Chloroplast acquisition without the gene transfer in kleptoplastic sea slugs, Plakobranchus ocellatus
Figures
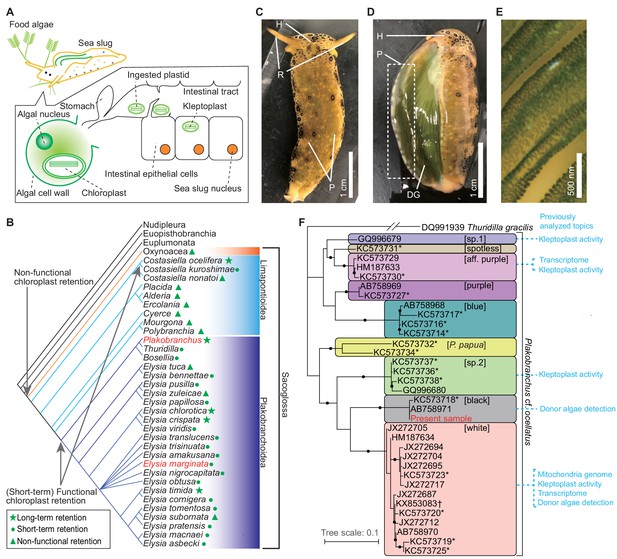
Kleptoplasty in sea slugs.
(A) Process of algal chloroplast retention by a sacoglossan sea slug (Pierce and Curtis, 2012). Sacoglossan sea slugs puncture the cell wall of food algae to suck out the protoplasm. The chloroplasts in the protoplasm are transported to the sea slug’s intestinal tract, and the intestinal epithelial cells sequester chloroplasts by phagocytosis. The sequestered chloroplasts (kleptoplasts) maintain the photosynthetic activity in the cell for days to months. The sacoglossan cell contains no algal nuclei. Kleptoplast has never been found in germ cells of sea slug. (B) Phylogenetic distribution of kleptoplasty in the order Sacoglossa. Phylogenetic analysis showed that a common ancestor of Sacoglossa acquired non-functional chloroplast retention phenomena (without the maintenance of photosynthetic function), and multiple sacoglossan groups subsequently acquired the ability to maintain photosynthetic activity. Phylogenetic tree and kleptoplasty states are simplified from Christa et al., 2015. Christa et al., 2014b defined functional chloroplast retention for less than 2 weeks as ‘short-term retention’, and for more than 20 days as ‘long-term retention’. Relationships within Heterobranchia are described according to Zapata et al., 2014. The red-colored taxa include the species used in the present study (P. ocellatus and E. marginata). (C–E) Photo images of PoB starved for 21 days. (C) Dorsal view. H, head; R, rhinophores; P, parapodia (lateral fleshy flat protrusions). Almost always, PoB folded parapodia to the back in nature. (D) The same individual of which parapodium was turned inside out (without dissection). The back of the sea slug and inside of the parapodia are green. This coloration is caused by the kleptoplasts in DG, which are visible through the epidermis. (E) Magnified view of the inner surface of parapodium. The diagonal green streaks are ridged projections on the inner surface of the parapodium. The cells containing kleptoplasts are visible as green spots. (F) Phylogeny of the P. cf. ocellatus species complex based on mitochondrial cox1 genes (ML tree from 568 nucleotide positions) from INSD and the whole mtDNA sequence. The sequence data for the phylogenetic analysis are listed in Figure 1—source data 1. Clade names in square brackets are based on Krug et al., 2013. Asterisks mark the genotypes from Krug et al., 2013. Study topics analyzed by previous researchers were described within the colored boxes for each cluster. Small black circles indicate nodes supported by a high bootstrap value (i.e., 80–100%). Thuridilla gracilis is an outgroup. Plakobranchus papua is a recently described species and previously identified as P. ocellatus (Meyers-Muñoz et al., 2016).
-
Figure 1—source data 1
Sequence ID list used for phylogenetic analysis on Figure 1B.
- https://cdn.elifesciences.org/articles/60176/elife-60176-fig1-data1-v1.xlsx
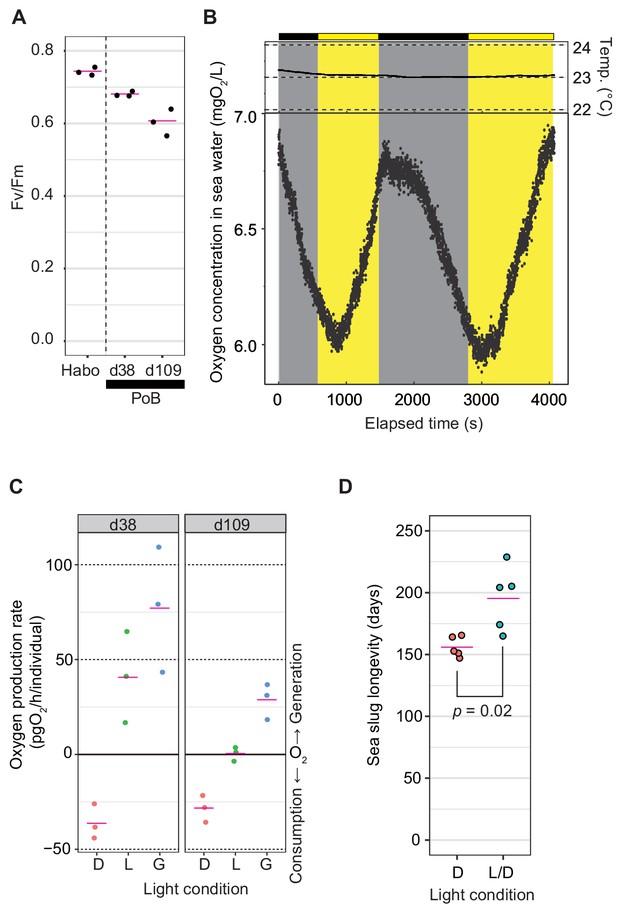
Photosynthetic activity of PoB.
(A) Jitter plot of Fv/Fm values indicating the photochemical efficiency of PSII. Habo, H. borneensis; d38, starved PoB for 38 days; d109, starved for 109 to 110 days (12 hr light/12 hr dark cycle; the light phase illumination was 10 µmol photons m−2 s−1). The magenta line indicates the mean, and the black dot indicates each individual’s raw value (n = 3 per group). (B) Dynamics of seawater oxygen concentration. To better demonstrate the PoB photosynthesis, a change of oxygen concentration every second was visualized. A PoB individual, one of the d38 samples (ID1), was put into the measurement chamber, and the light conditions were changed in tens of minutes. The measurements in other individuals are visualized in Figure 2—figure supplement 1, along with the diagrams of the measuring equipment. Each point represents the oxygen concentration value per second. Gray signifies a dark period, and yellow means an illuminated period (50 µmol photons m−2 s−1). Temp, water temperature. Although the values fluctuated by the noise due to ambient light and other factors, the oxygen concentration decreased in the dark conditions and increased in the light conditions. When the illumination condition was changed, the changing pattern kept the previous pattern for a few minutes. This discrepancy may reflect the distance between the organism and oxygen sensor and the time for adaptation to brightness by the kleptoplast. (C) Jitter plots of PoB oxygen consumption and generation. D, dark conditions; L, light conditions; G, gross rate of light-dependent oxygen generation (L minus D). (D) Jitter plots of PoB longevity (n = 5 per group). D, Continuous dark; L/D, 12 hr light/12 hr dark cycle. The p from Welch’s two-sample t-test was used. A source file of Fv/Fm jitter plot, time course of oxygen concentration, and longevity analysis are available in Figure 2—source data 1–3, respectively.
-
Figure 2—source data 1
Summarized data of Fv/Fm and oxygen generation activity analysis of P.
ocellatus used for Figure 2.
- https://cdn.elifesciences.org/articles/60176/elife-60176-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Raw data of oxygen concentration dynamics.
This zip archive contains raw data of oxygen concentration dynamics. The measurement data for each time series are stored in separate files, with the experiment ID (ID1–6) and types of the experiment (mock or P. ocellatus sample) indicated in the name.
- https://cdn.elifesciences.org/articles/60176/elife-60176-fig2-data2-v1.zip
-
Figure 2—source data 3
Summary of the longevity of analyzed P. ocellatus individuals used for Figure 2D.
- https://cdn.elifesciences.org/articles/60176/elife-60176-fig2-data3-v1.xlsx
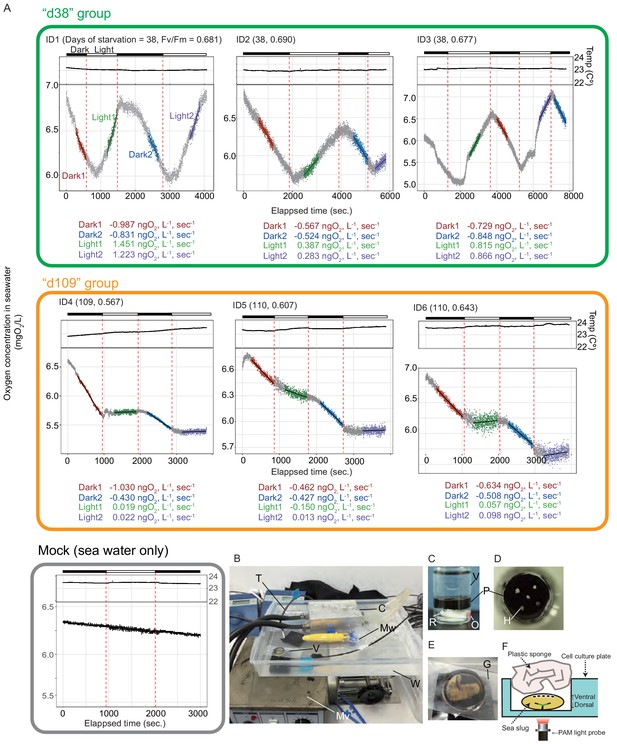
Light-dependent oxygen generation by P. ocellatus.
(A) Time-course changes of dissolved oxygen in the sea water in the vial containing the sea slug. Each plot indicates the data of a single PoB individual. Raw data are available in Figure 2—source data 2. d38 group, three PoBs starved for 38 days; d109 group, three PoBs starved for 109 days. (B–E) Oxygen measurement system. (B) Overview of the dissolved oxygen measurement system. C, iron chilling block; Mw, mixer for lagging water; Mv, mixer for sea slug rearing seawater; V, vial caging a sea slug; W, plastic tray filled with lagging water for the vial. (C) Horizontal view of the glass vial. O, oxygen-sensitive optode; P, rubber-made partition plate to separate sea slug from optode and rotor; R, rotor for seawater agitation. (D) Top view of the vial. H, holes for water ventilation. (E) Top vial view with a sea slug. G, glass plate sealing the top of the vial. (F) Diagram of PAM measurement method. Horizontal view of the sea slug in a well of the cell culture plate and positions of a sponge and a light probe. The sea slug was upside down facing the dorsal side to the PAM light probe.
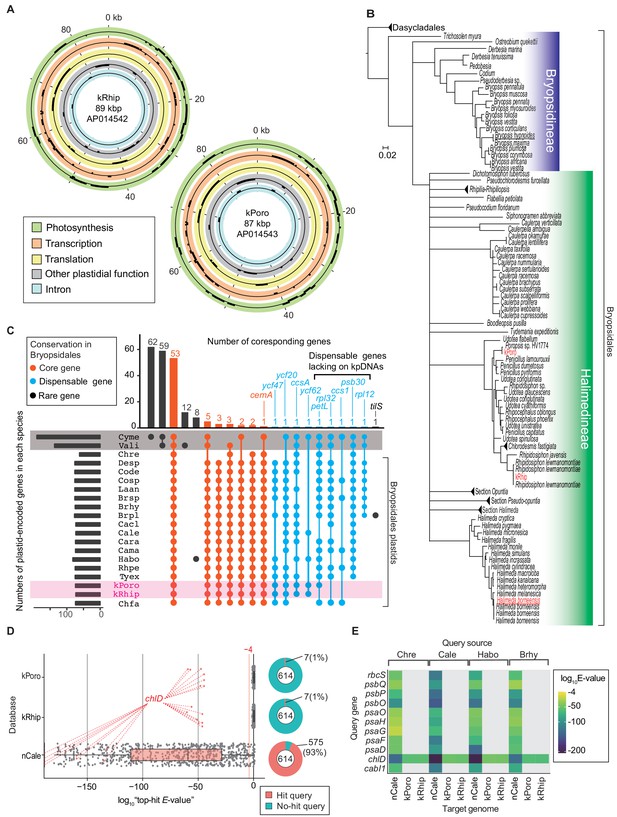
Gene composition of PoB kpDNAs.
