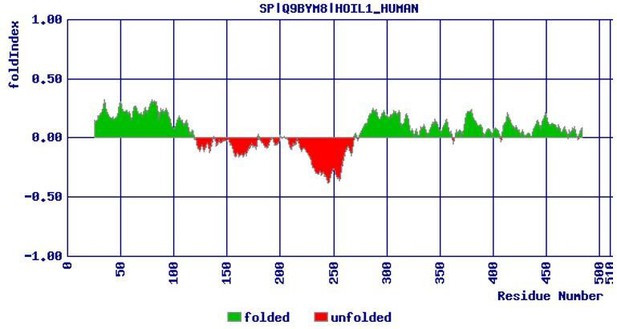
The linear ubiquitin chain assembly complex (LUBAC) generates heterotypic ubiquitin chains
Figures
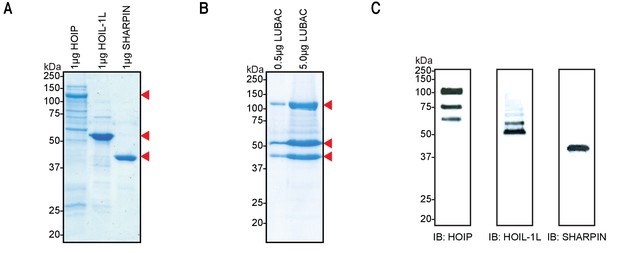
Co-expression and purification of linear ubiquitin chain assembly complex (LUBAC) yields high-quality protein.
(A) SDS-PAGE analysis of individually purified LUBAC components. (B) SDS-PAGE analysis of co-expressed and purified LUBAC. (C) Immunoblot analysis of co-purified LUBAC.
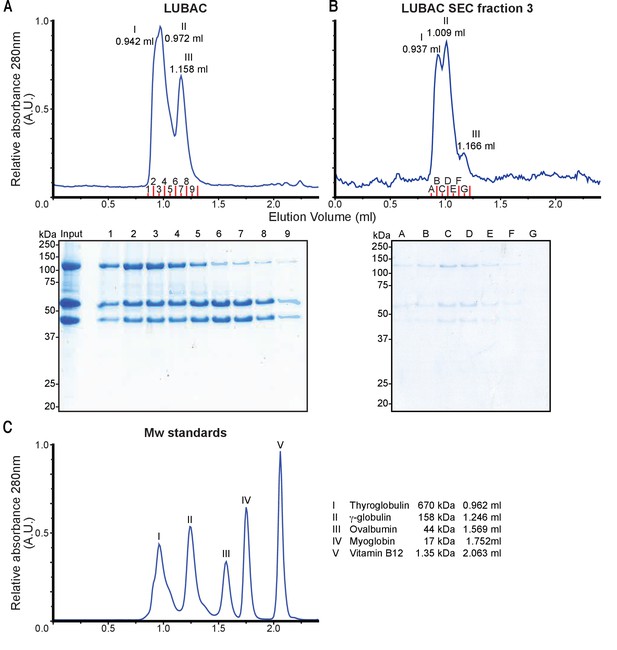
Gel filtration analysis of linear ubiquitin chain assembly complex (LUBAC) showing presence of multiple populations with different oligomeric states.
(A) Gel filtration profile of purified LUBAC separated over S200 column. (B) Tandem gel filtration separation of fraction 3 re-run over S200 column. (C) Molecular weight standards separated over S200 column.
