Induction of osteogenesis by bone-targeted Notch activation
Figures
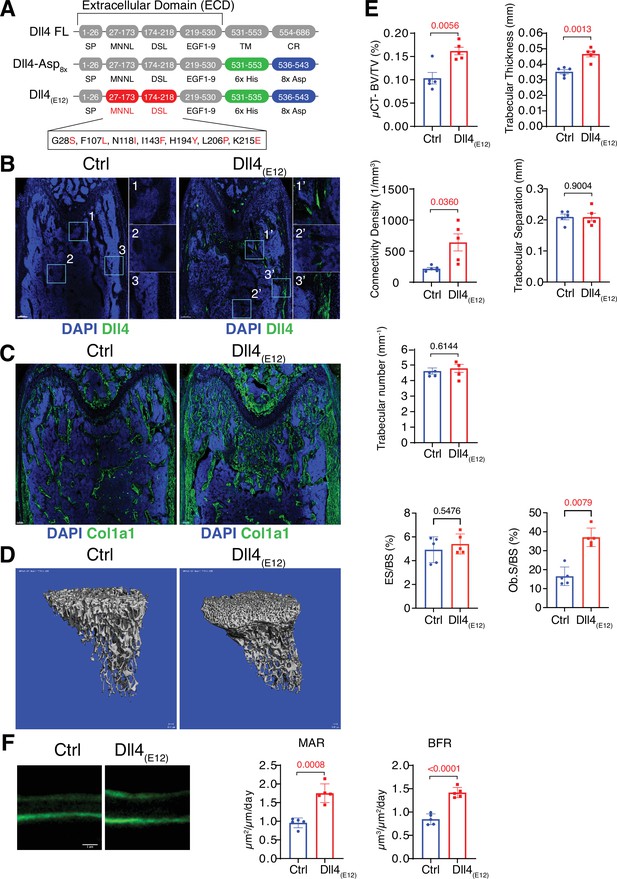
Recombinant Dll4(E12) increases bone formation in vivo.
(A) Schematic diagram showing the domain organization of murine Dll4 full-length protein and recombinant Dll4-Asp8x and Dll4(E12) fusion proteins. The latter contain 6x His epitope tags (green boxes) and negatively charged peptides consisting of eight Asp residues (8x Asp; blue boxes) instead of the transmembrane (TM) domain and cytoplasmic region (CR) of Dll4. Missense mutations were introduced in the MNNL (modulus at the N-terminus of Notch ligands) and DSL (Delta-Serrate-Lin) domains to generate Dll4(E12) with increased Notch-binding affinity. The resulting amino acid replacements are highlighted in red. SP, signal peptide. (B) Representative overview and high-magnification confocal images of Dll4 staining (green) on the femurs of pLIVE-Dll4(E12) and control-injected mice at the age of 11 weeks. Nuclei, DAPI (blue). Images on the right show higher magnifications of insets in metaphysis (1), bone marrow (2), and cortical bone (3). (C) Tile scan confocal images showing Collagen I alpha one chain (Col1a1) staining (green) in sections from pLIVE-Dll4(E12) and control femur. Nuclei, DAPI (blue). (D) Representative 3D reconstruction of micro-computed tomography (µ-CT) measurements for tibial metaphysis of 11-week-old pLIVE-Dll4(E12)- and control-injected mice. (E) Bone parameters measured by µ-CT analyses: bone volume/total volume (BV/TV) in percentage, trabecular thickness in millimeters, connectivity density in one per cubic millimeter, trabeculae number in one per millimeter, trabecular separation in millimeters, and trabecular number in one per millimeter. Data represent mean ± s.e.m. (n = 5 mice, except for trabecular number with n = 4 mice) (p-values determined by unpaired t-test with Welch’s correction). Graphs at the bottom represent quantitation of eroded surface over bone surface (ES/BS) in percentage and osteoblast surface overs total bone surface (Ob.S/BS) in percentage, calculated from histological HE-stained bone sections. Data represent mean ± s.e.m. (n = 5 mice) (p-values determined by Mann-Whitney U test). (F) Representative images of calcein double labeling (7-day time interval) in mineralized sections of the distal femur confirm increased bone formation after pLIVE-Dll4(E12) injection. Quantification of mineral apposition rate (MAR) and bone formation rate (BFR) (right panels). Data represent mean ± s.e.m. (n = 5 mice) (p-values determined by unpaired t-test with Welch’s correction).
-
Figure 1—source data 1
Source data for Figure 1E and F.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig1-data1-v2.xlsx
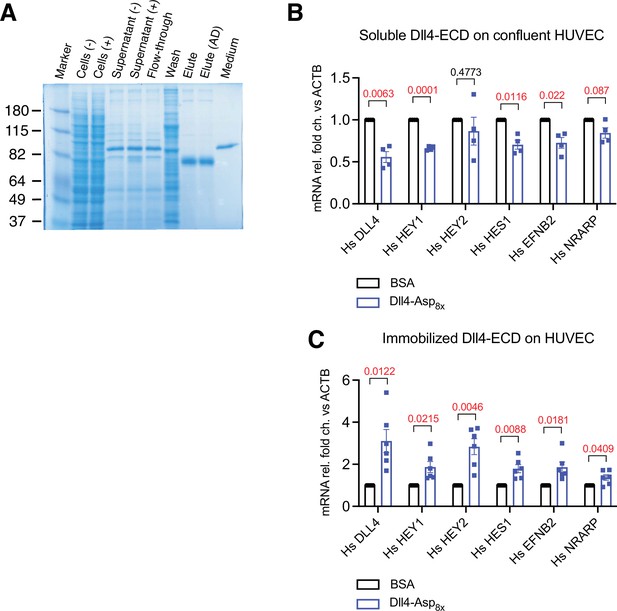
Generation of Dll4-Asp8x recombinant proteins.
(A) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of purified Dll4-Asp8x recombinant proteins. From left to right, protein molecular mass marker (Marker), extract of untransfected HEK293 cells (Cells (-)) OR pcDNA3.1-Dll4-Asp8x-transfected cells (Cells (+)), supernatant after culture of untransfected (Supernatant (-)) OR pcDNA3.1-Dll4-Asp8-transfected cells (Supernatant (+)), flow-through after loading of Supernatant (+) on His-tag protein purification column (Flow-through), wash buffer (Wash), elute (Elute), elute after dialysis (Elute (AD)), and Opti-MEM culture medium (Medium) were loaded. (B, C) RT-qPCR analysis of DLL4, HEY1, HEY2, HES1, EFNB2, and NRARP transcripts in confluent human umbilical vascular endothelial cells (HUVECs) stimulated with soluble (B) or in sub-confluent HUVECs stimulated with immobilized Dll4-Asp8x or BSA with poly-L-lysine coating (C). Data represent mean ± s.e.m. (n = 4 for (B) and n = 6 for (C)) (p-values determined by multiple unpaired t-test with Welch’s correction).
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1B,C.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig1-figsupp1-data1-v2.xlsx
-
Figure 1—figure supplement 1—source data 2
Images of uncropped blots and gels with the relevant bands labeled for Figure 1—figure supplement 1, Figure 1—figure supplement 4, and Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig1-figsupp1-data2-v2.pdf
-
Figure 1—figure supplement 1—source data 3
ZIP file containing the full raw unedited blots and gels for Figure 1—figure supplement 1, Figure 1—figure supplement 4, and Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig1-figsupp1-data3-v2.zip
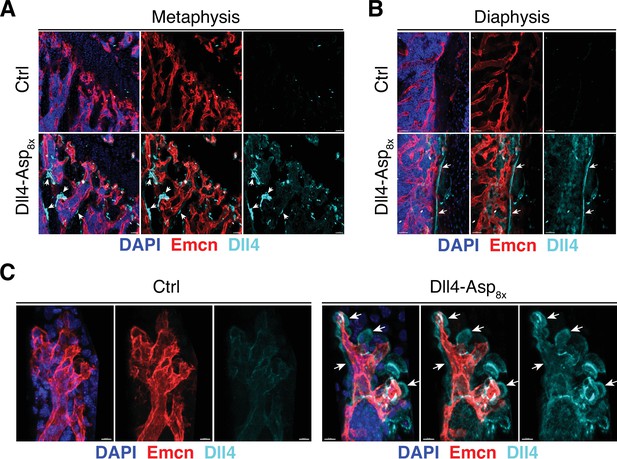
Dll4-Asp8x detection in bone.
(A, B) Confocal images showing Dll4 (cyan) and Emcn (red) staining in femoral metaphysis (A) and endosteum and cortical bone (B) of pLIVE-Dll4-Asp8x and control-injected 11-week-old mice. Nuclei, DAPI (blue). (C) Higher magnification images showing pronounced Dll4 signal (cyan, arrows) in proximity of Emcn+ (red) distal vessels in the pLIVE-Dll4-Asp8x metaphysis. Nuclei, DAPI (blue).
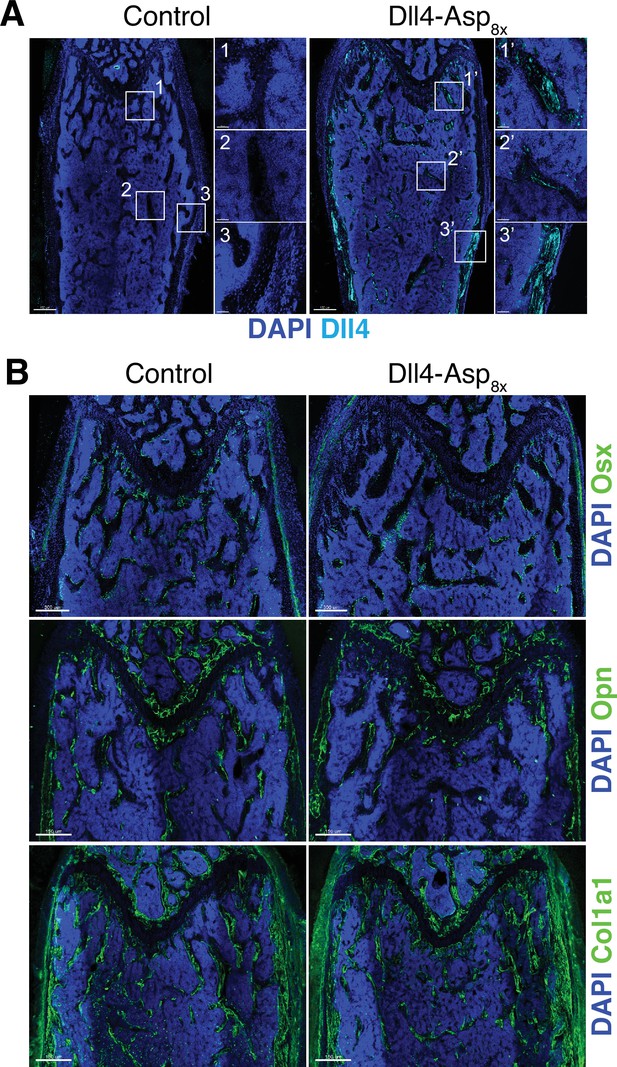
Bone formation is not increased by Dll4-Asp8x.