(A) Gene map of two kpDNAs from PoB. Gene positions are described in circles colored according to the gene’s functional category (see keys in the box). Genes on the outside and inside of each circle are transcribed in the clockwise and anticlockwise directions, respectively (for detailed maps, see Figure 3—figure supplements 1 and 2). (B) Phylogenetic positions of sequenced kleptoplasts among green algal plastids. The original ML tree (Figure 3—figure supplement 4) was created based on rbcL genes (457 positions) and converted to the tree. Red indicates sequenced kpDNA or cpDNA in the present study. Underlines indicate algal species used in RNA-Seq sequencing. (C) An UpSet plot of plastid gene composition. Species abbreviations are defined in Figure 3—source data 1. The horizontal bar chart indicates the gene numbers in each species. The vertical bar chart indicates the number of genes conserved among the species. Intersect connectors indicate the species composition in a given number of genes (vertical bar chart). Connections corresponding to no gene were omitted. Connectors were colored according to the gene’s conservation level in Bryopsidales: Core gene, conserved among all analyzed Bryopsidales species; Dispensable gene, retained more than two Bryopsidales species; Rare gene, determined from a single or no Bryopsidales species. Gray shading indicates non-Viridiplantae algae, and magenta shading indicates PoB kleptoplasts. Cyme (C. merolae) and Vali (V. litorea) had more than 100 genes that Bryopsidales does not have (e.g., left two vertical bars). (D) Box-plots of tblastn results. The vertical axis shows the database searched (kPoro and kRhip, PoB kpDNAs; nCale, nucDNA of C. lentillifera). Each dot represents the tblastn result (query is the A614 dataset). Red dots show the result using the chlD gene (encoding magnesium-chelatase subunit ChlD) as the query sequence; this sequence is similar to the kleptoplast-encoded chlL gene. The right pie chart shows the proportion of queries with hits (E-value < 0.0001). (E) Heat map of tblastn results of representative photosynthetic nuclear genes (a subset of data in D). The source species of the query sequences are described on the top. The source files of plastid gene composition and tblastn analysis are available in Figure 3—source data 1 and 2.
-
Figure 3—source data 1
Plastid gene composition.
This zip archive contains source files of plastid gene composition and of orthologous analysis.
- https://cdn.elifesciences.org/articles/60176/elife-60176-fig3-data1-v1.zip
-
Figure 3—source data 2
Tblastn analysis of kleptoplast/chloroplast DNA.
This zip archive contains source and result files of tblastn analysis of kleptoplast/chloroplast DNA.
- https://cdn.elifesciences.org/articles/60176/elife-60176-fig3-data2-v1.zip
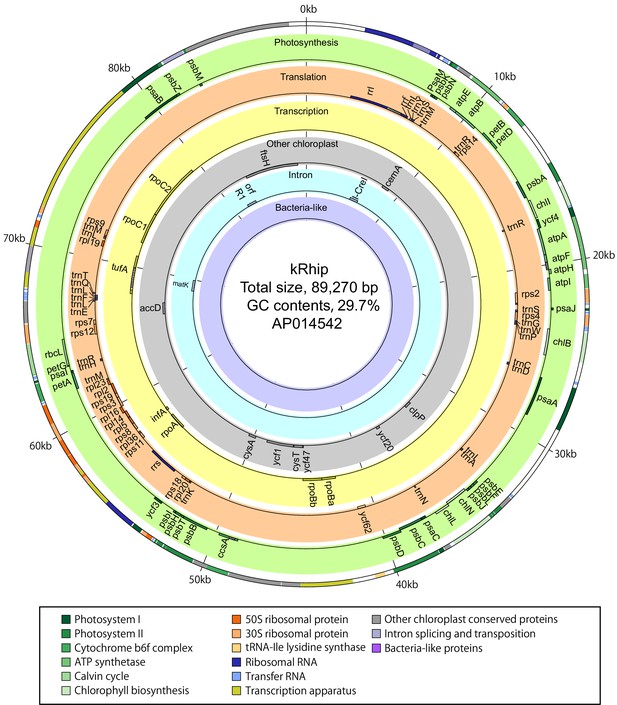
Gene map of a P. ocellatus kleptoplast DNA sequestered from Rhipidosiphon lewmanomontiae.
The outer circle shows the distribution of all genes on kpDNA. As shown in the box, colors indicate functional categories of the genes, respectively. Circles on different colored backgrounds show the names and positions of genes related to photosynthesis (second circle), translation (third), transcription (fourth), other conserved chloroplast genes (fifth), genes relating to intron-splicing and transposition (sixth), and bacteria-like genes (seventh), respectively. Genes on the outer-side of each circle are transcribed in the clockwise direction; genes on the inner-side are transcribed in the anticlockwise direction. Hatched boxes of genes on the circles are introns.
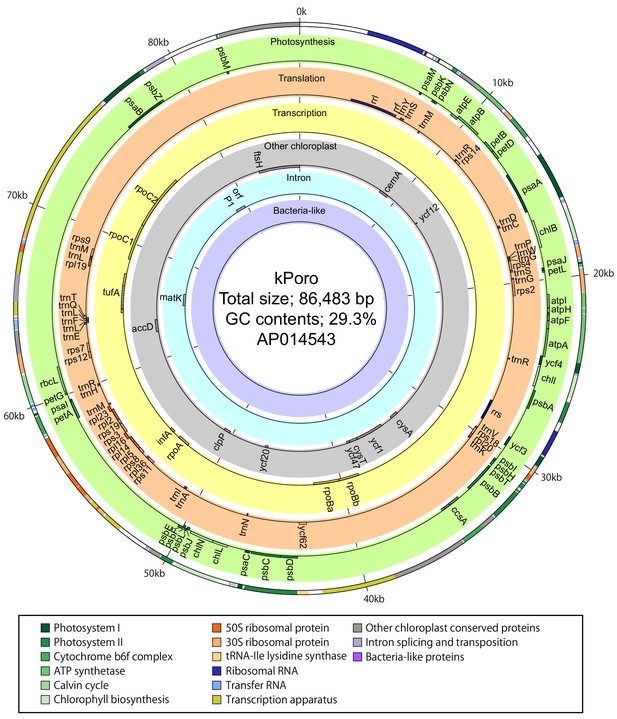
Gene map of a P. ocellatus kleptoplast DNA sequestered from Poropsis spp.
Circles and colors are the same as those in Figure 3—figure supplement 1.
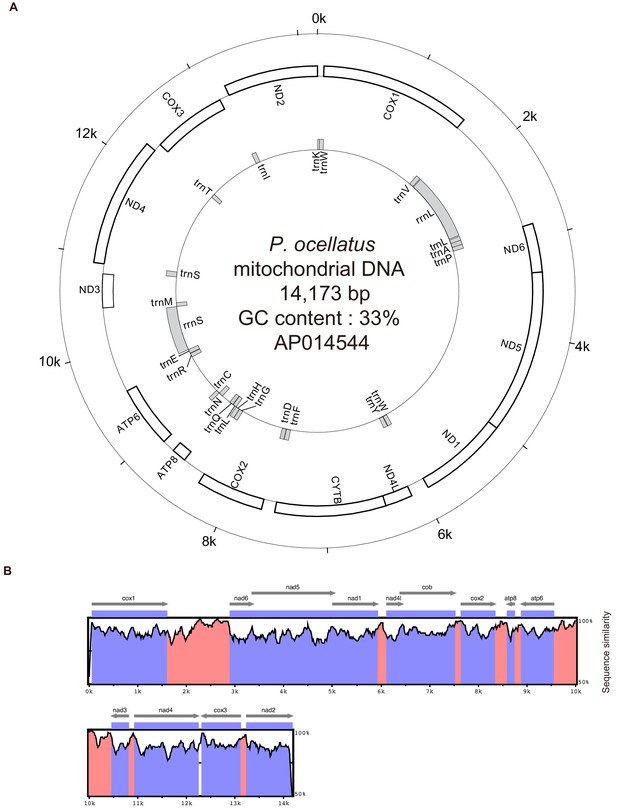
Mitochondrial gene map of the P. ocellatus type black.
(A) The outermost circle gives the scale in base pairs. Inner circles show the names and positions of protein-encoding genes (2nd circle) and structure RNA encoding genes (3rd). (B) Sequence similarity between our P. ocellatus type black mitochondrial DNA and previously sequenced ones from P. cf. ocellatus HW-2015 (KX853083). Genes on outside of each circle are transcribed clockwise and those on inside, anticlockwise. We used mVISTA software to calculate and visualize the similarity.
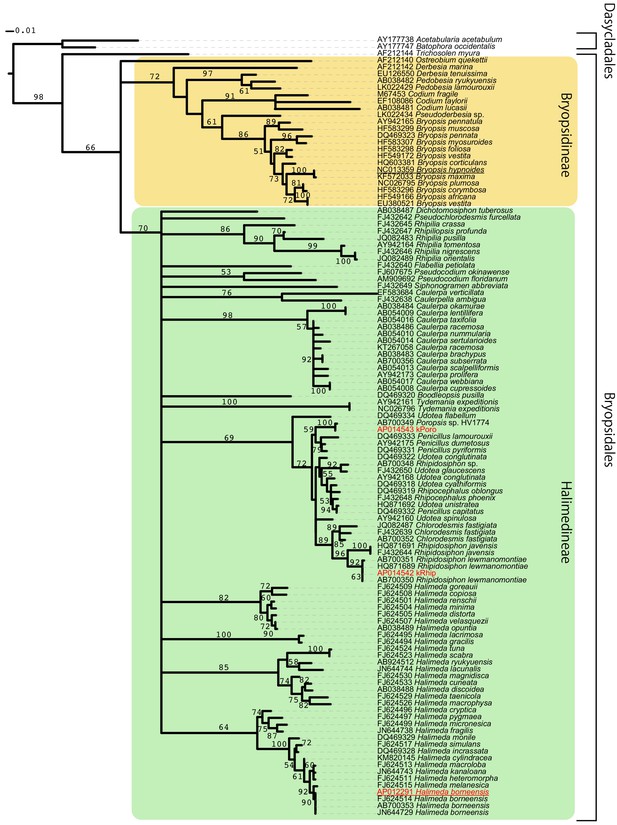
Bryopsidales ML tree made by RAxML (a -x 12345 p 12345 -# 300 m GTRGAMMAI) based on 457 positions of rbcL genes.
Bootstrap values are described at each node. We omitted the values less than 50. Scale bar represents the branch length. The color and underlines at the OTUs have the same meanings as in Figure 3B.
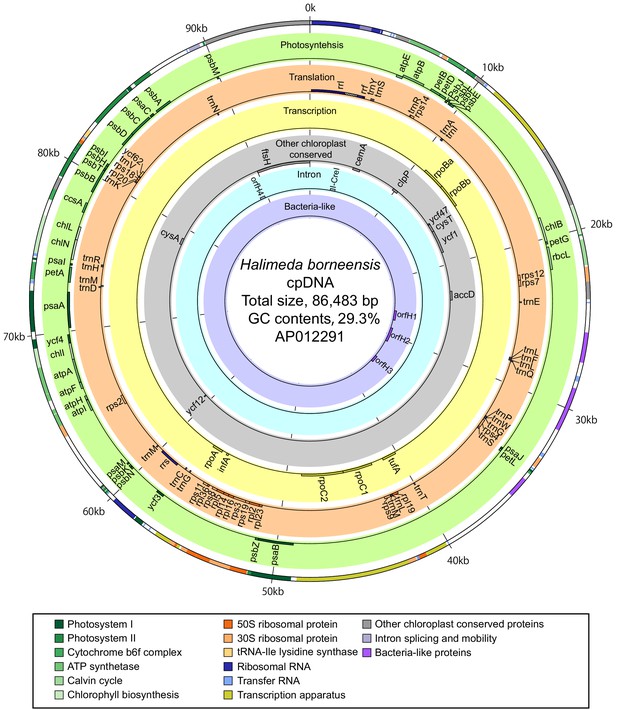
Gene map of the H. borneensis chloroplast DNA.