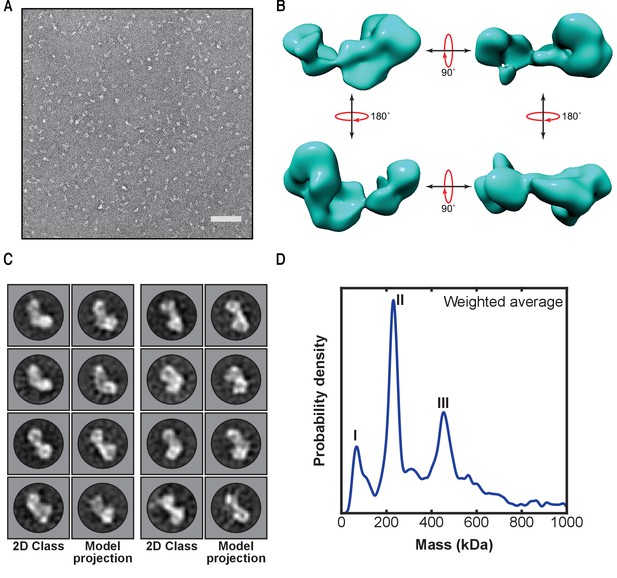
First low-resolution 3D map of linear ubiquitin chain assembly complex (LUBAC) obtained by negative staining electron microscopy of the recombinant complex.
(A) Representative negative stain transmission electron micrograph of recombinant LUBAC. Scale bar: 100 nm. (B) 3D refined model of LUBAC obtained by single particle analysis of negative stained electron micrographs. (C) LUBAC 2D class averages matched to projections made from 3D refined map. (D) Mass photometry measurements of LUBAC indicate formation of a ternary complex with 1:1:1 stoichiometry that can form dimers.
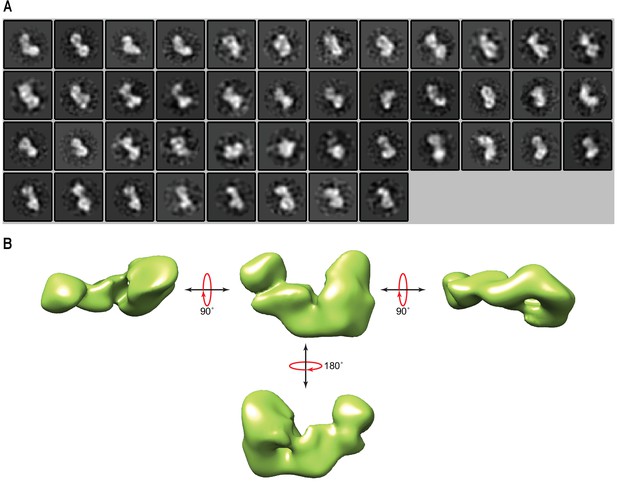
Modelling of the linear ubiquitin chain assembly complex (LUBAC) by negative staining electron microscopy.
(A) LUBAC 2D class averages obtained from negatively stained particles. (B) Initial 3D model of LUBAC made from particles picked in negatively stained electron micrographs.
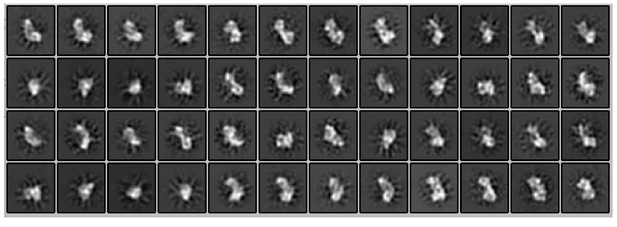
Projections made from 3D refined model of linear ubiquitin chain assembly complex (LUBAC).