(A) Tile scan images of Dll4 (cyan) staining in sections of pLIVE-Dll4-Asp8x and control femur. Nuclei, DAPI (blue). Images on the right show higher magnifications of insets in metaphysis (1), bone marrow (2), and cortical bone (3). (B) Osterix (OSX), Osteopontin, and Collagen I alpha one chain (Col1a1) (green) immunostaining, as indicated, in pLIVE-Dll4-Asp8x and control femur. Nuclei, DAPI (blue).
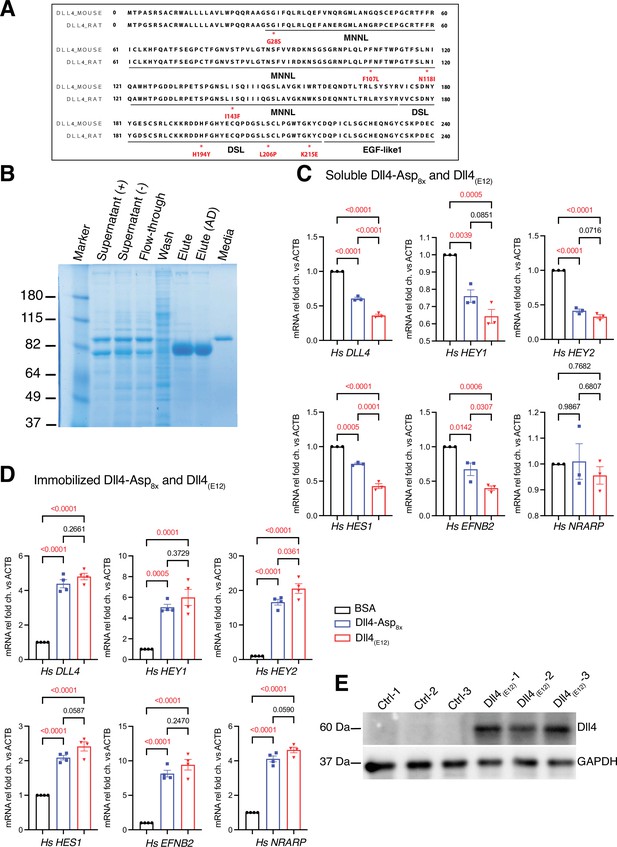
Generation of Dll4(E12).
(A) Diagram showing the amino acid sequence of mouse and rat Dll4 extracellular domain (ECD). Amino acid replacements shown in red were introduced into the murine sequence for the generation of Dll4(E12) with increased Notch-binding affinity. (B) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of purified Dll4(E12) recombinant protein. From left to right, protein molecular mass marker (Marker), supernatant after culture of untransfected (Supernatant (-)) OR pcDNA3.1-Dll4(E12)-transfected HEK293 cells (Supernatant (+)), flow-through after loading of Supernatant (+) on His-tag protein purification column (Flow-through), wash buffer (Wash), elute (Elute), elute after dialysis (Elute (AD)), and Opti-MEM culture medium (Medium) were loaded. (C, D) RT-qPCR analysis of DLL4, HEY1, HEY2, HES1, EFNB2, and NRARP transcripts in confluent human umbilical vascular endothelial cells (HUVECs) stimulated with soluble (C) or in sub-confluent HUVECs stimulated with immobilized Dll4(E12), Dll4-Asp8x, and BSA by poly-L-lysine coating (D). Data represent mean ± s.e.m. (n = 3 for (C) and n = 4 for (D)) (p-values determined by ordinary one-way ANOVA with Tukey’s multiple comparison test). (E) Western blot analysis of Dll4(E12) protein expression in liver lysate of pLIVE-Dll4(E12) and control-injected mice. GAPDH is used as loading control.
-
Figure 1—figure supplement 4—source data 1
Source data for Figure 1—figure supplement 1C,D.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig1-figsupp4-data1-v2.xlsx
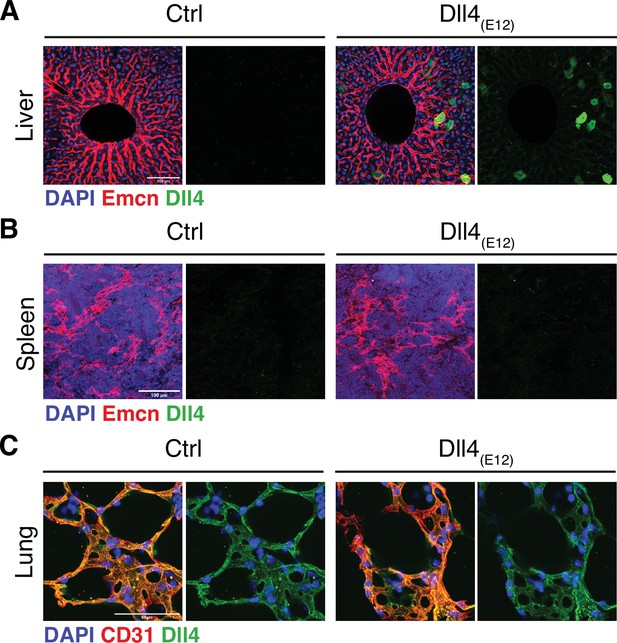
Dll4(E12) immunoreactivity in vivo.
(A–C) Representative confocal images showing Dll4(E12) immunoreactivity after pLIVE-Dll4(E12) or control injection in liver (A), spleen, (B) and lung (C). Dll4(E12) signal (green) is prominent in liver hepatocytes but undetectable in spleen and lung. Note absence of overt changes in the vasculature (red) of all three organs. Nuclei (DAPI, blue).
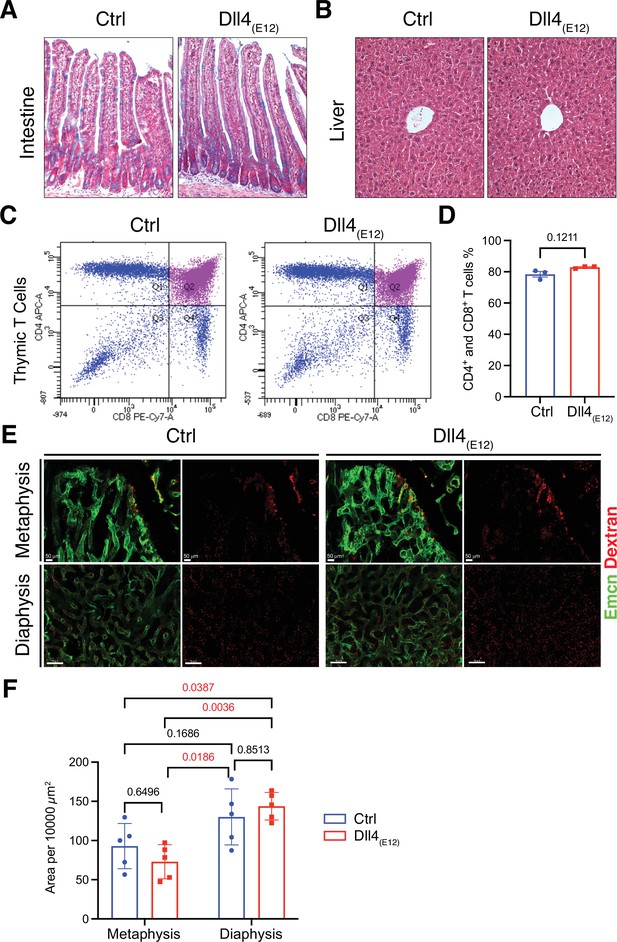
Notch-dependent processes are not blocked by Dll4(E12)in vivo.
(A) Alcian blue and nuclear fast red double staining of small intestine. Secretory goblet cells are labeled by Alcian blue. (B) Hematoxylin and eosin staining of liver sections of 11-week-old pLIVE-Dll4(E12) and control-injected mice. (C, D) Analysis of CD4+/CD8+ cells from pLIVE-Dll4(E12) and control thymi by flow cytometry (C) and corresponding quantification (D). Data represent mean ± s.e.m. (n = 3 mice) (p-values determined by unpaired t-test with Welch’s correction). (E) Confocal images showing comparable extravasation of fluorescent Dextran (70 kD) in femur of pLIVE-Dll4(E12) and control-injected mice. (F) Quantitation of Texas Red-labeled Dextran extravasation in femoral metaphysis and diaphysis of pLIVE-Dll4(E12) and control-injected mice. Data represent mean ± s.e.m. (n = 5 mice) (adjusted p-values determined by two-way ANOVA with Tukey’s multiple comparison test).
-
Figure 2—source data 1
Source data for Figure 2D and F.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig2-data1-v2.xlsx
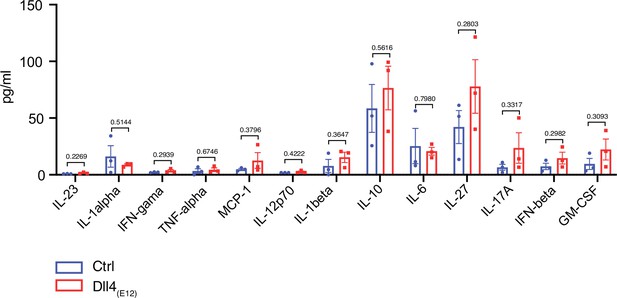
Inflammatory cytokines are not induced by Dll4(E12).
Quantification of inflammatory cytokines in the plasma of pLIVE-Dll4(E12)- and control-injected mice. Data represent mean ± s.e.m. (n = 3 mice) (p-values determined by multiple unpaired t-test with Welch’s correction).
