagatcctctagattaagcgtaatctggaacatcgtatgggtaGGCGTCGGCGAATCCCAG −3' | |
| Sequence-based reagent | PLCδ1-PH-EGFP forward | This paper | 5'- tcttatcctttacttcagGCGGCCGCATGGACTCGGGCCGGGAC-3' | |
| Sequence-based reagent | PLCδ1-PH-EGFP reverse | This paper | 5'- TTTCTAGATTACTTGTACAGCTCGTCCAT-3' | |
| Sequence-based reagent | YeMP-Nluc-Fwe-1D4 forward-1 | This paper | 5'- CCACTAGTATGGTCTTCACACTCGAAG-3' | |
| Sequence-based reagent | YeMP-Nluc-Fwe-1D4 forward-2 | This paper | 5'- AAAACTAGTGTCGACTCGTTTGCGGAAAAGATAACG −3' | |
| Sequence-based reagent | YeMP-Nluc-Fwe-1D4 Reverse-1 | This paper | 5'-AAAGTCGACCGAACCTCCGCCTCC-3' | |
| Sequence-based reagent | YeMP-Nluc-Fwe-1D4 Reverse-2 | This paper | 5'- AAAACGCGTTTACGCAGGCGCGACTTGG-3' | |
| Sequence-based reagent | YeMP-Nluc-Fwe[K29A/R33A]−1D4, YeMP-Nluc-Fwe[K95A/K100A/R105A/K146A/K147/R150A]−1D4, YeMP-Nluc-Fwe[K29A/R33A/K95A/K100A/R105A/K146A/K147A/R150A]−1D4 forward | This paper | 5'- AAAACTAGTGTCGACTCGTTTGCGGAAAAGATAACG-3' | |
| Sequence-based reagent | YeMP-Nluc-Fwe[K29A/R33A]−1D4, YeMP-Nluc-Fwe[K95A/K100A/R105A/K146A/K147/R150A]−1D4, YeMP-Nluc-Fwe[K29A/R33A/K95A/K100A/R105A/K146A/K147A/R150A]−1D4 Reverse | This paper | 5'- AAAACGCGTTTACGCAGGCGCGACTTGGCTGGTCTCTGTTGTGGGGCGCC-3' | |
| Software, algorithm | LSM Zen | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html | |
| Software, algorithm | Image J | https://imagej.net/contributors | https://imagej.nih.gov/ij/ | |
| Software, algorithm | pClamp 10.6 | Moleculardevices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref | |
| Software, algorithm | MetaMorph | Moleculardevices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref | |
| Software, algorithm | GraphPad Prism 8.0 | Prism | https://www.graphpad.com/ |