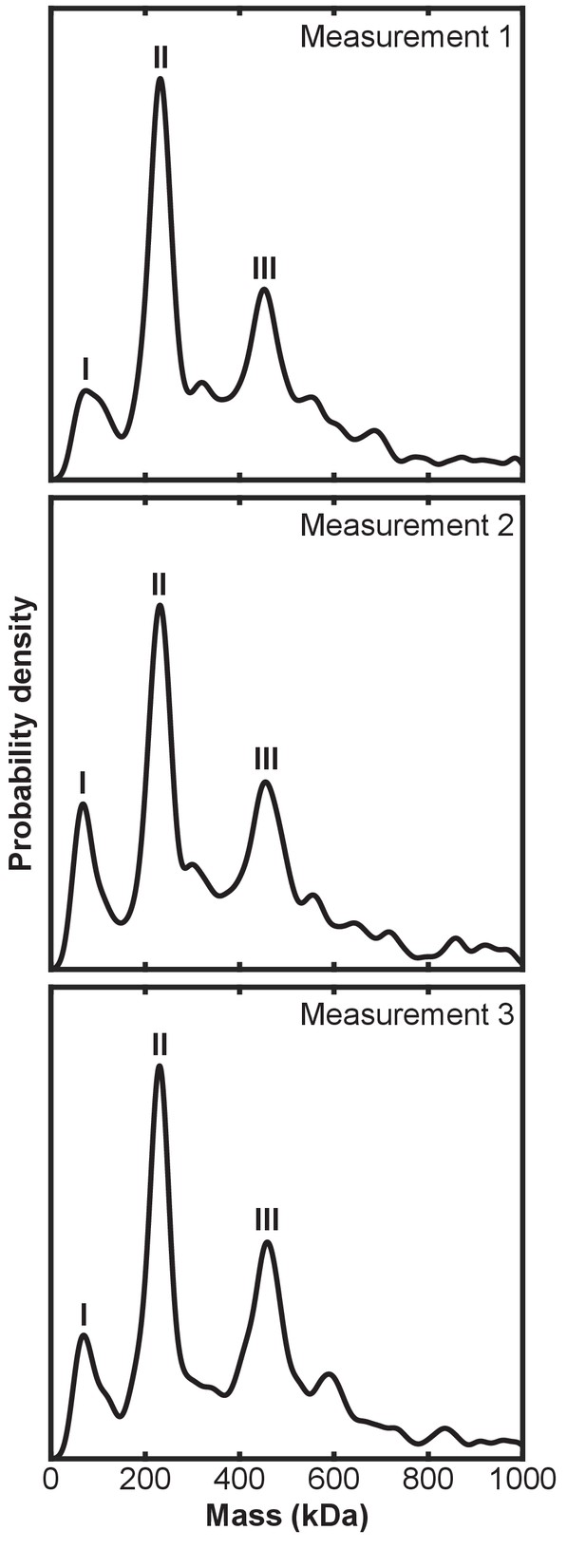
Independent mass photometry measurements of linear ubiquitin chain assembly complex (LUBAC).
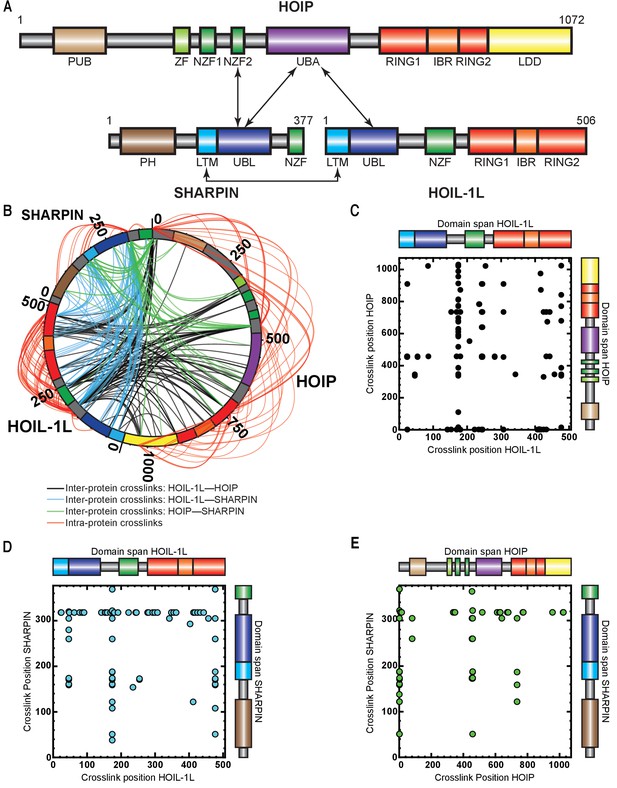
Cross-linking mass spectrometry (MS) analysis shows proximity between the catalytic domains of HOIL-1-interacting protein (HOIP) and heme-oxidized IRP2 ubiquitin ligase 1 (HOIL-1L).