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig2-figsupp1-data1-v2.xlsx
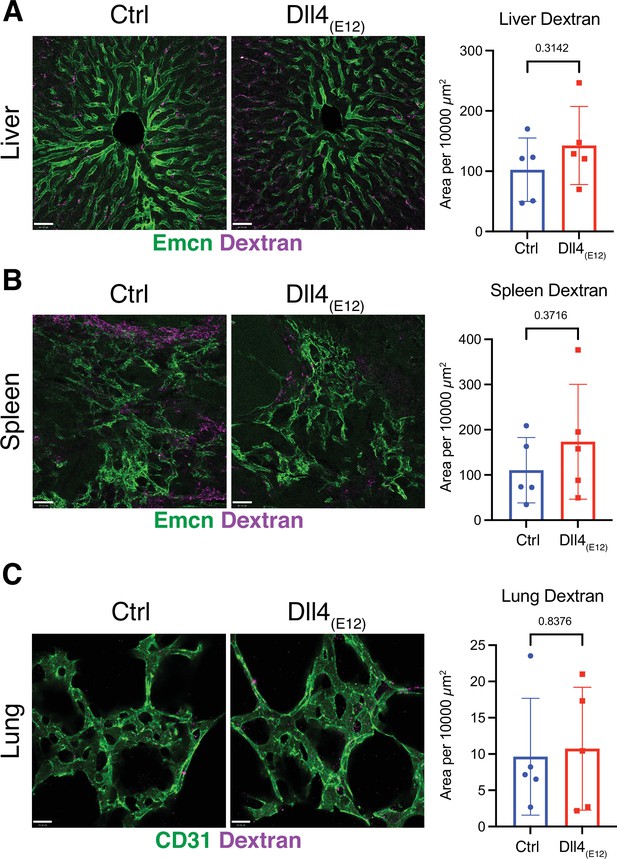
Dll4(E12) does not induce vascular leakage.
(A–C) Representative confocal images and corresponding quantitation showing comparable extravasation of Texas Red-coupled fluorescent Dextran (purple) in liver (A), spleen, (B) and lung (C) of pLIVE-Dll4(E12) and control-injected mice. Vasculature in liver and spleen (A, B) has been immunostained for Emcn (green) and in lung (C) for CD31 (green). Data represent mean ± s.e.m. (n = 5 mice) (p-values determined by unpaired t-test with Welch’s correction).
-
Figure 2—figure supplement 2—source data 1
Source data for Figure 2—figure supplement 2A-C.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig2-figsupp2-data1-v2.xlsx
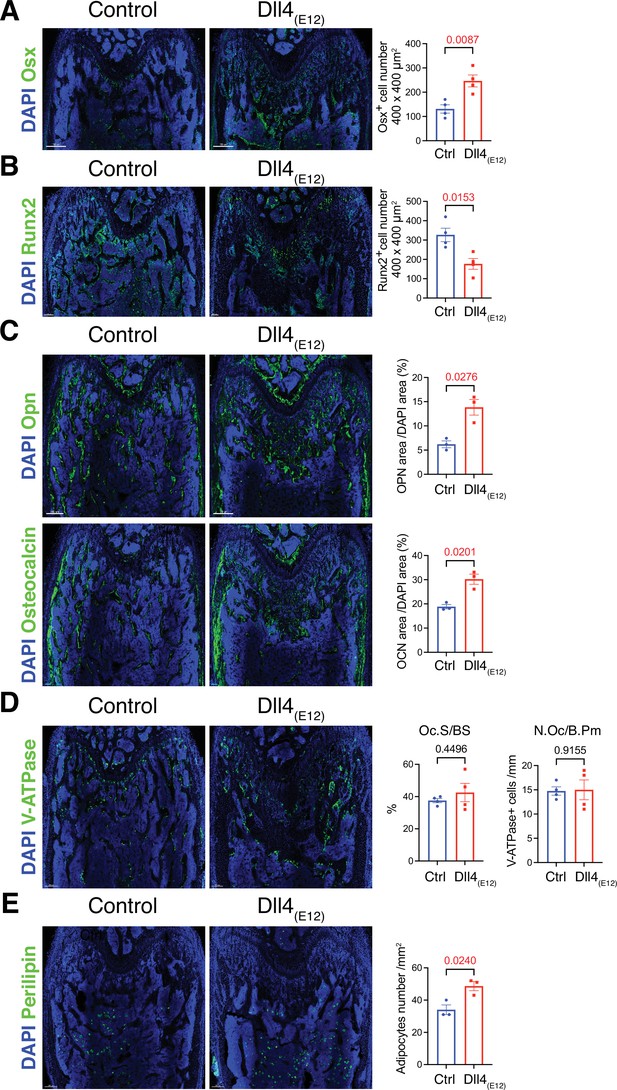
Recombinant Dll4(E12) increases osteogenesis.
(A) Tile scan confocal images of Osterix (OSX) staining (green) in femurs from pLIVE-Dll4(E12) and control-injected mice. Nuclei, DAPI (blue). Graph shows quantitation of OSX+ cells. Data represent mean ± s.e.m. (n = 4 mice) (p-values determined by two-tailed unpaired t-test). (B) Runx2 staining (green) of pLIVE-Dll4(E12) and control femurs. Nuclei, DAPI (blue). Graph shows quantitation of Runx2+ cells. Data represent mean ± s.e.m. (n = 4 mice) (p-values determined by two-tailed unpaired t-test). (C) Representative images showing Osteopontin (Opn) and Osteocalcin staining (green) in pLIVE-Dll4(E12) and control femur. Nuclei, DAPI (blue). Data represent mean ± s.e.m. (n = 3 mice) (p-values determined by unpaired t-test with Welch’s correction). (D) Tile scan images of ATP6V1B1+ ATP6V1B2 (V-ATPase) staining (green) in pLIVE-Dll4(E12) and control femur. Nuclei, DAPI (blue). Graphs show quantification of osteoclast surface/bone surface (Os. S/B S) and osteoclast number/bone perimeter (No. Oc./B. Pm). Data represent mean ± s.e.m. (n = 4 mice) (p-values determined by unpaired t-test with Welch’s correction). (E) Tile scan images of Perilipin staining (green) in pLIVE-Dll4(E12) and control femurs. Nuclei, DAPI (blue). Graph shows quantitation of Perilipin+ adipocytes. Data represent mean ± s.e.m. (n = 3 mice) (p-values determined by unpaired t-test with Welch’s correction).
-
Figure 3—source data 1
Source data for Figure 3A–E.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig3-data1-v2.xlsx
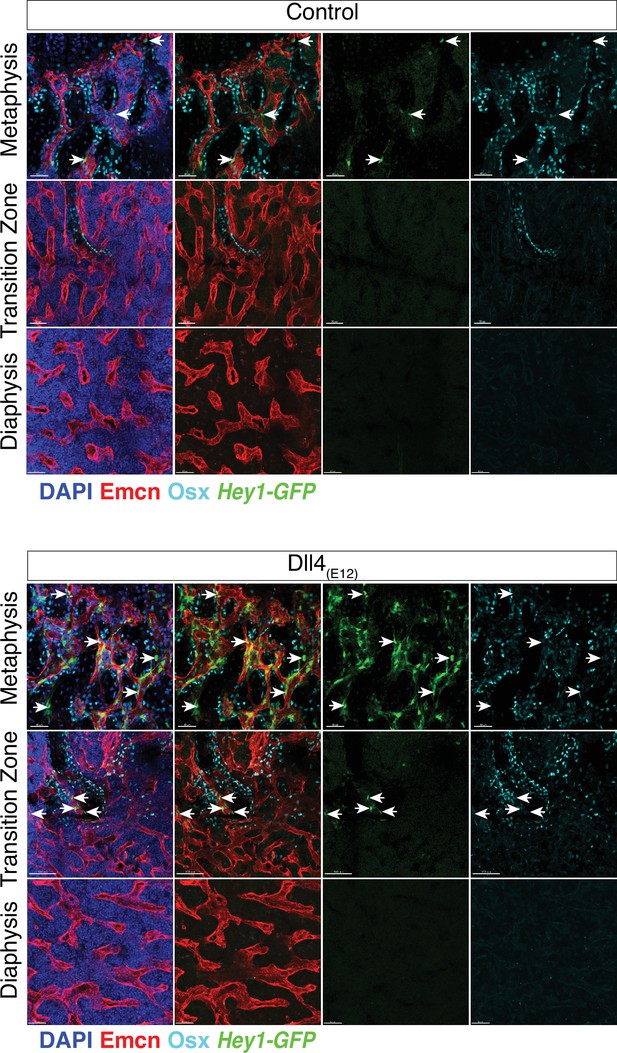
Activation of the Hey1-EGFP reporter by Dll4(E12).
Expression of the Tg(Hey1-EGFP)ID40Gsat reporter (green fluorescent protein [GFP], green) in 11-week-old pLIVE-Dll4(E12) and control femur. Note that Dll4(E12) increases the number of Osterix+ (OSX+) cells (cyan) and leads to induction of GFP expression in the metaphysis but also in the adjacent transition zone. GFP+ cells show no or weak nuclear OSX signals (arrows). Vessels, Emcn (red). Nuclei, DAPI (blue).
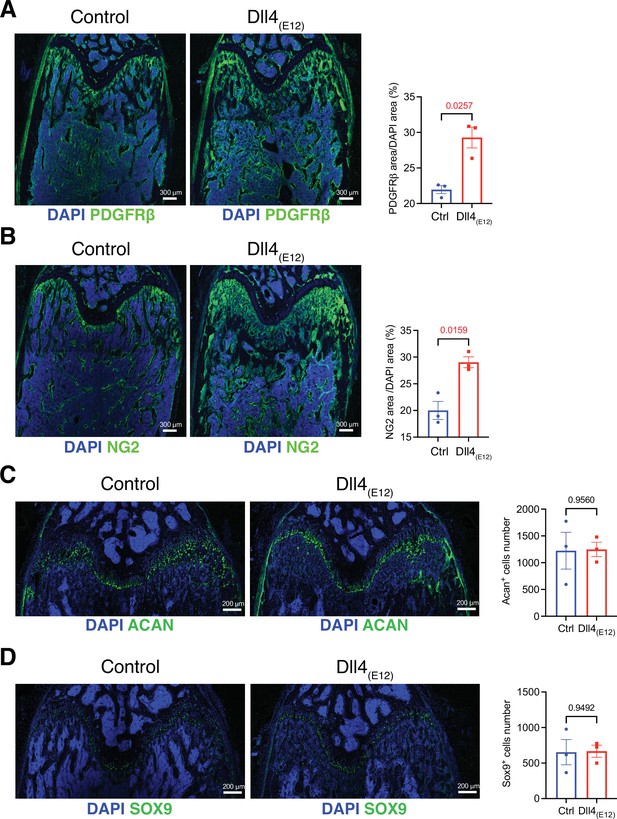
Effect of Dll4(E12) on metaphyseal mesenchymal stromal cells (MSCs) and chondrocytes.