Circles and colors are the same as those in Figure 3—figure supplement 1.
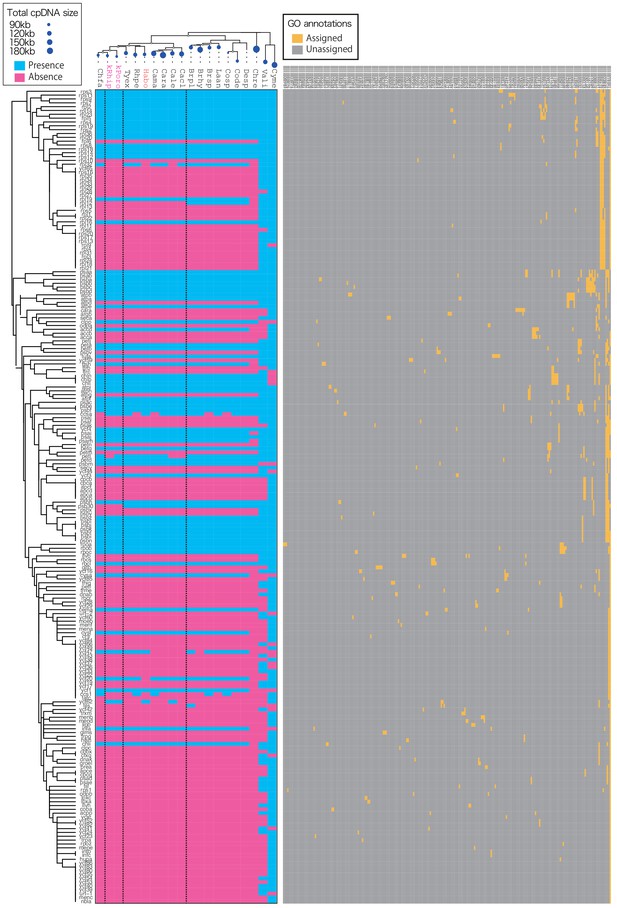
Gene compositions of 18 algal cpDNAs and P. ocellatus kpDNAs.
The presence (cyan) and absence (magenta) of the genes were visualized with the color tiles. The abscissa -axis shows the species, and the vertical axis shows the gene. kPoro, PoB kleptoplast derived from Poropsis sp.; kRhip, PoB kleptoplast derived from R. lewmanomontiae. Other abbreviations were explained in Figure 3—source data 1. Top tree shows the ML tree made by IQ-tree based on 51 conserved plastid genes (ycf4, ycf3, tufa, rps9, rps8, rps7, rps4, rps3, rps2, rps19, rps18, rps14, rps12, rps11, rpoa, rpl5, rpl36, rpl23, rpl20, rpl2, rpl16, rpl14, rbcL, psbZ, psbT, psbN, psbL, psbK, psbJ, psbI, psbH, psbF, psbE, psbD, psbC, psbB, psbA, psaJ, psaC, psaB, psaA, petG, petD, petB, petA, atpI, atpH, atpF, atpE, atpB, atpA, 9882 amino acid positions). Left side genes were clustered base on the GO annotation of each gene. GO annotations were described on the right-side boxes. Source files of gene compositions and annotations are available in Figure 3—source data 1.
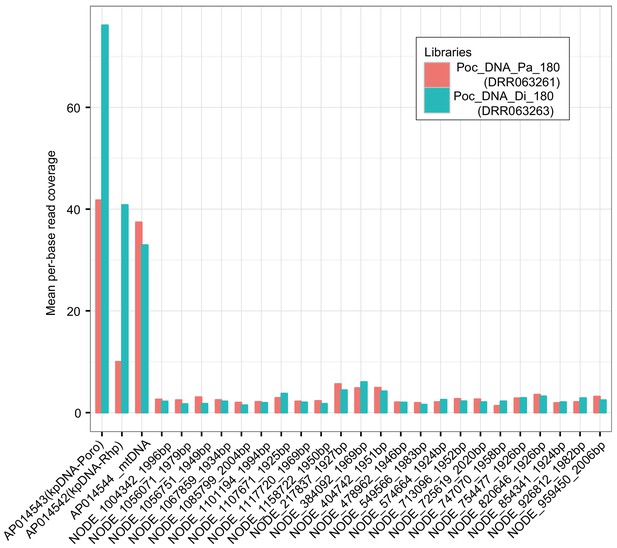
Read coverage depth of the constructed P. ocellatus genomic assemblies.
We mapped the two libraries to the constructed assemblies to determine the origin of each assembly. The ‘Poc_DNA_Pa_180’ is a kleptoplast-poor sample (Parapodium). The ‘Poc_DNA_Di_180’ is a kleptoplast-enriched sample (Digestive gland). Considerable change of depth on AP014543 and AP014542 reflects the different origin of the assemblies from the rest (mitochondria- or nuclear-derived assemblies).
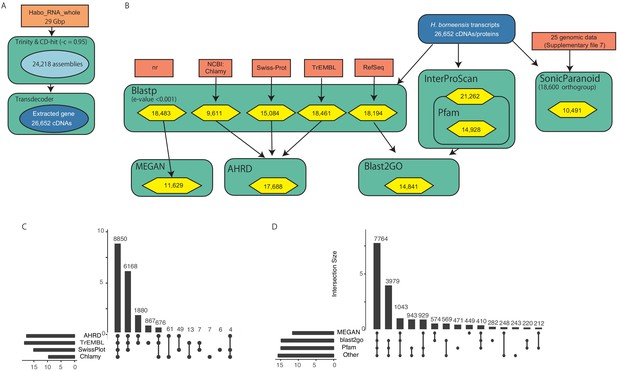
Annotation procedure of H. borneensis transcripts.
(A) De novo transcripts assembling process. (B) Annotation procedure of the constructed transcripts. Dark blue, submitted H. borneensis transcripts for DDBJ; light red, reference data; aquamarine, annotation method (for detail, see material and methods); yellow, annotated gene number for each step. (C, D) Upset plots of annotated gene number for each annotation step. The source file of the orthologous relationship of the kleptoplast/chloroplast-encoded genes is available in Figure 3—source data 1. We deposited detailed annotation data of H. borneensis transcripts in FigShare (DOI 10.6084/m9.figshare.13042016).
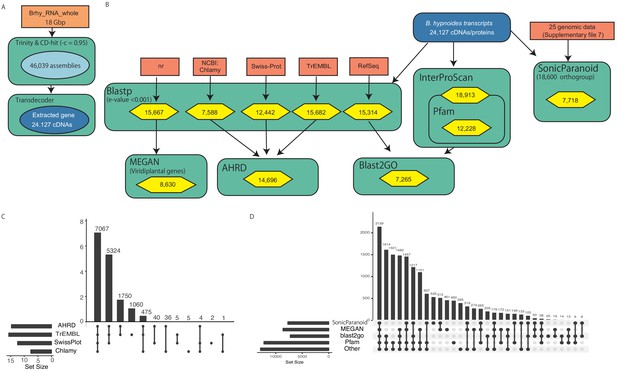
Annotation procedure of B. hypnoides transcripts.
(A) De novo transcripts assembling process. (B) Annotation procedure of the constructed transcripts. (C, D) Upset plots of annotated gene number for each annotation step. Colors and boxes are the same as those in Figure 3—figure supplement 8. Detailed annotation data were deposited in FigShare (DOI 10.6084/m9.figshare.13042016).
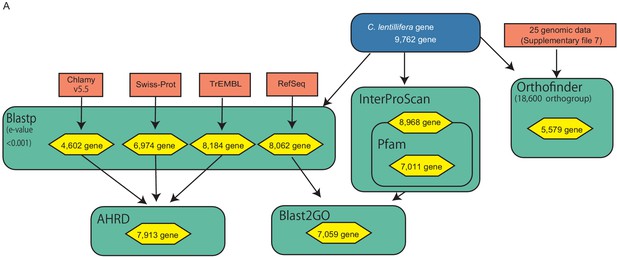
Annotation procedure of C. lentillifera.
Colors and boxes are the same as those in Figure 3—figure supplement 8. Detailed annotation data were deposited in FigShare (DOI 10.6084/m9.figshare.13042016).
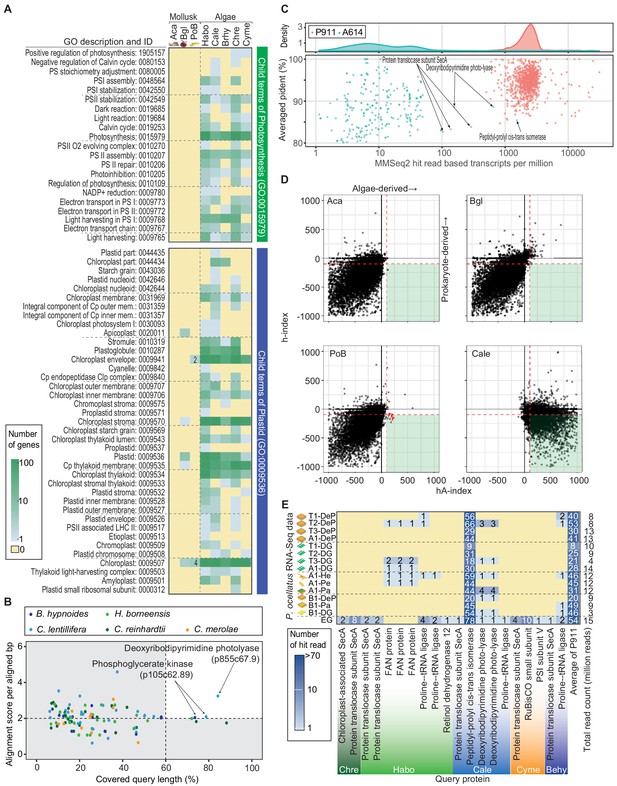
Search for horizontally transferred algal genes in the PoB genome.
(A) Heatmap of the number of genes assigned to photosynthesis- or plastid-related GO terms. The abundance of photosynthesis- or plastid-related genes was compared among PoB, two non-kleptoplastic mollusk species (Aca, A. californica; Bgl, B. glabrata), and five algal species (see abbreviations in Figure 3, Figure 3—source data 1). A color scale was visualized in the box on the left. (B) Scatter plot of the Exonerate results, the alignment of the A614 gene set (query sequences) to the PoB genome. The enlarged view with the tblastn results; Figure 4—figure supplement 5. The dot color shows the source algae of each query sequence (see keys in the box). The horizontal axis shows the percentage of the query length aligned to the hit sequences (PoB genome). The vertical axis shows the similarity of the aligned sequences between the query and PoB genome; alignment score (the sum of the substitution matrix scores and the gap penalties) divided by aligned length (bp). Dashed lines are thresholds for a credible query hit (i.e., a hit covering >60% of the query sequence and a normalized Exonerate alignment score of >2). (C) Scatter plot of MMseq2 results for the A614 dataset (algal photosynthetic genes, red) and P911 reference dataset (PoB single-copy genes, blue) used as query sequences against the database of pre-assembled read sets from paired-end DNA libraries of PoB. The top panel shows the probability density distribution of the number of hit reads (normalized with TPM: transcripts per kilobase million; x-axis) versus the averaged ‘pident’ value (percentage of identical matches) from the hit reads (y-axis). (D) Scatter plot of HGT indices (hA-index versus h-index) for genes in PoB, the two non-kleptoplastic mollusk species (Aca and Bgl), and one algae species (C. lentillifera [Cale]). Each dot represents a gene. A high hA-index or h-index value means the possibility of algal or prokaryote origin, respectively. Dashed red lines represent the conventional threshold for HGT (−100 for h-index and 100 for hA-index). Three red arrowheads indicate PoB genes exceeding the thresholds. (E) Heatmap of the results of searches for algae-like RNA fragments in the PoB RNA-Seq data. Pa, parapodium; EG, egg; He, head; Pe, pericardium. The blue gradient indicates the number of RNA-Seq reads assigned as algae-like fragments (see key). The y-axis labels show the RNA-Seq library name and analyzed tissue types. The x-axis labels indicate the query protein; the queries having no corresponding reads were omitted from the figure. For queries using the P911 reference dataset, the mean of the hit-read counts from each library was described. The total number of reads for each library is given on the far right. The source files of homology-based algal gene searching analysis using Exonerate, MMseq2, and blastp are available in raw data Figure 4—source data 1.