(A) Schematic representation of linear ubiquitin chain assembly complex (LUBAC) components with their domains and known interactions. (B) Circos plot of inter-protein cross-links formed between LUBAC components. (C) Detected inter-protein cross-links formed between HOIL-1L and HOIP. (D) Detected inter-protein cross-links formed between HOIL-1L and Shank-associated RH domain-interacting protein (SHARPIN). (E) Detected inter-protein cross-links formed between HOIP and SHARPIN.
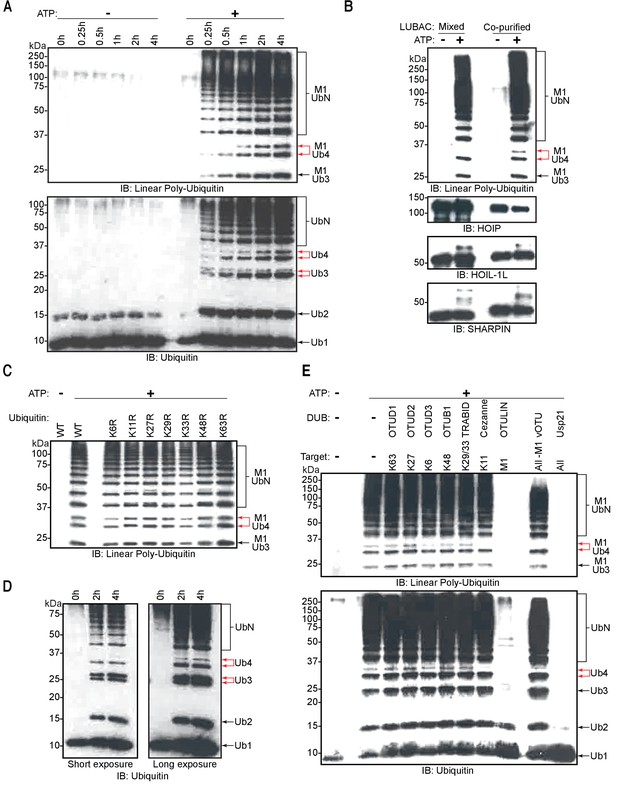
Linear ubiquitin chain assembly complex (LUBAC) assembles heterotypic poly-ubiquitin chains containing M1 and non-Lys linkages in vitro.
(A) Time course of co-purified LUBAC in vitro ubiquitin chain assembly reaction. (B) Comparison of in vitro chain assembly between HOIL-1-interacting protein (HOIP), heme-oxidized IRP2 ubiquitin ligase 1 (HOIL-1L), and Shank-associated RH domain-interacting protein (SHARPIN) mixed at 1:1:1 molar ratio versus co-purified LUBAC. (C) LUBAC in vitro chain assembly using different ubiquitin K to R mutants. (D) LUBAC in vitro chain assembly using K0 ubiquitin. (E) Ubiquitin chain restriction (UbiCRest) analysis of poly-ubiquitin chains assembled by LUBAC in vitro. All experiments were performed in triplicate representative results are shown.

Anti-linear ubiquitin antibody validation.
(A) Detection of mono-ubiquitin and di-ubiquitin of different linkages by anti-linear ubiquitin antibody. (B) Detection of longer Lys48 and Lys63-linked ubiquitin chains by anti-linear ubiquitin antibody.
Independently purified HOIL-1-interacting protein (HOIP), heme-oxidized IRP2 ubiquitin ligase 1 (HOIL-1L), and Shank-associated RH domain-interacting protein (SHARPIN) mixed at an equimolar ratio cannot reconstitute the trimeric linear ubiquitin chain assembly complex (LUBAC).