(A, B) Tile scan confocal images showing that PDGFRβ (A) and NG2 (B) immunostaining in the adult metaphysis is increased by recombinant Dll4(E12). Nuclei, DAPI (blue). Graph shows quantitation of PDGFRβ area/DAPI area (A) and NG2 area/DAPI area in percentage. Data represent mean ± s.e.m. (n = 3 mice) (p-values determined by unpaired t-test with Welch’s correction). (C, D) Representative confocal images of chondrocyte staining in adult metaphysis showing that Acan+ (C) and Sox9+ (D) chondrocytes are not affected by pLIVE-Dll4(E12) injection. Nuclei, DAPI (blue). Graphs show quantitation of Acan+ cells number (C) and Sox9+ cells number (D). Data represent mean ± s.e.m. (n = 3 mice) (p-values determined by unpaired t-test with Welch’s correction).
-
Figure 3—figure supplement 2—source data 1
Source data for Figure 3—figure supplement 2A-D.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig3-figsupp2-data1-v2.xlsx
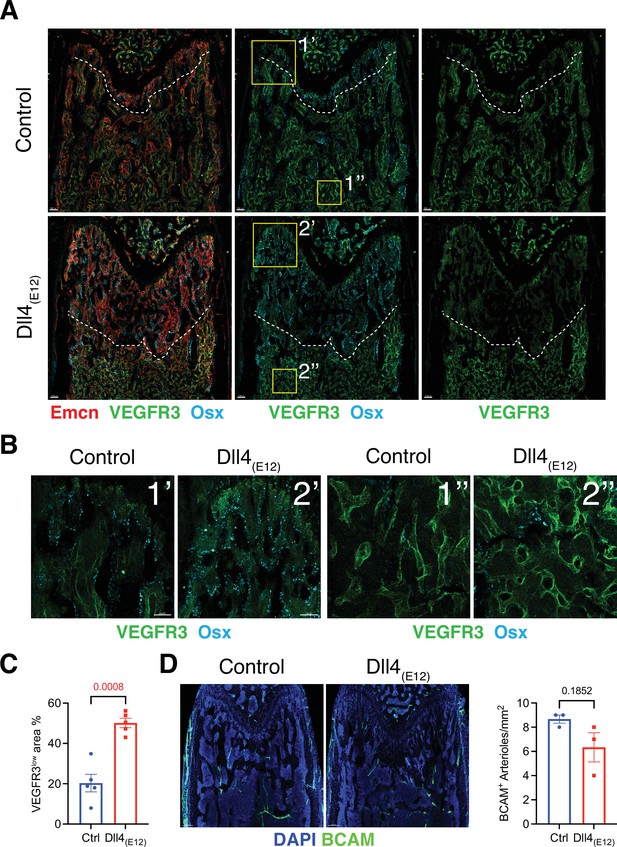
Effect of Dll4(E12) on bone vasculature.
(A) Tile scan images of Emcn (red), VEGFR3 (green), and Osterix (OSX)- (cyan) stained sections of pLIVE-Dll4(E12) and control femur at 11 weeks of age. Dashed line indicates the border of VEGFR3high and VEGFR3low area. (B) Higher magnifications of insets in (A) showing the metaphysis close to growth plate (1’ and 2’) and bone marrow close to transition zone (1’’ and 2’’). Stainings show VEGFR3 (green) and OSX (cyan). Note increase in OSX+ cells and presence of OSX-stained nuclei close to VEGFR3high vessels after Dll4(E12) treatment. (C) Quantification of VEGFR3low area per region of interest (ROI) in the metaphysis. Data represent mean ± s.e.m. (n = 5 mice) (p-values determined by unpaired t-test with Welch’s correction). (D) Tile scan images of BCAM (green) stained sections of pLIVE-Dll4(E12) and control femur. Graph shows quantification of the number of BCAM+ arteries per ROI in the metaphysis. Data represent mean ± s.e.m. (n = 3 mice) (p-values determined by unpaired t-test with Welch’s correction).
-
Figure 4—source data 1
Source data for Figure 4C and D.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig4-data1-v2.xlsx
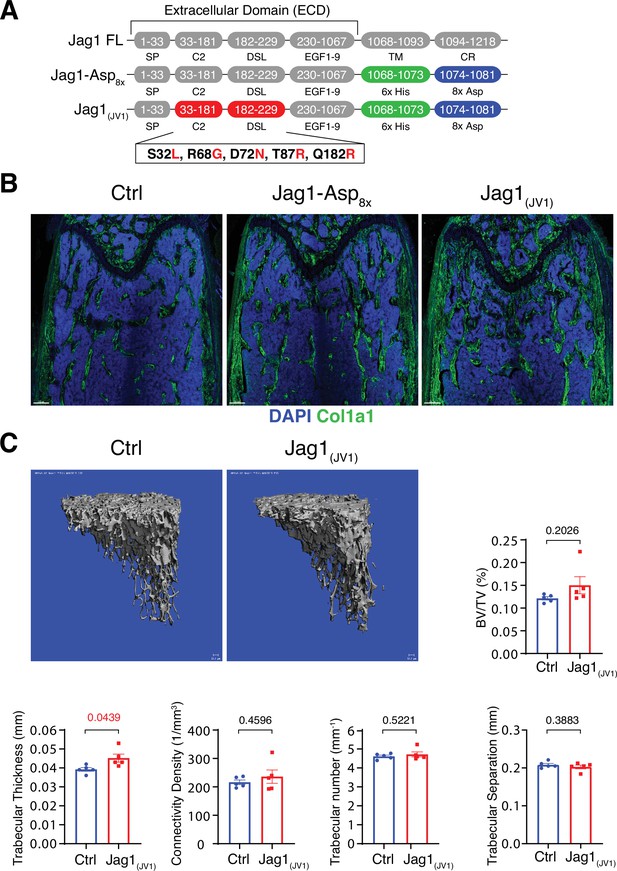
Bone formation is not changed by recombinant Jag1-Asp8x and Jag1(JV1).
(A) Schematic diagram showing the domain organization of murine Jag1 full-length protein and recombinant Jag1-Asp8x and Jag1(JV1) fusion proteins. Fusion proteins contain 6x His epitope tag (green boxes) and Asp8x negatively charged peptide motif (blue boxes) instead of the transmembrane (TM) domain and cytoplasmic region (CR) of Jag1. Missense mutations were introduced in the calcium-binding C2 and DSL (Delta-Serrate-Lin) domains to generate Jag1(JV1) with increased Notch binding affinity. The resulting amino acid replacements are highlighted in red. SP, signal peptide. (B) Tile scan images of Collagen I alpha one chain (Col1a1) (green) staining on the femur sections of control, pLIVE-Jag1-Asp8x and pLIVE-Jag1(JV1)-injected mice at the age of 11 weeks. Nuclei, DAPI (blue). (C) Representative 3D reconstruction from micro-computed tomography (µ-CT) measurements of tibial metaphysis of pLIVE-Jag1(JV1) and control-injected mice. Diagrams show bone parameters measured in µ-CT analyses: bone volume/total volume (BV/TV) in percentage, trabecular thickness in millimeters, connectivity density in one per cubic millimeter, trabeculae number in one per millimeter, and trabecular separation in millimeters. Data represent mean ± s.e.m. (n = 5 mice), (p-values determined by unpaired t-test with Welch’s correction).
-
Figure 5—source data 1
Source data for Figure 5C.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig5-data1-v2.xlsx
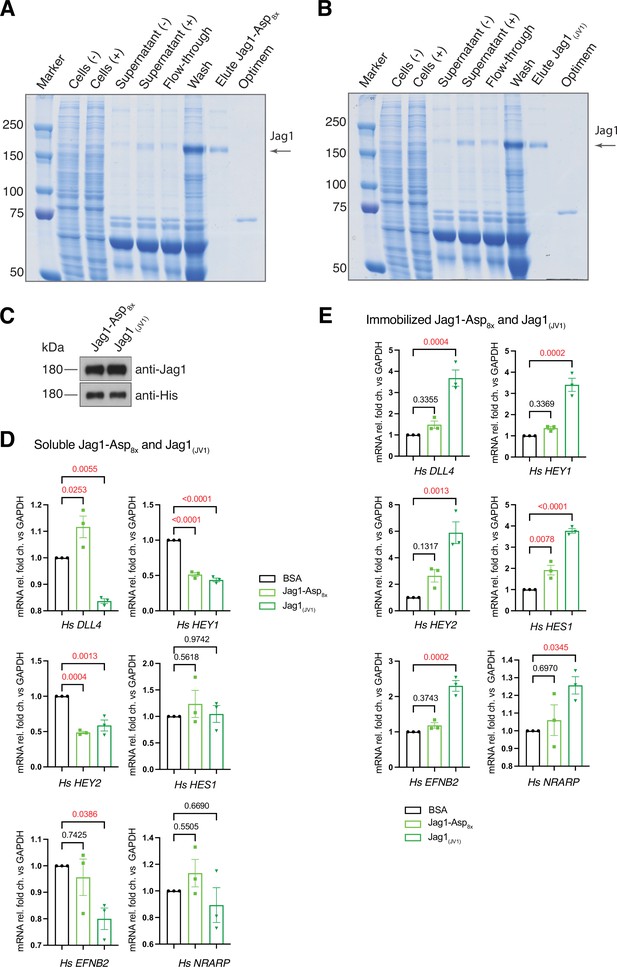
Generation of Jag1-Asp8x and Jag1(JV1) recombinant proteins.
(A) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of purified Jag1-Asp8x recombinant protein. From left to right, protein molecular mass marker (Marker), extract of untransfected HEK293 cells (Cells (-)) OR pcDNA3.1-Jag1-Asp8x-transfected cells (Cells (+)), supernatant after culture of untransfected (Supernatant (-)) OR pcDNA3.1-Jag1-Asp8x-transfected cells (Supernatant (+)), flow-through after loading of Supernatant (+) on His-tag protein purification column (Flow-through), wash buffer (Wash), elution of Jag1-Asp8x (Elute), and Opti-MEM culture medium (Medium) were loaded. (A, B) SDS-PAGE analysis of purified Jag1-Asp8x (A) and Jag1(JV1) (B) recombinant proteins. From left to right, protein molecular mass marker (Marker), extract of untransfected (Cells (-)) or transfected HEK293 cells (Cells (+)), supernatant after culture of untransfected (Supernatant (-)) or transfected cells (Supernatant (+)), flow-through after loading of Supernatant (+) on His-tag protein purification column (Flow-through), wash buffer (Wash), elution of recombinant Jag1 (Elute) and Opti-MEM culture medium (Medium) were loaded. (C) Western blot analysis of eluted Jag1-Asp8x and Jag1(JV1) recombinant proteins used for in vitro stimulation experiments (D, E). Proteins detected at the expected molecular weight with anti-Jag1 and anti-His1 antibodies. (D, E) RT-qPCR analysis of human (HS) DLL4, HEY1, HEY2, HES1, EFNB2, and NRARP transcripts in confluent human umbilical vascular endothelial cells (HUVECs) after stimulation with soluble (D) or immobilized (poly-L-lysine coating) Jag1-Asp8x, Jag1(JV1), and BSA (E). Data represent mean ± s.e.m. (n = 3) (p-values determined by ordinary one-way ANOVA with Tukey’s multiple comparison test).