-
Figure 4—source data 1
Algal gene detection from PoB genome.
This zip archive contains source files of algal gene searching from PoB genome and pre-assembled reads.
- https://cdn.elifesciences.org/articles/60176/elife-60176-fig4-data1-v1.zip
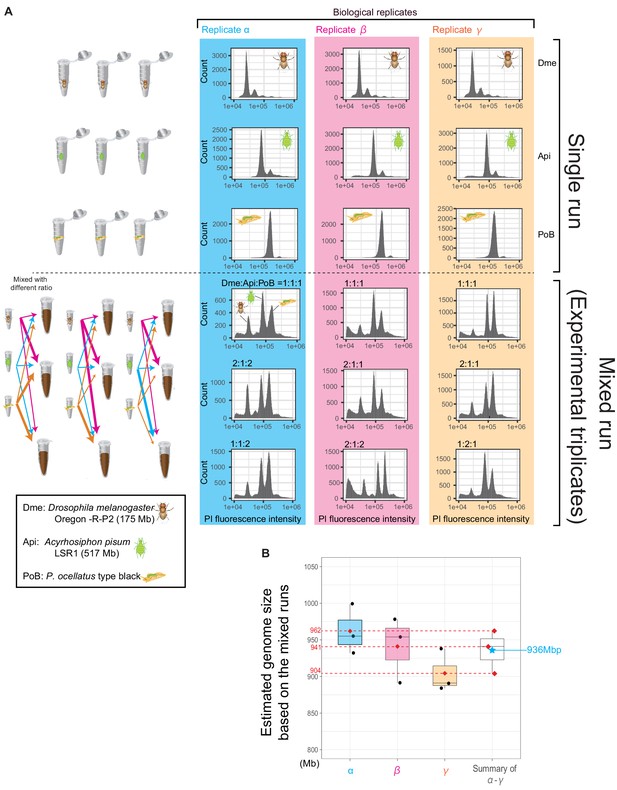
Genome size estimation of P. ocellatus type black by flow cytometry.
(A) Histograms showing the fluorescence intensities of the analyzed nuclei (x-axis) and count of the nuclei (y-axis). Studied species were represented with pictograms and abbreviations as in the left bottom box. Each nuclear suspension was analyzed for each species to confirm that the peak (particle) was free from contamination (Upper panels). On lower panels, we mixed the suspensions of three species to estimate the PoB genome size. The mixed ratio of the suspensions is shown on the upper-side of each histogram. We changed the mixing ratio to identify the respective peaks derived from the species. (B) Box-plots of genomic size of P. ocellatus estimated from the mixed flowcytometry with other genome size standards. The black dots are the values of PoB genome size estimated from triplicate experiments (runs). Red diamond, an averaged genome size estimated of the experimental triplicates (estimated genome size for each biological triplicate). A blue star means an averaged genome size of the biological triplicates. We made biological triplicates from different individuals of each species.
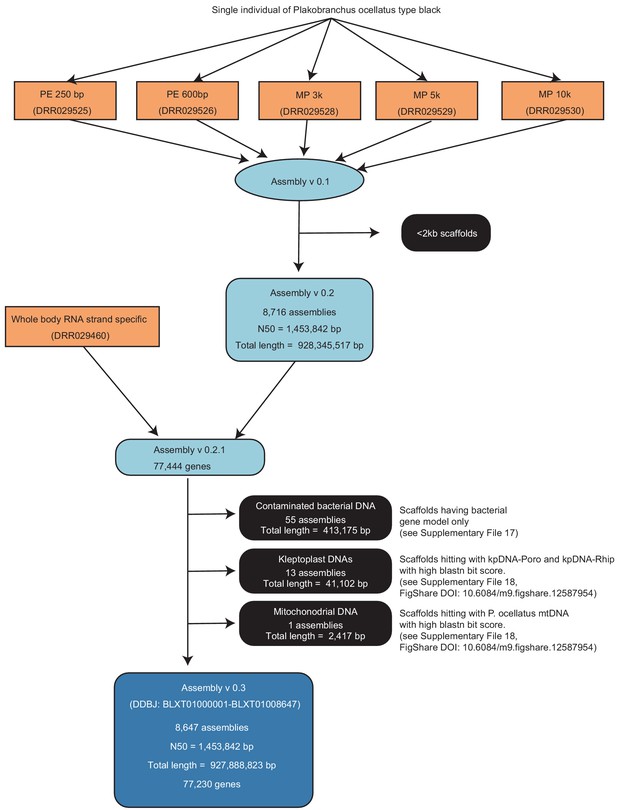
Genome assembling and gene modeling approach for the P. ocellatus genome analysis.
Light orange, raw read data; light blue, intermediate assembly data; dark blue, a final assembly for DDBJ submission; black, eliminated assemblies. Information on the raw read data are available in Supplementary file 1.
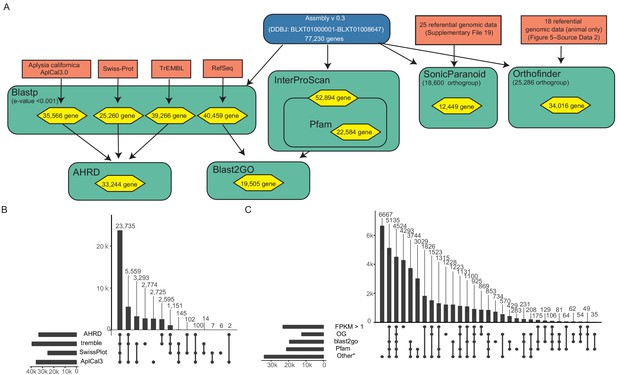
Annotation procedure for the genomic gene models of P. ocellatus type black.
(A) Overvew of the annotation procedure. Dark blue, P. ocellatus genome assembly for DDBJ submission; light red, reference data; aquamarine, annotation method (for detail, see Materials and methods); yellow, annotated P. ocellatus gene number. Other colors and boxes are the same as those in Figure 3—figure supplement 8. (B, C)Upset plots of annotated gene number for each annotation step.
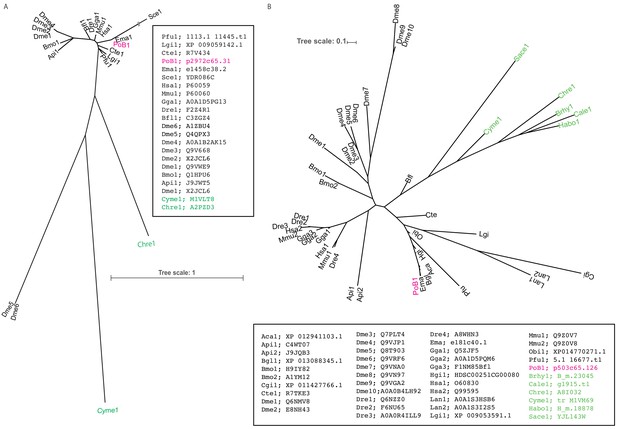
ML tree for the candidate HGT genes with algal and molluscan homologous genes.
The abbreviations represent the species from which the genes are derived (see Supplementary file 19, Figure 5—source data 1). The full gene ID (INSDC ID, OIST Marine Genomics Unit database ID, and our annotation ID) were noted in the boxes. Green, algal gene; magenta, P. ocellatus gene; black, other molluscan genes. Our GO analysis found six PoB genes assigned with the child terms of ‘Plastid’ (GO:0009536). In these PoB genes, p2972c65.3 and p503c65.126 (annotated as ‘Chloroplast envelope’ GO:0009941) made orthogroups ‘4058’ and ‘2017’ with several algal genes by the SonicParanoid analysis, respectively. Rest four genes (gene ID: p197c68.18, p234c64.89, p45387c41.1, and p466c59.83) showed no orthologous relationships with algal gene and were omitted from the phylogenetic analysis. (A) ML tree for the orthologous group ‘4058’. (B) ML tree for the orthologous group ‘2017’. The p2972c65.31 and p503c65.126 formed a monophyletic clade with molluscan orthologs rather than algal gene, indicating this gene is of molluscan origin (no HGT). Source files are available in Figure 4—source data 1.
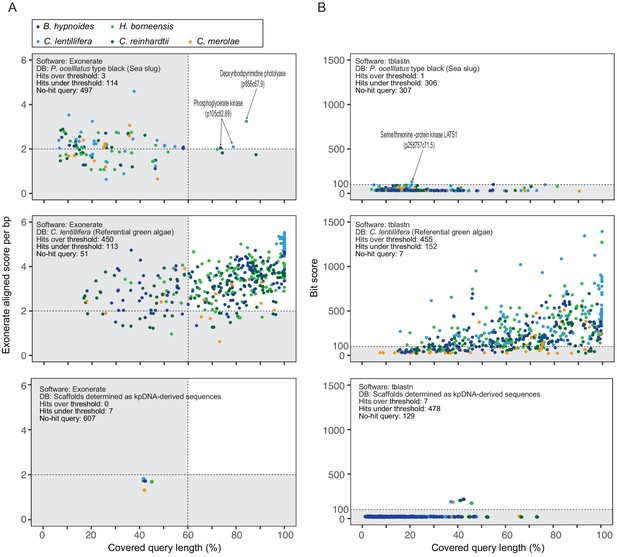
Summary of Exonerate and tblastn results.
Scatter plots of the analyzed results by Exonerate (A) and tblasn (B). Adopted databases were P. ocellatus genome (top plate), C. lentillifera genome (middle plate), or kpDNA-derived scaffolds (bottom plate). The query sequence was the algal A614 gene set. The top box shows the source algae of each query. Query hit statistics were described on the left shoulder of each plate. (A) Dashed lines are thresholds for the credible query hit (covering over 60% of the query sequence and over two normalized aligned scores for aligned length). All seven hit queries on kpDNAs were algal chlD gene (encoding magnesium-chelatase subunit ChlD) resembling the kleptoplast-encoded chlL gene. (B) Dashed lines are thresholds (bit score = 100) for the credible query hit. All seven significant hit queries on the kpDNA-derived scaffolds were algal chlD gene (encoding magnesium-chelatase subunit chlD) resembling the kleptoplast-encoded chlL gene (see Supplementary file 20 and Figure 4—source data 1).
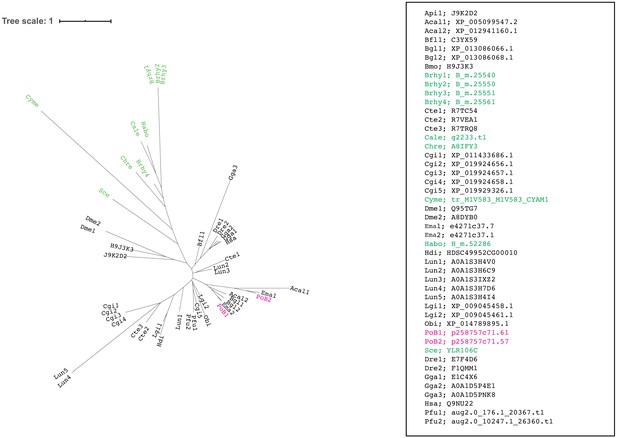
ML tree for the orthologous group ‘65’ by SonicParanoid.
Colors are the same as those in Figure 4—figure supplement 4. Source files are available in Figure 4—source data 1.