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1D,E.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig5-figsupp1-data1-v2.xlsx
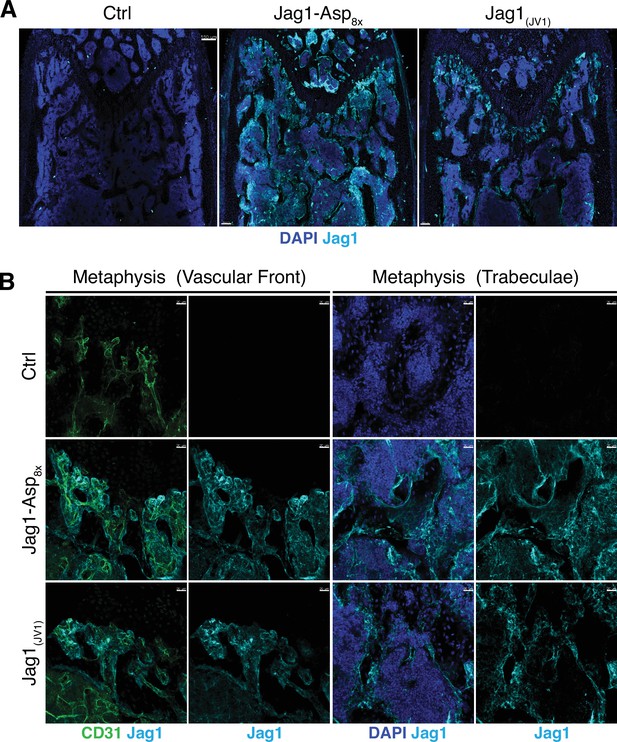
Detection of Jag1-Asp8x and Jag1(JV1) in the bone.
(A) Tile scan confocal images of Jag1 (cyan) immunostaining in sections from control, pLIVE-Jag1-Asp8x and pLIVE-Jag1(JV1) femur. Nuclei, DAPI (blue). (B) Representative higher magnification images of CD31 (green) and Jag1 (cyan) staining in the metaphysis of control, pLIVE-Jag1-Asp8x and pLIVE-Jag1(JV1)-injected mice. Nuclei, DAPI (blue).
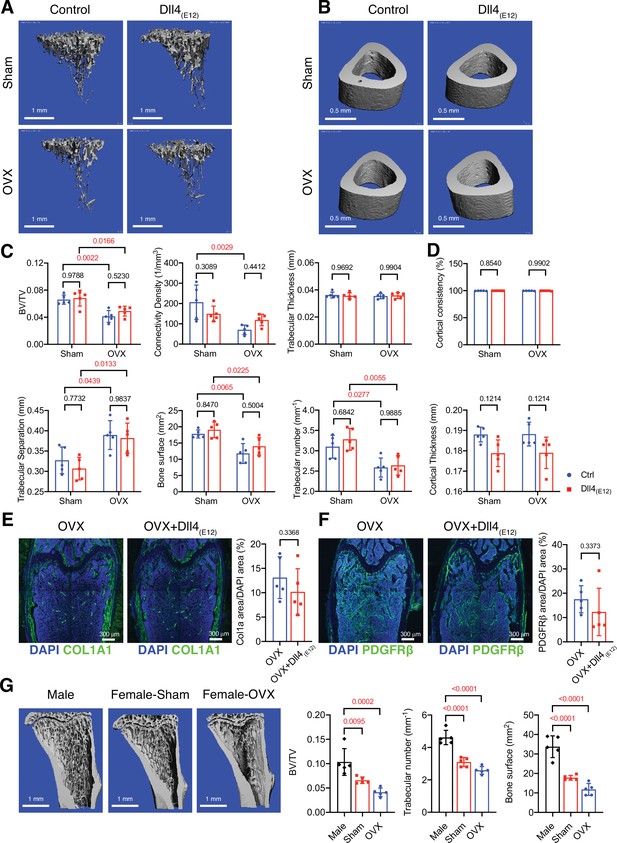
Bone formation is not increased by Dll4(E12) in ovariectomized female mice.
(A, B) Representative 3D micro-computed tomography (μ-CT) images of trabecular bone in the distal tibial metaphysis (A) and cortical bone in the mid-tibial diaphysis (B) of sham or ovariectomized mice treated with vehicle or pLIVE-Dll4(E12). (C) Bone parameters measured by μ-CT analysis of trabecular bone volume/total volume, trabecular connectivity density, trabecular thickness, trabecular separation, bone surface, and trabecular number in the distal tibial metaphysis. Data represent mean ± SD. (n = 5 mice) (p-values determined by two-way ANOVA with Tukey’s multiple comparisons test). (D) Quantitation of μ-CT analysis of cortical bone consistency and cortical thickness in the mid-tibial diaphysis. Data represent mean ± SD. (n = 5 mice) (p-values determined by two-way ANOVA with Tukey’s multiple comparisons test). (E, F) Tile scan confocal images showing Collagen I alpha one chain (Col1a1) staining (green; E) and PDGFRβ staining (green; F) in femoral sections from ovariectomized female mice treated with vehicle or pLIVE-Dll4(E12). Nuclei, DAPI (blue). Graphs show relative ratio (percentage) of Col1a area/DAPI area and PDGFRβ area/DAPI area. Data represent mean ± SD. (n = 5 mice) (p-values determined by two-tailed unpaired t-test (E) with Welch’s correction (F)). (G) Representative μ-CT images of trabecular bone in the distal tibial metaphysis of male, female sham, and ovariectomized mice. Graphs show quantitation of the trabecular bone volume/total volume, trabecular number, and bone surface. Data represent mean ± SD. (n = 5 mice), (p-values determined by one-way ANOVA followed by Sidak’s multiple comparisons test).
-
Figure 6—source data 1
Source data for Figure 6C–G.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig6-data1-v2.xlsx
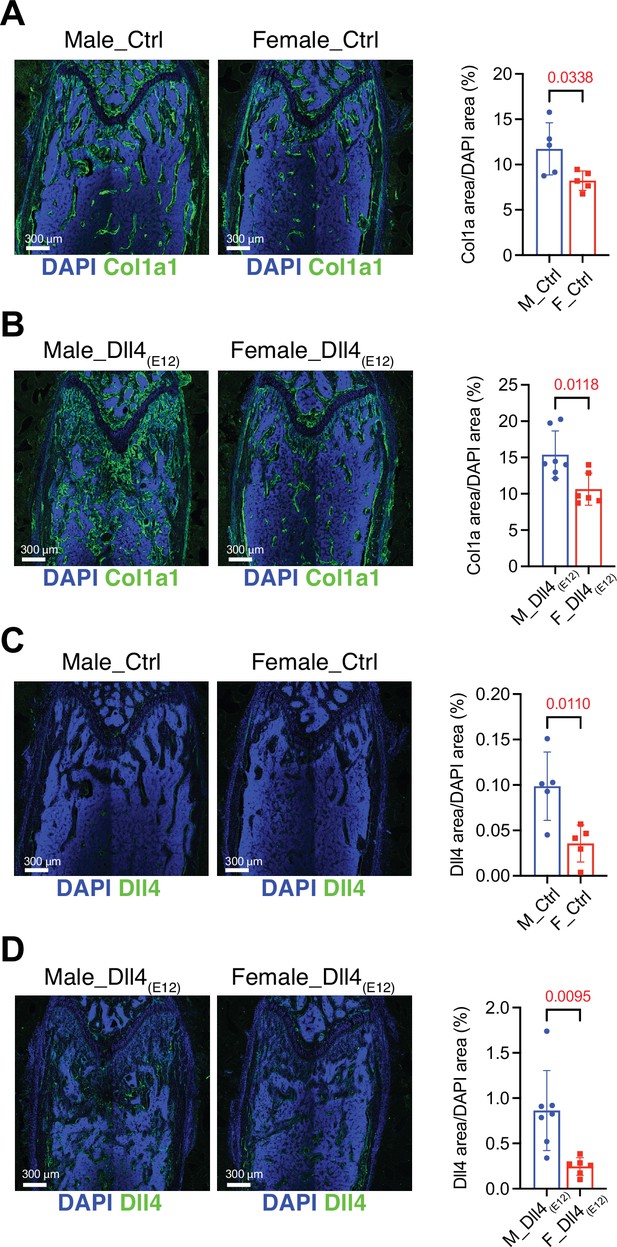
Reduced Dll4(E12) immunoreactivity and trabecular bone in female mice compared to male mice.
(A, B) Representative overview confocal images of Collagen I alpha one chain (Col1a1) staining (green) in the femurs of wild-type male and female mice treated with vehicle (A) or pLIVE-Dll4(E12) (B). Nuclei, DAPI (blue). Data represent mean ± SD. (n = 5–7 mice) (p-values determined by two-tailed unpaired t-test (control) or unpaired t-test). (C, D) Representative overview confocal images of Dll4 staining (green) in the femurs of wild-type male and female mice treated with vehicle (C) or pLIVE-Dll4(E12) (D). Nuclei, DAPI (blue). Diagrams represent percentage of Dll4 area/DAPI area. Data represent mean ± SD. (n = 5–7 mice) (p-values determined by two-tailed unpaired t-test (control) or unpaired t-test with Welch’s correction (Dll4(E12))).
-
Figure 6—figure supplement 1—source data 1
Source data for Figure 6—figure supplement 1A-D.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig6-figsupp1-data1-v2.xlsx
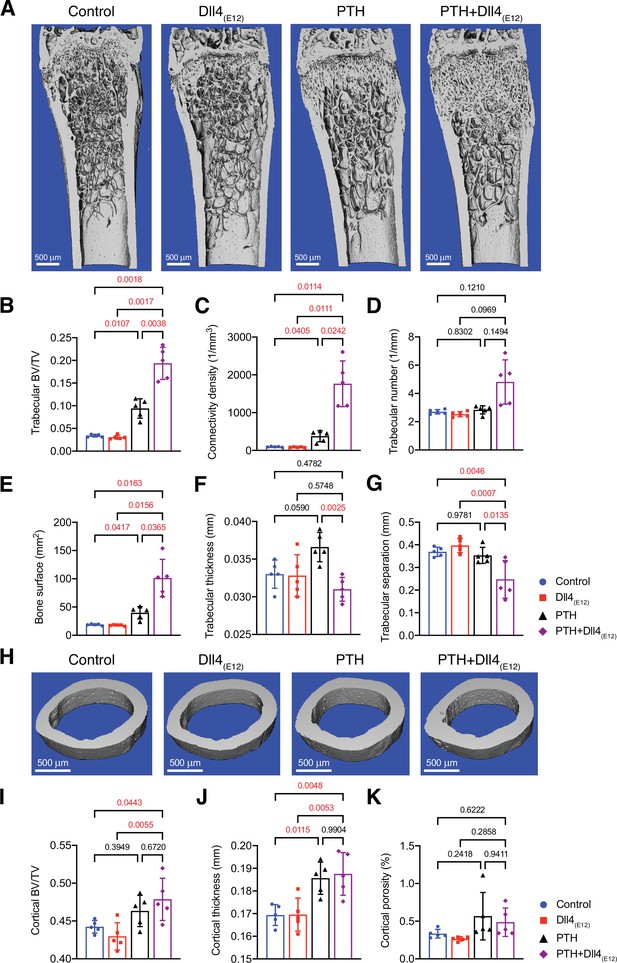
Synergistic action of recombinant Dll4(E12) and parathyroid hormone (PTH).
(A) Representative micro-computed tomography (μ-CT) images of trabecular bone in the distal femoral metaphysis of female wild-type mice treated with vehicle, pLIVE-Dll4(E12), PTH, or the combination of both for 3 weeks. (B–G) Quantitative analysis of μ-CT data on trabecular bone volume/total volume (B), trabecular connectivity density (C), trabecular number (D), bone surface (E), trabecular thickness (F), trabecular separation (G) in the distal femoral metaphysis. Data represent mean ± SD. (n = 5 mice) (p-values determined by one-way ANOVAs (F, G) followed by Sidak’s multiple comparisons test or Brown-Forsythe and Welch’s ANOVAs (B, C, D, E) followed by Dunnett’s T3 multiple comparisons test). (H) Representative μ-CT images of cortical bone in the female midfemoral diaphysis. (I–K) Graphs from μ-CT analysis of the cortical bone volume/total volume (I), cortical thickness (J), and cortical porosity (K) in the midfemoral diaphysis. Data represent mean ± SD. (n = 5 mice) (p-values determined using one-way ANOVAs followed by Sidak’s multiple comparisons test).
-
Figure 7—source data 1
Source data for Figure 7B–G,I-K.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig7-data1-v2.xlsx
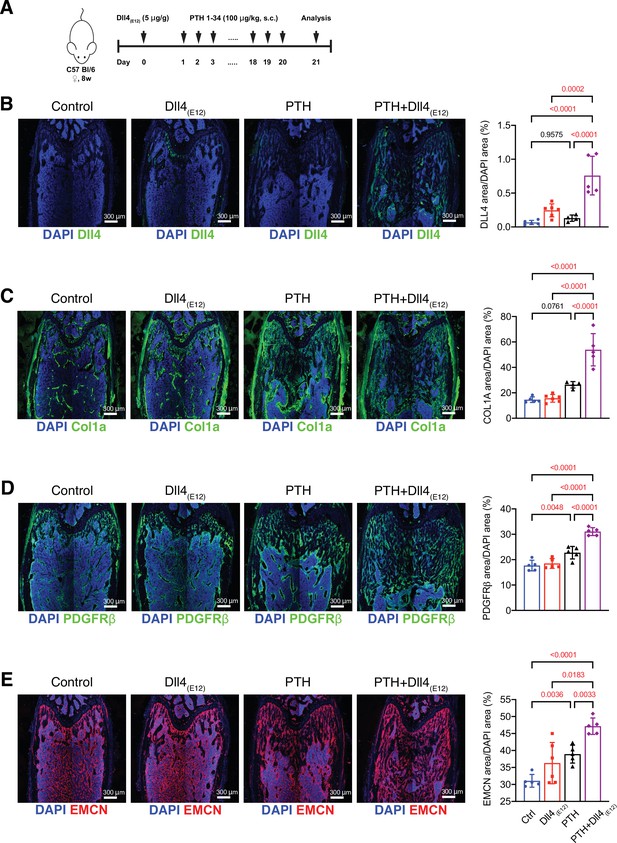
Effects of combined parathyroid hormone (PTH) and Dll4(E12) administration.
(A) Time course of pLIVE-Dll4(E12) and PTH administration to adult wild-type female mice. (B–E) Representative overview confocal images and corresponding quantification of immunostainings for Dll4 (B, green), Collagen I alpha one chain (Col1a1) (C, green), PDGFRβ (D, green), Endomucin (E, red) in the femurs treated with vehicle, pLIVE-Dll4(E12), PTH, or the combination of both. Nuclei, DAPI (blue). Data represent mean ± SD. (n = 4–5 mice). Statistical significance was assessed using one-way ANOVAs (B, C, D) followed by Sidak’s multiple comparisons test or Brown-Forsythe and Welch’s ANOVA (E) followed by Dunnett’s T3 multiple comparisons test.
-
Figure 7—figure supplement 1—source data 1
Source data for Figure 7—figure supplement 1B-E.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig7-figsupp1-data1-v2.xlsx
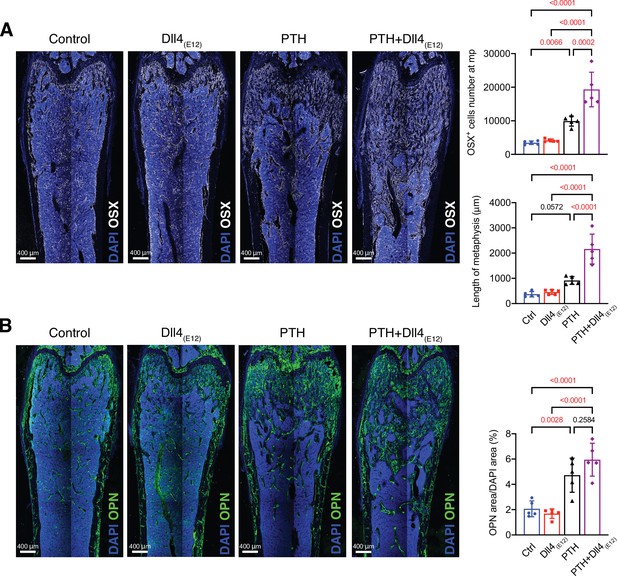
Synergistic effects of Dll4(E12) and parathyroid hormone (PTH) on osteogenesis.
Representative overview confocal images and corresponding quantifications for Osterix (OSX) (A, white) and Opn (B, green) immunostaining in wild-type female femur after treatment with vehicle, pLIVE-Dll4(E12), PTH, or the combination of both. Nuclei, DAPI (blue). Data represent mean ± SD. (n = 5 mice) (p-values determined using one-way ANOVA followed by Sidak’s multiple comparisons test).
-
Figure 7—figure supplement 2—source data 1
Source data for Figure 7—figure supplement 2A,B.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig7-figsupp2-data1-v2.xlsx
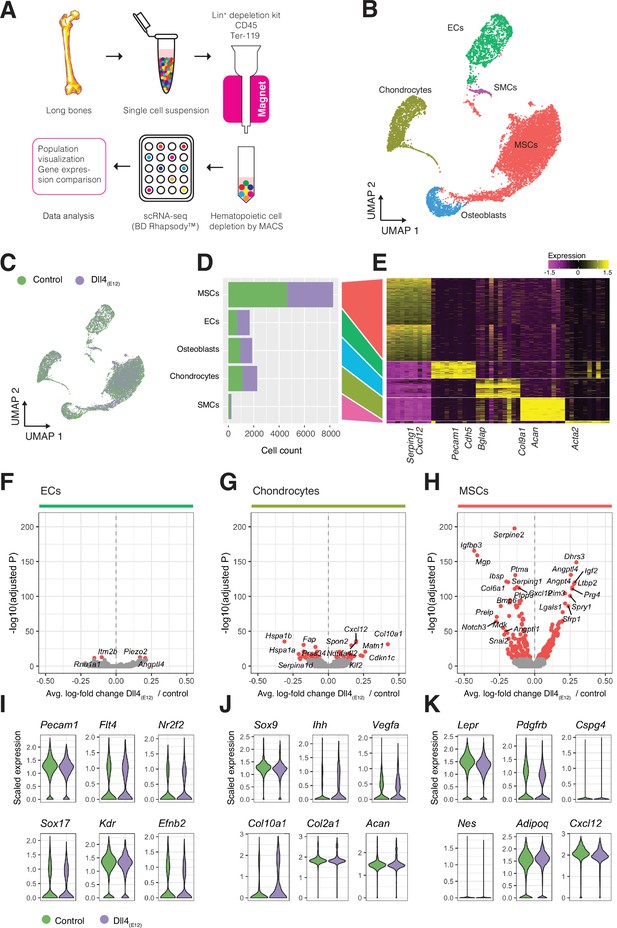
Single cell RNA sequencing (scRNA-seq) analysis of control and Dll4(E12)-treated bone.
(A) Overview of the sample processing and scRNA-seq procedure. (B) UMAP projection of all cells in pLIVE-Dll4(E12) and control, colored by Louvain clusters. Endothelial cells (ECs), mesenchymal stromal cells (MSCs), smooth muscle cells (SMCs), chondrocytes, and osteoblasts are indicated. (C) UMAP projection of all cells colored by experimental condition (green = control, purple = Dll4(E12)). (D, E) Barplot showing absolute numbers of cells (D) and scaled expression heatmap of the top 10 marker genes (E) for each of the clusters shown in (B). (F–H) Differential expression volcano plots showing -log10(adjusted p-value) against average log-fold change of pLIVE-Dll4(E12) vs. control in ECs (F), chondrocytes (G), and MSCs (H). Genes with adjusted p-values smaller than 1e-10 are colored in red. (I–K) Selected cell population relevant gene expression shown as violin (density) plots for EC (I), chondrocyte, (J) and MSC (K) subpopulations.