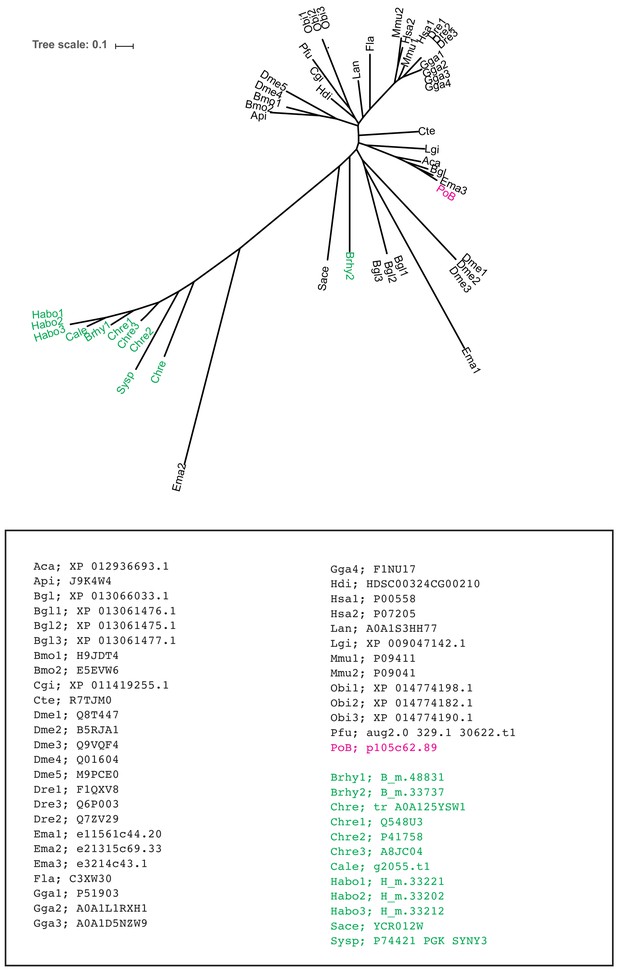
ML tree for the orthologous group ‘501’ by SonicParanoid.
Colors are the same as those in Figure 4—figure supplement 4. Source files are available in Figure 4—source data 1.
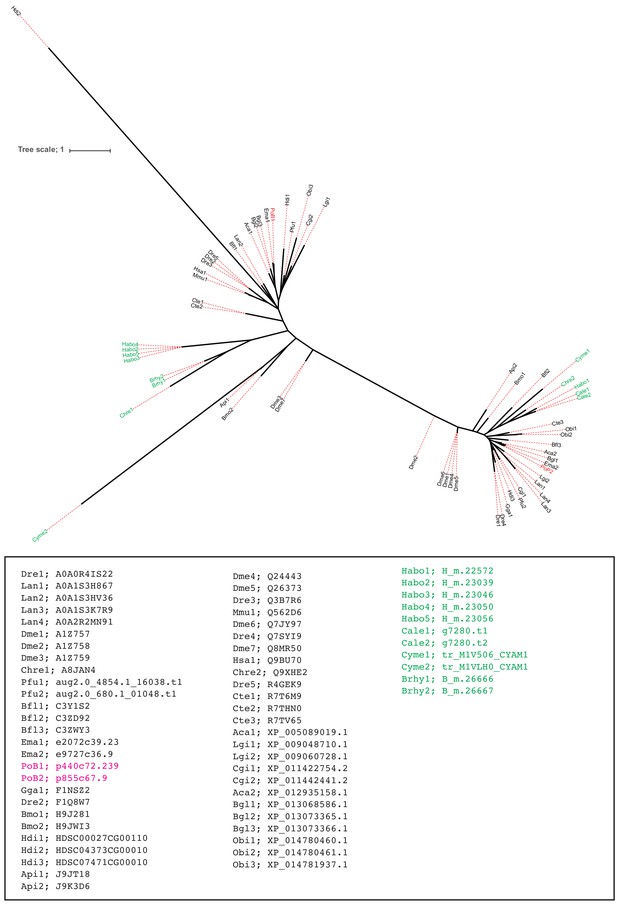
ML tree for the orthologous group ‘456’ by SonicParanoid.
Colors are the same as those in Figure 4—figure supplement 4. Source files are available in Figure 4—source data 1.
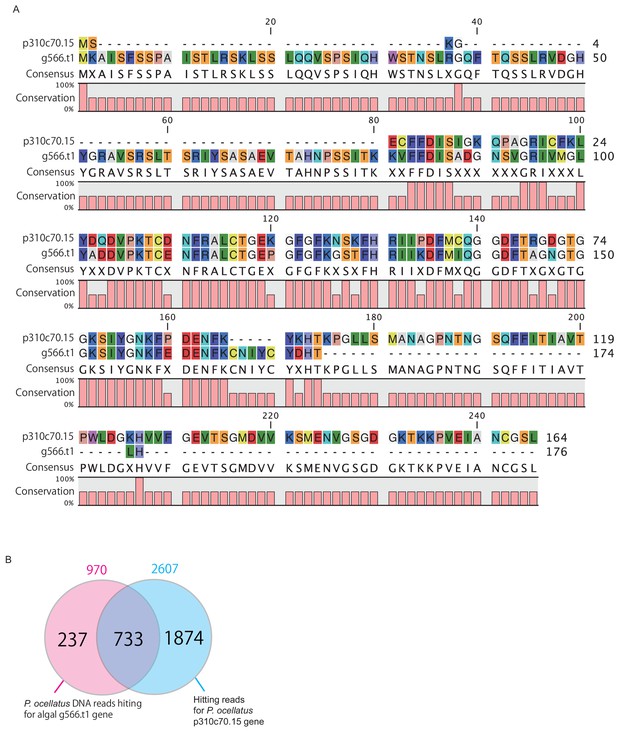
Sequence similarity between g566.t1 (C. lentillifera) and p310c70.15 (P. ocellatus).
(A) Sequence alignment and similarity. (B) Venn’s plot of number of hit reads by MMseq2 analysis. The adopted database is pre-assembling Illumina DNA sequencing read (DRR029525 and DRR029526).
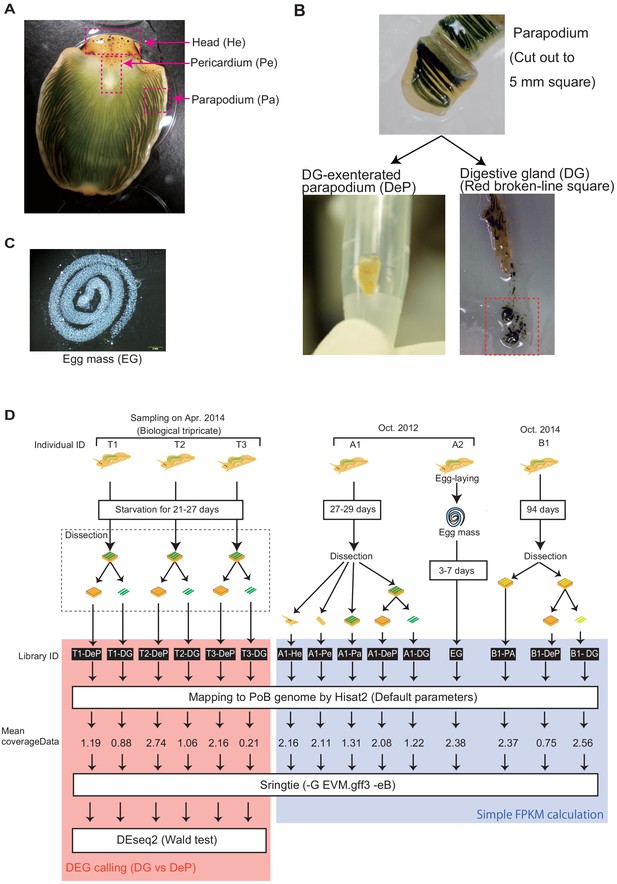
Overview of the sample preparation for P. ocellatus type black transcriptomic analysis.
(A–C) Photographs of the PoB dissection and the positions of sample tissues. (A) Dorsal views of a fresh PoB individual with spread (or inside out) parapodia. (B) The upper plate shows an enlarged view of the tissue dissected from the parapodium (-PA sample). The plate on the lower left shows the parapodium, excluding the digestive gland part, collected in a plastic tube (-DeP sample). The lower-right plate shows the digestive gland cut out from the parapodium using a razor (-DG sample). (C) Egg mass of PoB. (D) Incubation states of the dissected sea slugs and the relationships among the samples, libraries, and RNA-seq analysis. ‘Mean coverageData’ means the averaged depth of coverage of each library to the PoB genome. The libraries from three individuals (T1–3) were used for DEG calling using the Wald test.
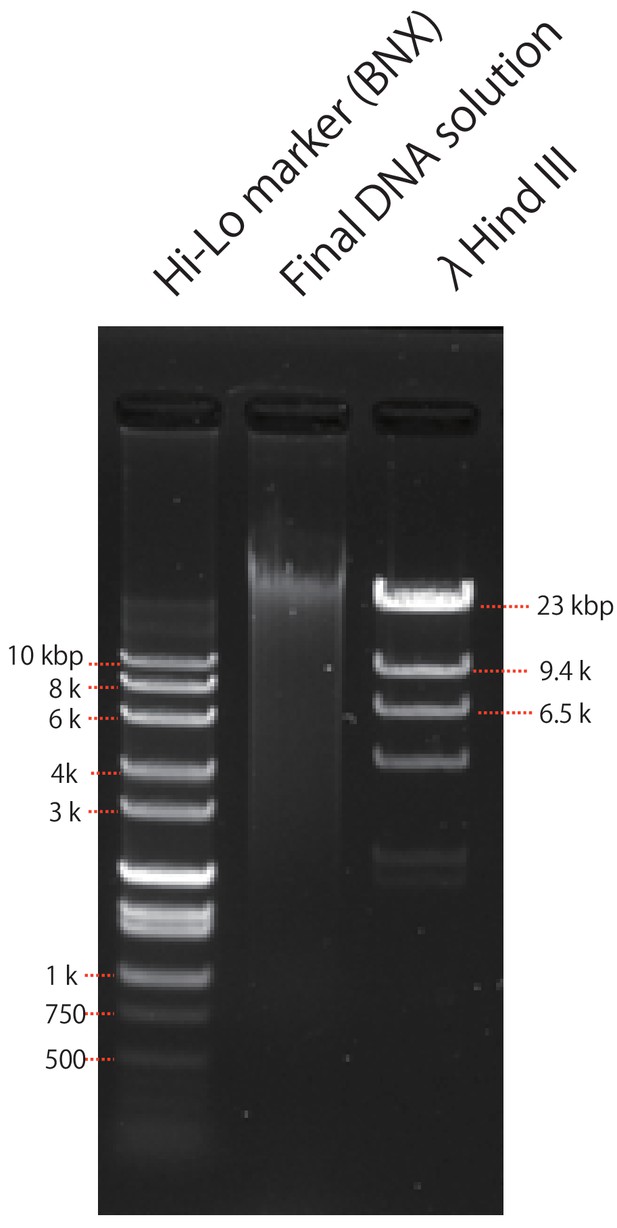
Agarose gel electrophoresis image displaying extracted P. ocellatus DNA for the nuclear genome sequencing with two fragment size markers.
The extracted total DNA from P. ocellatus was mixed with gel loading buffer, electrophoresed at 50V in 0.7% (w/v) agarose gel in TAE buffer, and stained by SYBR safe (Thermo).
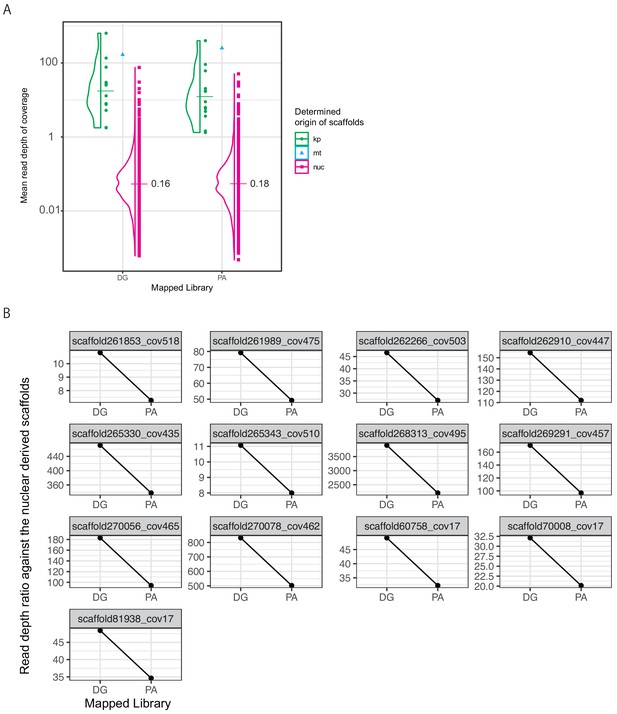
Read depth of the P. ocellatus genome assemblies.