-
Figure 8—source data 1
Source data for Figure 8F–H.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig8-data1-v2.xlsx
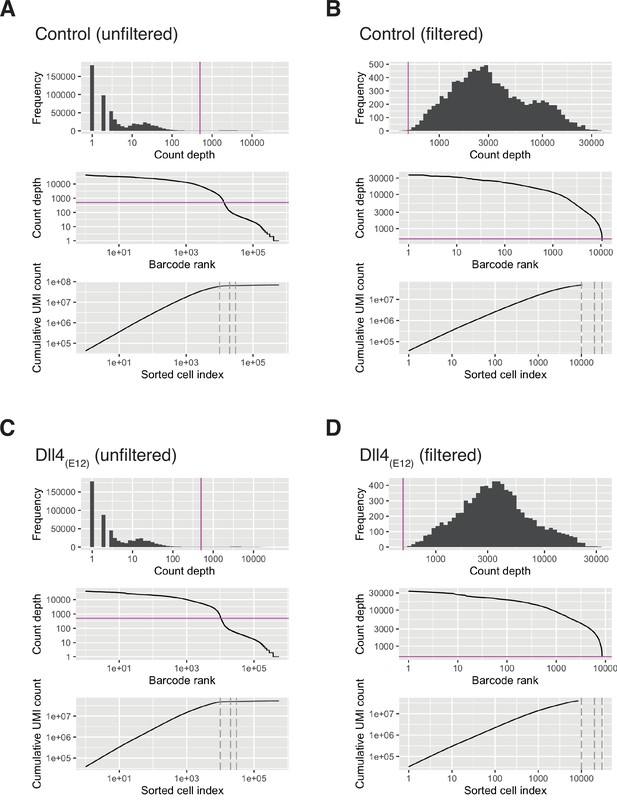
Single cell RNA sequencing (scRNA) quality control statistics.
(A–D) Quality control plots. In each panel, the top plot shows distribution of count depths, middle plot shows knee plot, and the bottom plot shows cumulative count depth plotted as a function of cell barcodes sorted by count depth. Plots show control sample before filtering (A), control sample after filtering (B), pLIVE-Dll4(E12) sample before filtering (C), and pLIVE-Dll4(E12) sample after filtering (D).
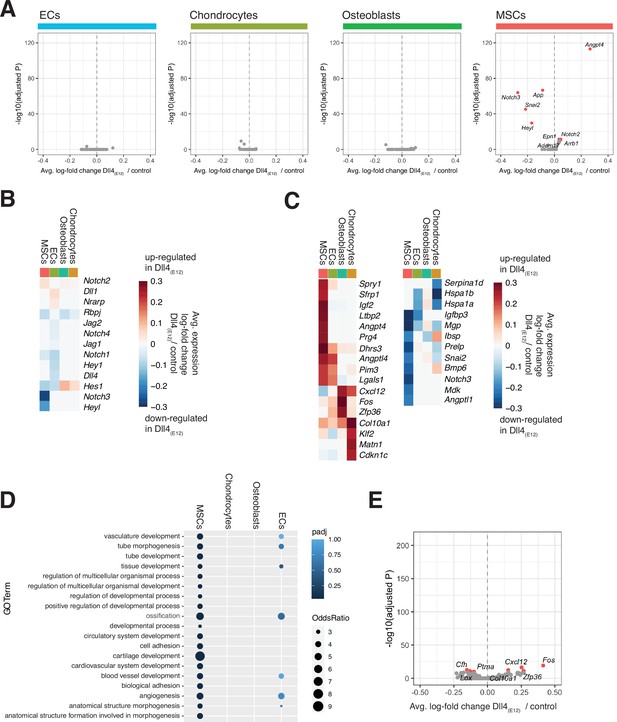
Extended analysis of differentially expressed genes.
(A) Differential expression volcano plots showing -log10(adjusted p-value) against average log-fold change of pLIVE-Dll4(E12) vs. control for Notch pathway related genes (parent Gene Ontology [GO] term ‘Notch signaling pathway’ GO:0007219) for endothelial cell (EC), chondrocyte, osteoblast, and mesenchymal stromal cell (MSC) populations. (B) Heatmap of the average expression log-fold change of selected Notch pathway genes with a p-value < 1e-10 and log-fold change > 0.2 across the four major cell populations. (C) Heatmap of the average expression log-fold change of genes with a p-value < 1e-10 and log-fold change >0.2 across the four major cell populations. (D) Dot plot of GO analysis results showing all enriched GO terms with a p-value below 1e-5 across all four major cell populations. Color encodes hypergeometric test p-value, size of each dot is proportional to the odds ratio (OR). (E) Differential expression volcano plot for the osteoblast population showing -log10(adjusted p-value) against average log-fold change of pLIVE-Dll4(E12) vs. control. Genes with an adjusted p-value less than 1e-10 are colored in red.
-
Figure 8—figure supplement 2—source data 1
Source data for Figure 8—figure supplement 2A-E.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig8-figsupp2-data1-v2.xlsx
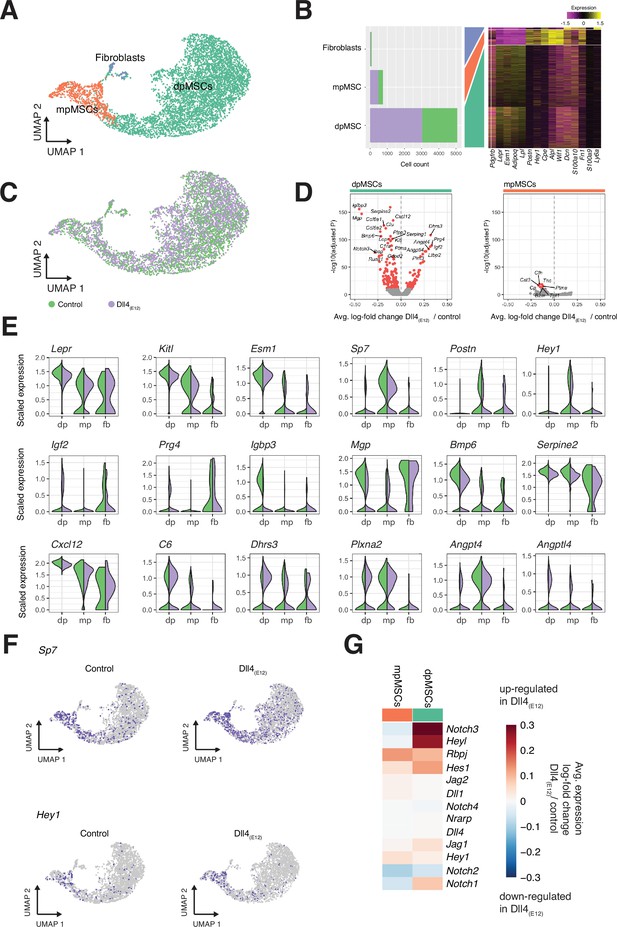
Single cell RNA sequencing (scRNA-seq) analysis of mesenchymal stromal cell (MSC) subclusters.
(A) UMAP projection of MSCs in both control and pLIVE-Dll4(E12) samples colored by Louvain clusters. Metaphyseal MSCs (mpMSCs), diaphyseal MSCs (dpMSCs), and fibroblasts are indicated. (B) Barplot showing absolute numbers of cells per sample (left), and scaled expression heatmap of the top 10 marker genes (right) in each of the clusters shown in (A). (C) UMAP projection of all cells colored by experimental condition green = control, purple = Dll4(E12). (D) Differential expression volcano plots showing -log10(adjusted p-value) against average log-fold change of treatment/control for dpMSCs (left) and mpMSCs (right). Genes with an adjusted p-value less than 1e-10 are colored in red. (E) Violin plots showing selected differentially expressed genes in the three MSC subpopulations. Each violin is split along the vertical axis into control and pLIVE-Dll4(E12). (F) UMAP projections of control and Dll4(E12)-treated cells colored for the expression of Sp7 and Hey1. (G) Heatmap of the average expression log-fold change of selected Notch pathway genes with a p-value < 1e-10 and log-fold change >0.2 in dpMSCs and mpMSCs.
-
Figure 9—source data 1
Source data for Figure 9D–G.
- https://cdn.elifesciences.org/articles/60183/elife-60183-fig9-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus, C57BL/6JRj) | WT | Janvier Labs | ||
| Genetic reagent (Mus musculus) | Tg(Hey1-EGFP)ID40Gsat | GENSAT | MGI:4847129 | |
| Recombinant DNA reagent | pcDNA3.1- Dll4(ECD)- His(6x)-Asp(8x)(plasmid) | This paper | Dll4-Asp(8x) | |
| Recombinant DNA reagent | pcDNA3.1- Dll4(ECD)- Variant- His(6x)-Asp(8x) (plasmid) | This paper | Dll4(E12) | |
| Recombinant DNA reagent | pLIVE-Dll4(ECD)-His(6x)-Asp(8x) (plasmid) | This paper | pLIVE-Dll4-Asp(8x) | |
| Recombinant DNA reagent | pLIVE-Dll4(ECD) Variant- His(6x)-Asp(8x) (plasmid) | This paper | pLIVE-Dll4(E12) | |
| Recombinant DNA reagent | pcDNA3.1- Jag1 (ECD)- His(6x)-Asp(8x)(plasmid) | This paper | Jag1- Asp(8x) | |
| Recombinant DNA reagent | pcDNA3.1- Jag1 (ECD)- JV1- His(6x)-Asp(8x)(plasmid) | This paper | Jag1(JV1) | |
| Recombinant DNA reagent | pLIVE- Jag1 (ECD)- His(6x)-Asp(8x) (plasmid) | This paper | pLIVE-Jag1- Asp(8x) | |
| Recombinant DNA reagent | pLIVE- Jag1 (ECD)- JV1- His(6x)-Asp(8x) (plasmid) | This paper | pLIVE-Jag1(JV1) | |
| Cell line (Homo sapiens) | Human Umbilical Vein Endothelial Cells (HUVEC) | ThermoFisher | Cat# C0035C | Cell identity and absence of mycoplasma contamination or human pathogens were certified by the supplier |
| Cell line (Homo sapiens) | Human Embryonal kidney –293(HEK293) | DSMZ | Cat# ACC 305 | Cell identity and absence of mycoplasma contamination were certified by the supplier |
| Antibody | Anti-Endomucin (Rat, monoclonal) | Santa Cruz | Cat# SC-65495 | IF (1:100) |
| Antibody | Anti-PECAM-1(Rat, monoclonal) | Pharmigen | Cat# 553,370 | IF (1:100) |
| Antibody | Anti-CD31(Goat, polyclonal) | R&D Systems | Cat# FAB3628 | IF (1:100) |
| Antibody | Anti-Pdgfrβ (Goat, polyclonal) | R&D Systems | Cat# AF1042 | IF (1:100) |
| Antibody | Anti-NG2 (Rabbit, polyclonal) | Millipore | Cat# AB5320 | IF (1:100) |
| Antibody | Anti-BCAM (Goat, polyclonal) | R&D Systems | Cat# AF8299 | IF (1:50) |
| Antibody | Anti-ATP6V1B1+ ATP6V1B2 (Rabbit, polyclonal) | Abcam | Cat# ab200839 | IF (1:100) |
| Antibody | Anti-Aggrecan (Rabbit, polyclonal) | Millipore | Cat# AB1031 | IF (1:100) |
| Antibody | Anti-Sox9 (Goat, polyclonal) | R&D Systems | Cat# AF3075 | IF (1:100) |
| Antibody | Anti-Perilipin (Rabbit, polyclonal) | Cell Signaling | Cat# 9349 | IF (1:100) |
| Antibody | Anti-Osterix (Rabbit, polyclonal) | Abcam | Cat# ab22552 | IF (1:1000) |
| Antibody | Anti-Collagen Type I (Rabbit, polyclonal) | Millipore | Cat# AB765P | IF (1:200) |
| Antibody | Anti-Osteopontin (Goat, polyclonal) | R&D Systems | Cat# AF808 | IF (1:100) |
| Antibody | Anti-Osteocalcin (Rabbit, polyclonal) | LifeSpan BioSciences | Cat# LS-C17044 | IF (1:100) |
| Antibody | Anti-Runx2 (Rabbit, monoclonal) | R&D Systems | Cat# MAB2006 | IF (1:50) |
| Antibody | Anti-Dll4 (Goat, polyclonal) | R&D Systems | Cat# AF1389 | IF (1:100)WB (1:200) |
| Antibody | Anti-Jag1 (Goat, polyclonal) | Sigma | Cat# J4127 | IF (1:100)WB (1:500) |
| Antibody | Anti-GAPDH (Rabbit, monoclonal) | Ambion | Cat# AM4300 | WB (1:1000) |
| Antibody | Anti-CD45-FITC (Rat, monoclonal) | eBioscience | Cat# 11–0451 | |
| Antibody | Anti-CD8-Biotin (Rat, monoclonal) | eBioscience | Cat# 13–0081 | |
| Antibody | Anti-CD4-APC (Rat, monoclonal) | eBioscience | Cat# 17-0042-82 | |
| Antibody | Streptavidin PE/Cy7 | Thermo Scientific | Cat# SA1012 | |
| Antibody | Lineage Cell Depletion Kit | Miltenyi Biotec | Cat# 130-090-858 | |
| Antibody | CD45 Microbeads | Miltenyi Biotec | Cat# 130-052-301 | |
| Antibody | CD117 Microbeads | Miltenyi Biotec | Cat# 130-091-224 | |
| Antibody | Ter-119 Microbeads | Miltenyi Biotec | Cat# 130-049-901 | |
| Antibody | Anti-rabbit Alexa Fluor-488 (Donkey, polyclonal) | Invitrogen | Cat# A21206 | IF (1:500) |
| Antibody | Anti-rabbit Alexa Fluor-594 (Donkey, polyclonal) | Invitrogen | Cat# A21207 | IF (1:500) |
| Antibody | Anti-rabbit Alexa Fluor-647 (Donkey, polyclonal) | Invitrogen | Cat# A31573 | IF (1:500) |
| Antibody | Anti-rat Alexa Fluor-488 (Donkey, polyclonal) | Invitrogen | Cat# A21208 | IF (1:500) |
| Antibody | Anti-rat Alexa Fluor-594 (Donkey, polyclonal) | Invitrogen | Cat# A21209 | IF (1:500) |
| Antibody | Anti-rat Alexa Fluor-647 (Donkey, polyclonal) | Jackson ImmunoResearch | Cat# 712-605-153 | IF (1:500) |
| Antibody | Anti-rat Alexa Fluor-647 (Donkey, polyclonal) | Invitrogen | Cat# A21247 | IF (1:500) |
| Antibody | Anti-goat Alexa Fluor-488 (Donkey, polyclonal) | Invitrogen | Cat# A11055 | IF (1:500) |
| Antibody | Anti-goat Alexa Fluor-647 (Donkey, polyclonal) | Invitrogen | Cat# A21447 | IF (1:500) |
| Antibody | Anti-goat Alexa Fluor-647 (Donkey, polyclonal) | Thermo Scientific | Cat# A32849 | IF (1:500) |
| Antibody | Anti-rabbit IgG HRP-linked (Goat) | Cell Signaling | Cat# 7074 | WB (1:15000) |
| Antibody | Anti-goat IgG HRP-linked (Donkey) | Antibody online | Cat# ABIN1536502 | WB (1:15000) |
| Antibody | Anti-mouse IgG 656G HRP-linked (Sheep) | GE Healthcare | Cat# NA931 | WB (1:40000) |
| Antibody | Anti-goat IgG (H + L) Peroxidase AffiniPure Bovine | Jackson ImmunoResearch | Cat# 805-035-180 | WB (1:15000) |
| Sequence-based reagent | Human GAPDH Endogenous Control (VIC/MGB probe, primer limited) | Applied Biosystems | Cat# 4326317E | TaqMan probe Hs99999905_m1 |
| Sequence-based reagent | Human ACTB Endogenous Control (VIC/MGB probe, primer limited) | Applied Biosystems | Cat# 4326315E | TaqMan probe Hs99999903_m1 |
| Sequence-based reagent | Human HEY1 TaqMan Gene Expression Assay (FAM) | Applied Biosystems | Cat# 4331182 | TaqMan probe Hs00232618_m1 |
| Sequence-based reagent | Human DLL4 TaqMan Gene Expression Assay (FAM) | Applied Biosystems | Cat# 4331182 | TaqMan probe Hs00184092_m1 |
| Sequence-based reagent | Human HEY2 TaqMan Gene Expression Assay (FAM) | Applied Biosystems | Cat# 4331182 | TaqMan probe Hs00232622_m1 |
| Sequence-based reagent | Human HES1 TaqMan Gene Expression Assay (FAM) | Applied Biosystems | Cat# 4331182 | TaqMan probe Hs00172878_m1 |
| Sequence-based reagent | Human EFNB2 TaqMan Gene Expression Assay (FAM) | Applied Biosystems | Cat# 4331182 | TaqMan probe Hs00187950_m1 |
| Sequence-based reagent | Human NRARP TaqMan Gene Expression Assay (FAM) | Applied Biosystems | Cat# 4331182 | TaqMan probe Hs01104102_s1 |
| Peptide, recombinant protein | Recombinant Human PTH (1-34) | BACHEM | Cat# H-4835-GMP, 4033364 | |
| Commercial assay or kit | BCA Protein Assay Kit | Pierce | Cat# 23225 | |
| Commercial assay or kit | RNeasy Plus Micro Kit | QIAGEN | Cat# 74034 | |
| Commercial assay or kit | iScript cDNA Synthesis Kit | BIO-RAD | Cat# 170–8891 | |
| Commercial assay or kit | SsoAdvanced Universal Probes Supermix | BIO-RAD | Cat# 172–5284 | |
| Commercial assay or kit | LEGENDplex Mouse Inflammation Panel (13-plex) with V-bottom plates | BioLegend | Cat# 740446 | |
| Commercial assay or kit | Anticoagulant EDTA-treated Microvettes | Sarstedt | Cat# 20.1341 | |
| Commercial assay or kit | BD Rhapsody Whole Transcriptome Analysis (WTA) Amplification kit | BD Biosciences | Cat# 633,801 | |
| Commercial assay or kit | Agencourt AMPure XP magnetic beads | Beckman Coulter Life Sciences | Cat# A638880 | |
| Commercial assay or kit | Disposable polystyrene columns | Thermo Scientific | Cat# 29922 | |
| Commercial assay or kit | Amicon Ultra-0.5 Centrifugal Filter | Millipore | Cat# UFC500396 | |
| Commercial assay or kit | Pierce Slide-A-Lyzer 10K MWCO Dialysis Cassettes | Thermo Scientific | Cat# 66380 | |
| Chemical compound, drug | Ni-NTA agarose resin | Qiagen | Cat# 302010 | |
| Chemical compound, drug | Sucrose | Sigma | Cat# S0389 | |
| Chemical compound, drug | cOmplete ULTRA Tablets Protease Inhibitor Cocktail | Roche | Cat# 05892970001 | |
| Chemical compound, drug | Phosphatase inhibitor cocktail set V | EMD Millipore | Cat# 524629 | |
| Chemical compound, drug | SimplyBlue SafeStain | Invitrogen | Cat# LC6060 | |
| Chemical compound, drug | Gelatine | Sigma | Cat# G1890 | |
| Chemical compound, drug | Polyvinylpyrrolidone | Sigma | Cat# P5288 | |
| Chemical compound, drug | Trypsin-EDTA solution | Sigma | Cat# T3924 | |
| Chemical compound, drug | Paraformaldehyde | Sigma | Cat# P6148 | |
| Chemical compound, drug | 70 kDa Dextran, Texas Red Lysine fixable | Molecular Probes | Cat# D1864 | |
| Chemical compound, drug | Calcein | Sigma | Cat# C0875 | |
| Chemical compound, drug | Fluoromount-G | Southern Biotech | Cat# 0100–01 | |
| Chemical compound, drug | ECL Prime Western Blotting Detection Reagent | GE-Healthcare | Cat# RPN2236 | |
| Chemical compound, drug | 4-Hydroxy tamoxifen | Sigma | Cat# H7904 | |
| Chemical compound, drug | Hematoxilin | Sigma | Cat# MHS16 | |
| Chemical compound, drug | Dimethyl sulfoxide | Sigma | Cat# D8418 | |
| Chemical compound, drug | MEM200 endothelial cells medium | ThermoFisher | Cat# M200500 | |
| Chemical compound, drug | LSGS | ThermoFisher | Cat# S00310 | |
| Chemical compound, drug | EBM-2 endothelial cells medium | Lonza | Cat# CC-3156 | |
| Chemical compound, drug | EGM-2 Single Quots | Lonza | Cat# CC-4176 | |
| Chemical compound, drug | Opti-MEM | Gibco | Cat# 31985–047 | |
| Chemical compound, drug | Poly-L-lysine | Sigma | Cat# P6282 | |
| Chemical compound, drug | HEPES | Sigma | Cat# H3537 | |
| Chemical compound, drug | Ketamine | Zoetis | Cat# 344771 | |
| Chemical compound, drug | Rompum | Bayer HealthCare | Cat# D-51368 | |
| Chemical compound, drug | Collagenase I | Gibco | Cat# 17100–017 | |
| Chemical compound, drug | Collagenase IV | Gibco | Cat# 17104–019 | |
| Chemical compound, drug | Dispase | Gibco | Cat# 17105–041 | |
| Software, algorithm | ImageJ (v2.0.0 Fiji) | Schindelin et al., 2012 | ||
| Software, algorithm | Volocity (v6.3) | Perkin Elmer | ||
| Software, algorithm | Illustrator (vCC2018) | Adobe | ||
| Software, algorithm | GraphPad Prism7 | GraphPad Software | ||
| Software, algorithm | FlowJo (v10.3) | BD Life Sciences | ||
| Other | DAPI stain | Sigma | Cat# D9542 | (1 mg/mL) |