To determine the scaffolds from kleptoplasts and mitochondria, we mapped the Illumina libraries of P. ocellatus (DRR063261 and DRR063263, Supplementary file 1) to the genomic reference data and compared the coverage depth. We used two types of P. ocellatus DNA (DG = digestive gland, PA = parapodium) to construct the Illumina libraries. The genomic reference data contained 13 kleptoplastic (kp), one mitochondrial (mt), and 8647 nuclear (nuc) scaffolds. (A) Violin and scatter plots of the mapped read depths. Each dot indicates a scaffold. A horizontal bar indicates the averaged depth values of coverage. The right boxes show the legend of coloration. (B) Line graphs of the relative values of the read depths of the kleptoplastic scaffolds against the averaged depth of all nuclear scaffolds in libraries. Kleptoplastic scaffold IDs were described on the tops of respective graphs.
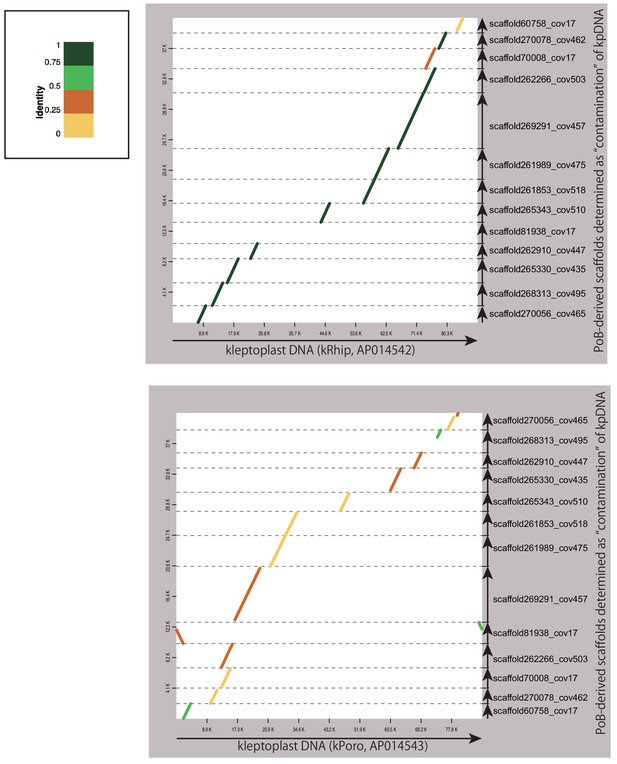
Dot-plot of the referential kleptoplast DNAs and PoB genomic scaffolds determined as the kleptoplast DNA (kpDNA).
Abscissa-axis, referential kleptoplast DNAs (kPoro and kRhip); horizontal axis, 13 scaffolds selected as kpDNA sequences. The diagonals indicate contiguously aligned regions. The coloration means the identity of the aligned regions. The top left box indicates the legend of similarity coloration. Arrows indicate the aligned sequences and direction. We used D-GENIES software to make the dot-plots. Both referential kpDNAs indicated high similarity with the 11 scaffolds, supporting kpDNA-origin of the 11 of 13 scaffolds. For the rest two scaffolds, ‘scaffold81938_cov17’ and ‘scaffold268313_cov495’ indicated a similarity to kPoro and kRhip, respectively. This discrepancy may be due to the sequence diversity of plastidial DNA among the species (donor algae). Overall, these results support that the sequence of 13 scaffolds are not derived from anything other than plastidial (kleptoplast) DNA.
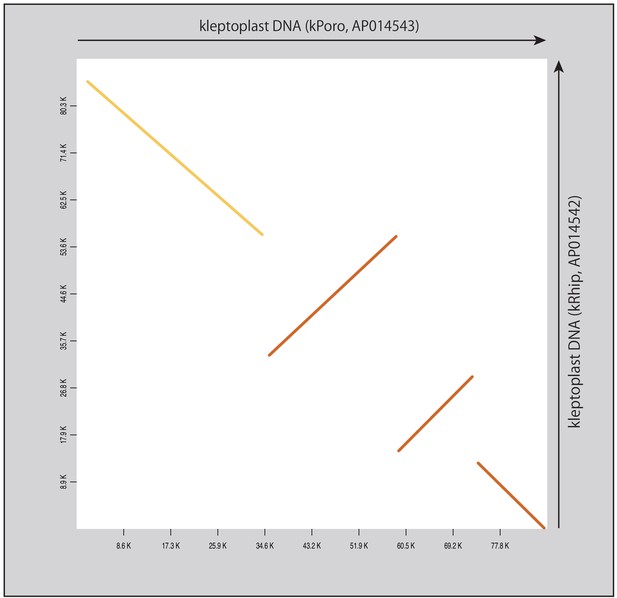
Dot plot of the two referential kleptoplast DNAs.
Arrows and colors are the same as those in Figure 4—figure supplement 13.
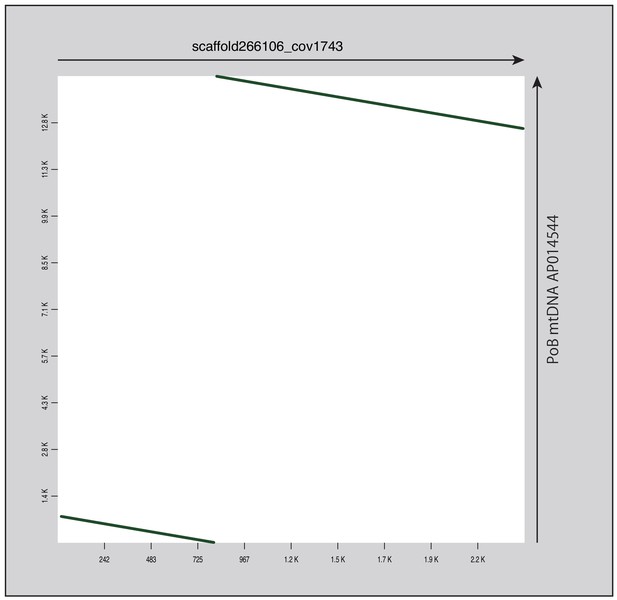
Dot plot of the referential mtDNAs and one scaffold determined as mtDNA sequence.
Arrows and colors are the same as those in Figure 4—figure supplement 13.
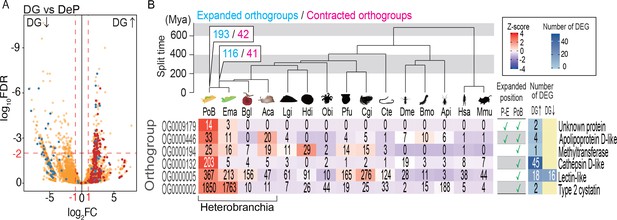
Probable KRM genes in PoB.
(A) Volcano plot comparing gene expressions in DG and DeP tissues of PoB. Red, orthogroup OG0000132 (cathepsin D-like protease genes); blue, orthogroup OG0000005 (lectin-like genes); orange, other orthogroups (for details, see Figure 5—figure supplement 2). Up and down arrows signify up- and down-regulation, respectively, in DG. (B) Orthogroups expanded on the P. ocellatus lineage and containing DG-up-regulated genes. Ema, E. marginata; Bgl, B. glabrata; Lgi, Lottia gigantea; Hdi, Haliotis discus; Obi, Octopus bimaculoides; Pfu, Pinctada fucata; Cgi, Crassostrea gigas; Cte, Capitella teleta; Dme, D. melanogaster; Bmo, Bombyx mori; Api, A. pisum; Hsa, Homo sapiens; Mmu. Mus musculus. The phylogenetic tree was scaled to divergence time based on 30 conserved single-copy genes. Mya, million years. The numbers of rapidly expanded (blue) and contracted (magenta) orthogroups on the lineages to PoB are provided at the nodes (for details, see Figure 5—figure supplements 4 and 5). Below the tree is information for the six expanded orthogroups that contained DG-up-regulated genes. The left-side heatmap shows the gene numbers (number in boxes) and Z-score of gene numbers (color gradient) for each orthogroup. The table shows the expanded/not expanded status of each orthogroup (P–E, Plakobranchus-Elysia node; PoB, Plakobranchus node). The right-side heat map indicates the number of DEGs between DG and DeP tissue in each orthogroup. Representative gene products are given on the far right. The source files of RNA-Seq analysis and comparative genomic analysis are available in raw data Figure 5—source data 1 and 2, respectively.
-
Figure 5—source data 1
KRM gene searching using RNA-Seq.
This zip archive contains source files of kleptoplasty relating molluscan (KRM) genes searching based on RNA-Seq analysis.
- https://cdn.elifesciences.org/articles/60176/elife-60176-fig5-data1-v1.zip
-
Figure 5—source data 2
KRM gene searching using ortholog analysis.
This zip archive contains source files of KRM gene searching based on the comparative genomic (orthologous) analysis.
- https://cdn.elifesciences.org/articles/60176/elife-60176-fig5-data2-v1.zip
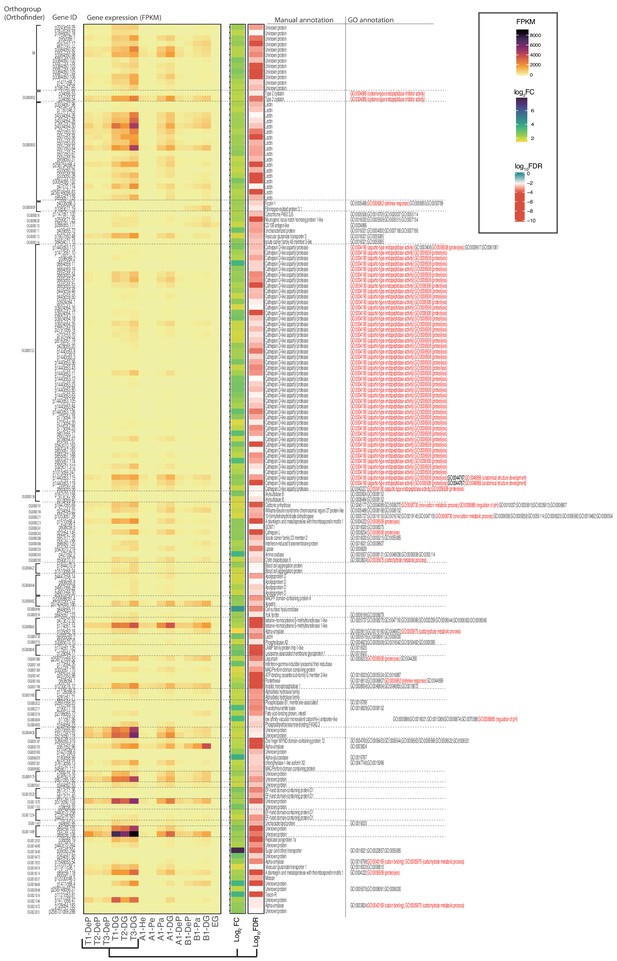
DG-up-regulated genes by the RNA-seq comparison with -DeP samples.
Differentially expressed gene calling among the libraries from DG (digestive gland) and DeG (DG-exenterated parapodium) found 162 significantly DG-up-regulated genes. The expression levels and annotation information were summarized in Figure 5—source data 1. Fold changes (FC) and false discovery rate (FDR) were calculated between the DG and DeP samples and visualized after logarithmic transformation.
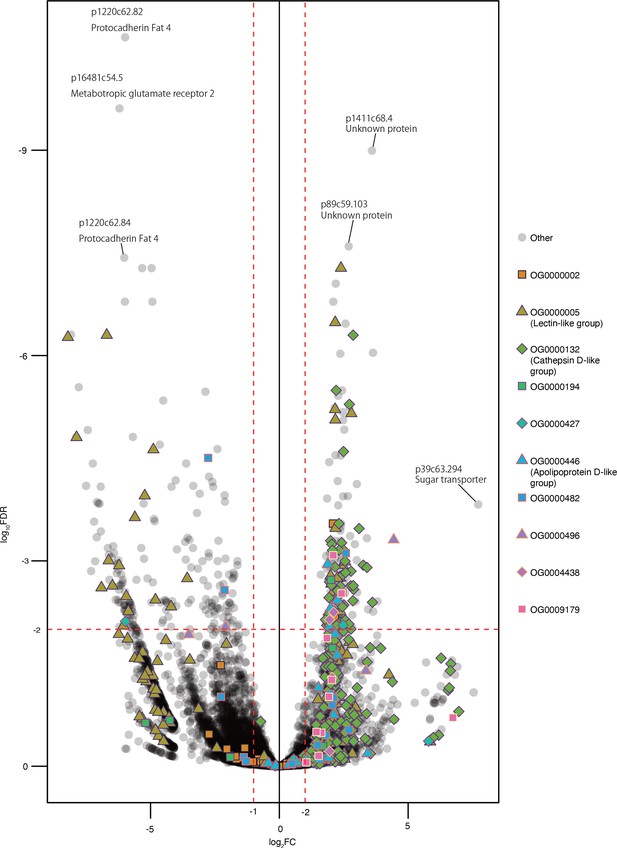
Volcano plot of the cross-tissue comparison between DG and DeP samples.
Each dot represents a gene. Colors and dot shapes mean the belonging orthogroups of genes. Source files are available in Figure 5—source data 1.
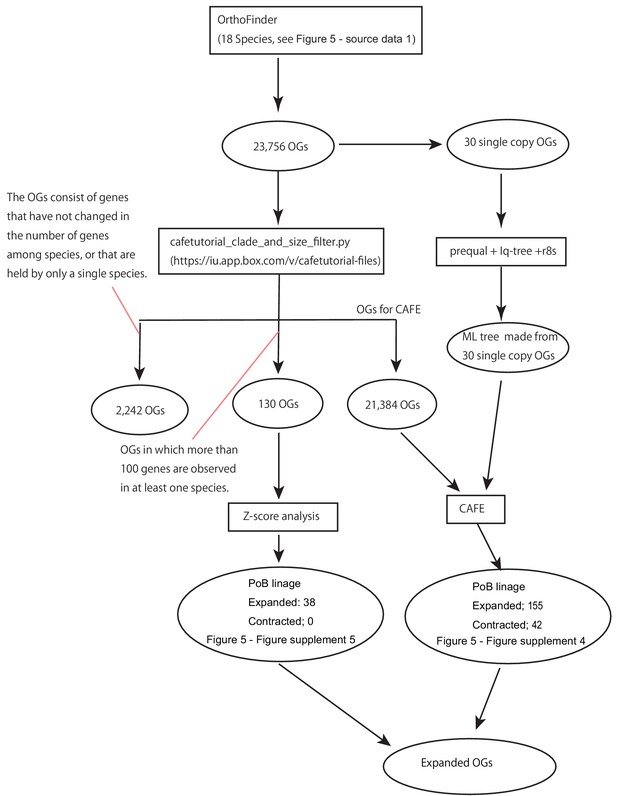
The procedure of gene family history analyses.
The square box and the circle mean the adopted analysis and the obtained orthogroup (OG) datasets, respectively.
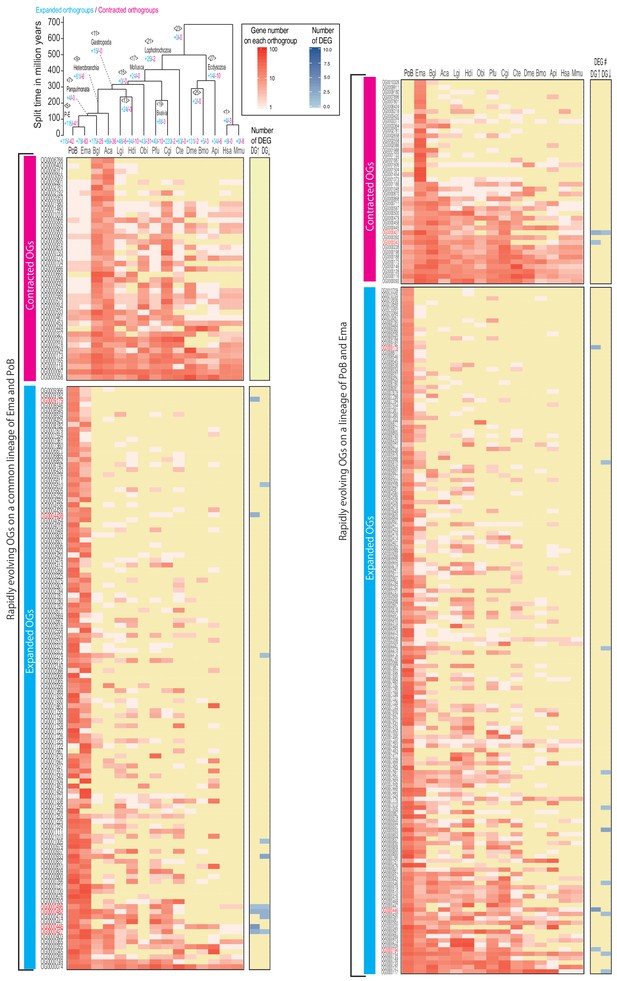
CAFE-based all rapidly expanding/contracting genes on the sacoglossan linage.
The gene count of all rapidly expanding/contracting orthogroups on P. ocellatus (left side) and the common node between P. ocellatus and E. marginata (right side) were plotted with a heat map. The left top tree is an ultrametric tree made by r8s based on 30 genes. The red color means the number of genes in each species. The blue color indicates the number of differentially expressed genes between DG and DeP tissue. The up and down arrows mean up- and down-regulation on the DG sample, respectively. Source files are available in Figure 5—source data 1 and 2.
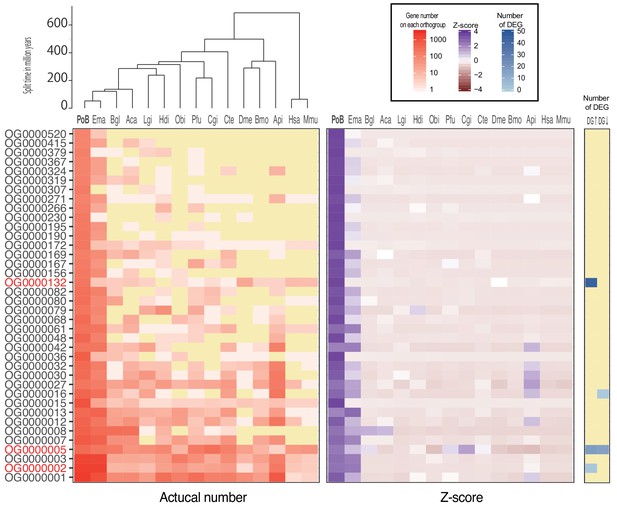
Z-score-based rapidly expanding/contracting genes on the P. ocellatus linage (Threshold: Z-score > 2).
Color legends were described in the top right box.
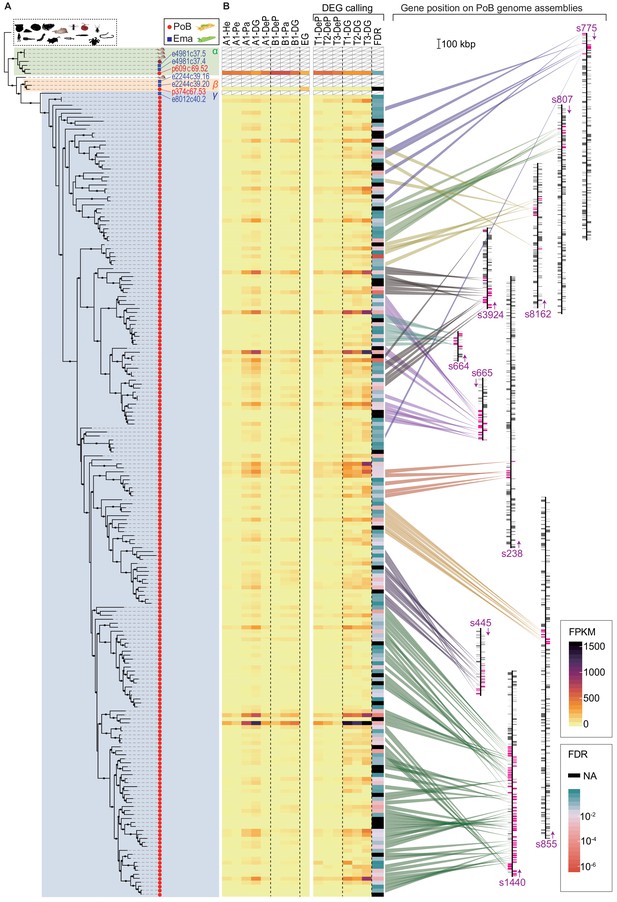
Evolutional process of KRM gene candidates of OG0000132.
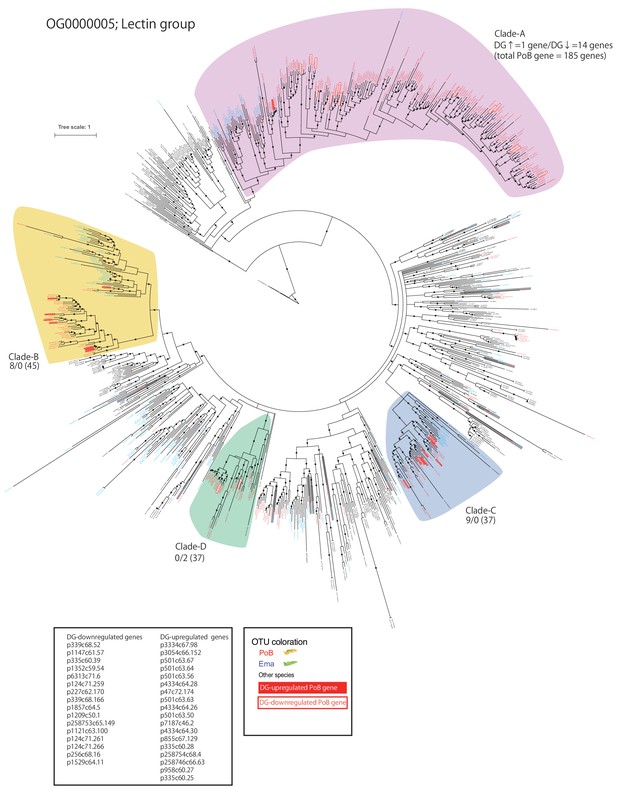
(A) ML phylogenetic tree for OG0000132 genes (for details, see Figure 6—figure supplement 1). Red circle, PoB gene; blue square, E. marginata (Ema) gene. Genes mentioned in the text are indicated with their IDs beside the circles. Pictograms represent non-sacoglossan species defined in Figure 5—source data 2. (B) Expression of each PoB gene and gene position on the genomic assemblies. The left-side heat plot indicates normalized expression degree (FPKM) of the PoB genes in various tissues; tissue abbreviations are defined in Figure 4, Figure 4—figure supplement 10. The vertical gene order corresponds with the position in (A). The white box with a diagonal line means a non-PoB gene (no expression data is available). Tissue samples for the FDR calculation (DG versus DeP) were enclosed with square brackets. Color scales for FPKM and FDR are visualized on the right-side boxes. The FDR’s black color indicates that the value could not be calculated (NA) due to its undetectable gene expression. The right panel shows the genomic positions of the genes. The bands’ color indicates the correlation between the gene and position, purple text and arrows indicate the scaffold ID and direction, and colored boxes on the scaffolds mean the genes’ positions (magenta, OG0000132 gene; gray box, other protein-encoding genes). Scaffolds having less than five OG0000132 genes were omitted from the figure.
-
Figure 6—source data 1
Outputs of a prediction of protein subcellular localization via DeepLoc.
- https://cdn.elifesciences.org/articles/60176/elife-60176-fig6-data1-v1.zip
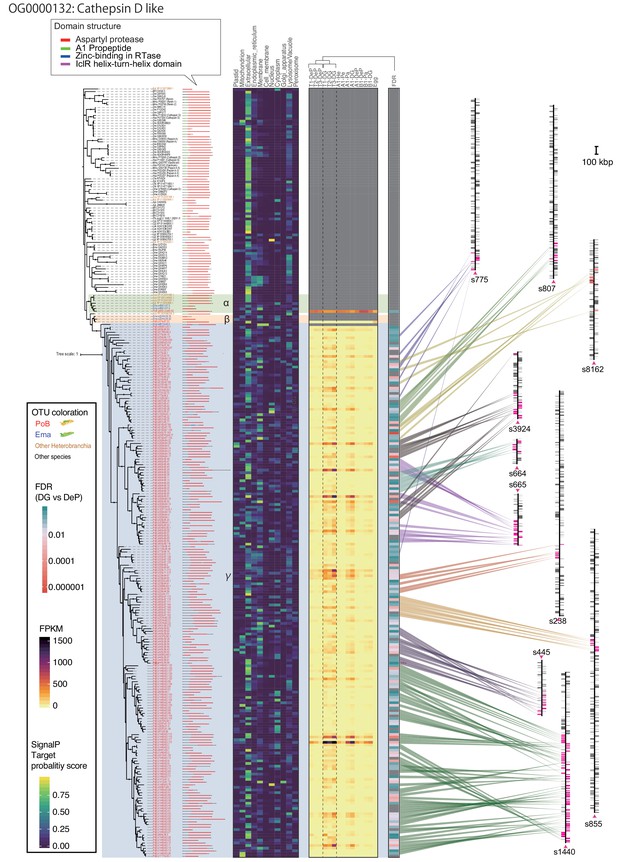
Enlarged view of gene tree and annotation data for all member of OG0000132 (cathepsin D-like).
From left, gene tree, domain structure, predicted intracellular location with DeepLoc-1.0, expression profiles in cross-tissue RNA-Seq, and the gene position on P. ocellatus genome. Color legends are shown in the left box.
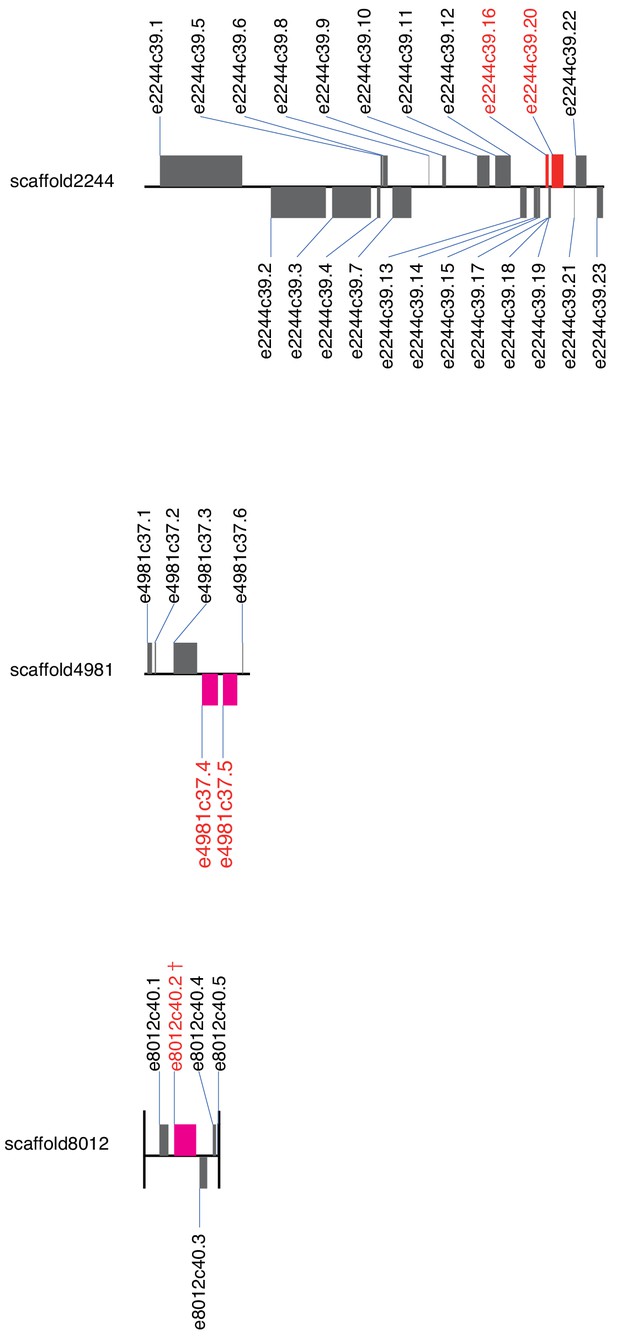
Positions of genes belonging to OG0000132 on the E. marginata genome.
Genes on the upperside are transcribed from left to right; genes on the bottom side are transcribed from right to left. Magenta, OG0000132 genes; gray, other genes. †Clade-γ gene.
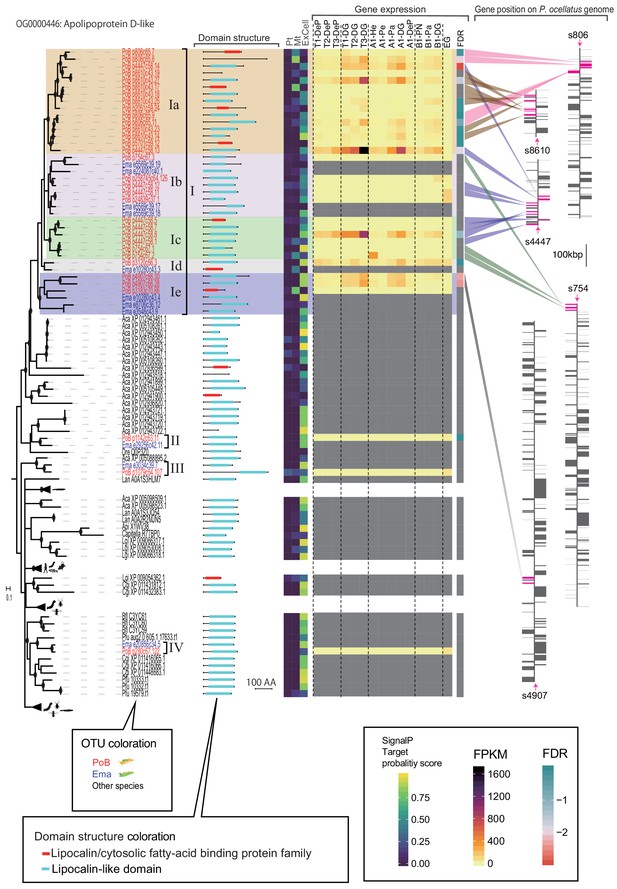
Gene tree and annotation data for all genes of OG0000446 (apolipoprotein D-like).
From left, gene tree, domain structure, predicted intracellular location with DeepLoc-1.0, expression profiles in cross-tissue RNA-Seq, and the gene position on P. ocellatus genome. Color legends are shown in the bottom box.
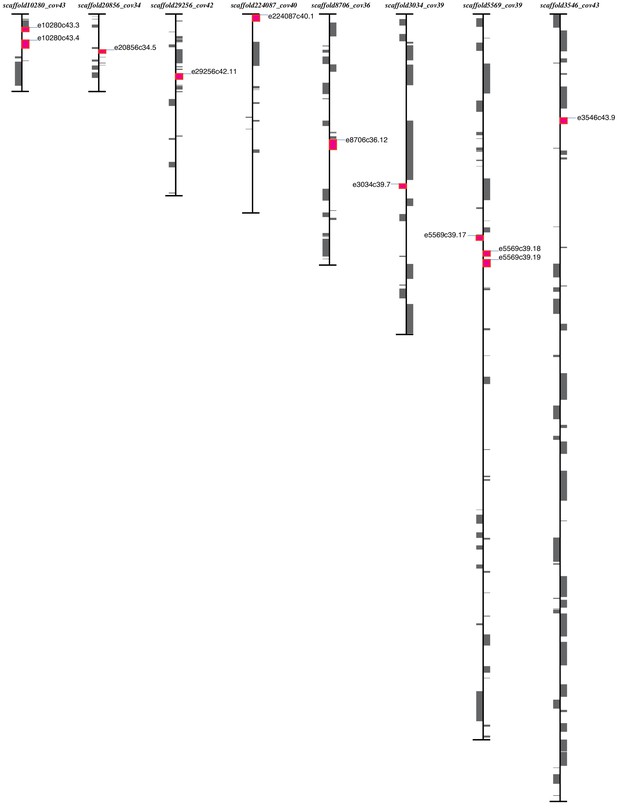
Gene positions of OG0000446 on E. marginata genome.
Genes on the right side are transcribed from top to bottom; genes on the leftside are transcribed from bottom to top. Magenta, OG0000446 genes; gray, other genes.
Additional files
-
Supplementary file 1
Summary of the raw sequencing data.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp1-v1.xlsx
-
Supplementary file 2
Assembly statistics of de novo RNA-seq of donor algae.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp2-v1.xlsx
-
Supplementary file 3
Algal genes used for referential query (A614 dataset).
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp3-v1.xlsx
-
Supplementary file 4
Assembly statistics of Plakobranchus ocellatus and Elysia marginata genome.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp4-v1.xlsx
-
Supplementary file 5
PoB genes having algal top-hit results of blastp.
This zip archive contains a file for statistics of PoB genes having algal top-hit results of blastp analysis and a file for detailed annotation of the algal top-hit PoB genes.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp5-v1.xlsx
-
Supplementary file 6
Detailed result of MEGAN analysis.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp6-v1.xlsx
-
Supplementary file 7
Annotation data of the six PoB genes assigned with the child terms of ‘Plastid(GO:0009536)’.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp7-v1.xlsx
-
Supplementary file 8
Applied species for HGT index analysis.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp8-v1.xlsx
-
Supplementary file 9
Gene annotation of the DG-up-regulated PoB genes.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp9-v1.xlsx
-
Supplementary file 10
Enriched GOs of the significantly up-regulated genes on the digestive gland of PoB.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp10-v1.docx
-
Supplementary file 11
Cross-tabulation table for the significant enrichment analysis of DG-upregulated gene number on the PoB-expanded orthogroup orthologous group.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp11-v1.xlsx
-
Supplementary file 12
Summary of the results of dN/dS-based selection test.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp12-v1.zip
-
Supplementary file 13
E. marginata scaffolds determined as bacterial contaminants.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp13-v1.xlsx
-
Supplementary file 14
E. marginata scaffolds selected as kleptoplast/mitochondrial DNA.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp14-v1.xlsx
-
Supplementary file 15
OrthoFinder result with additional data from E. chlorotica gene set (gene counts).
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp15-v1.docx
-
Supplementary file 16
Composition information of DNA extraction solutions for P. ocellatus genomic DN.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp16-v1.xlsx
-
Supplementary file 17
Scaffolds determined as bacterial contaminants during the PoB genome assembling and the result of MEGAN analysis before the removing of the bacterial scaffolds.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp17-v1.zip
-
Supplementary file 18
Scaffolds determined as kleptoplast/mitochondrion sequences.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp18-v1.xlsx
-
Supplementary file 19
Analyzed protein sequences by SonicParanoid analysis.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp19-v1.xlsx
-
Supplementary file 20
Result of the Exonerate search.
Database, the 13 PoB-derived scaffolds determined as kpDNA; query, algal photosynthetic genes (A614).
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp20-v1.xlsx
-
Supplementary file 21
Sequence similarity among the genes belonging OG0000005, OG0000132, and OG0000446.
- https://cdn.elifesciences.org/articles/60176/elife-60176-supp21-v1.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60176/elife-60176-transrepform-v1.docx