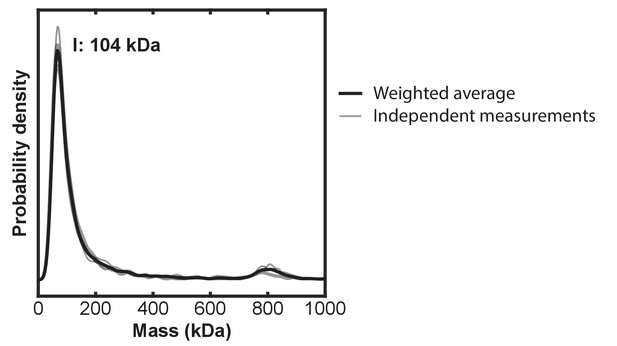
Independent mass photometry (MP) measurements and weighted average made from HOIP, HOIL-1L, and SHARPIN mixed to a final concentration of 4 pM of each protein.
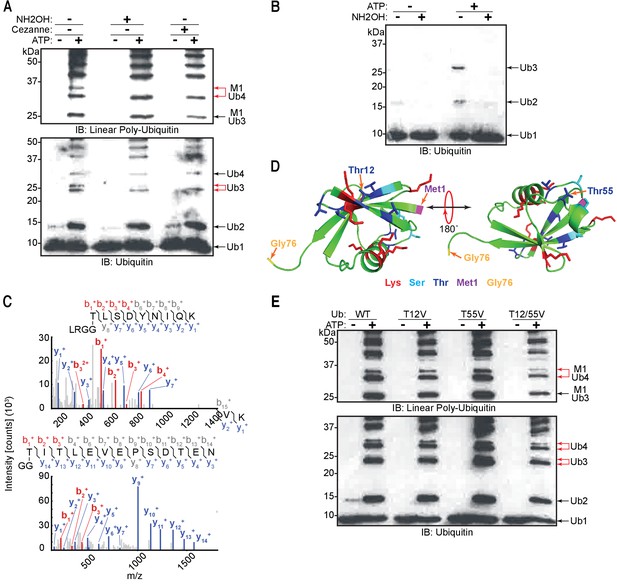
Linear ubiquitin chain assembly complex (LUBAC) assembles heterotypic poly-ubiquitin chains containing M1 and ester bond linkages at T12 and T55.
(A) Treatment of LUBAC-assembled heterotypic poly-ubiquitin chains with hydroxylamine. (B) Hydroxylamine treatment of ubiquitin polymers assembled by LUBAC using N-terminally blocked ubiquitin. (C) MS/MS spectra of ubiquitin polymerized at T55 (top) and T12 (bottom). Poly-ubiquitin chains assembled by LUBAC were separated by SDS-PAGE, bands were cut from the gel and subjected to mass spectrometry analysis. (D) Positions of Thr12 and Thr55 on structure of ubiquitin (PDB:1UBI). (E) Assembly of ubiquitin chains by LUBAC using different ubiquitin Thr to Val point mutants as substrates. All experiments were performed in triplicate representative results are shown.
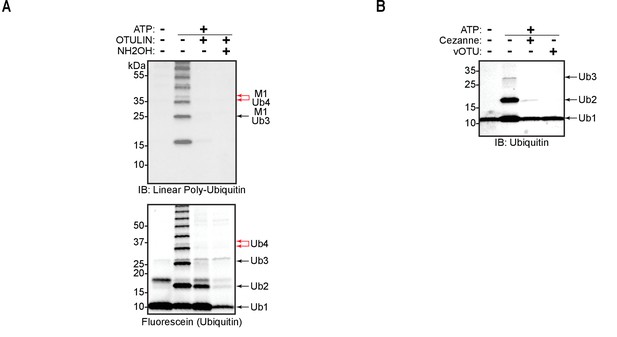
Cezanne, vOTU, and hydroxylamine can cleave oxyester bonds in linear ubiquitin chain assembly complex (LUBAC)-assembled heterotypic ubiquitin chains.
(A) Combined OTULIN and hydroxylamine treatment of LUBAC-assembled poly-ubiquitin chains. (B) Cezanne and vOTU ubiquitin chain restriction (UbiCRest) analysis of ubiquitin polymers assembled by LUBAC using N-terminally blocked ubiquitin.
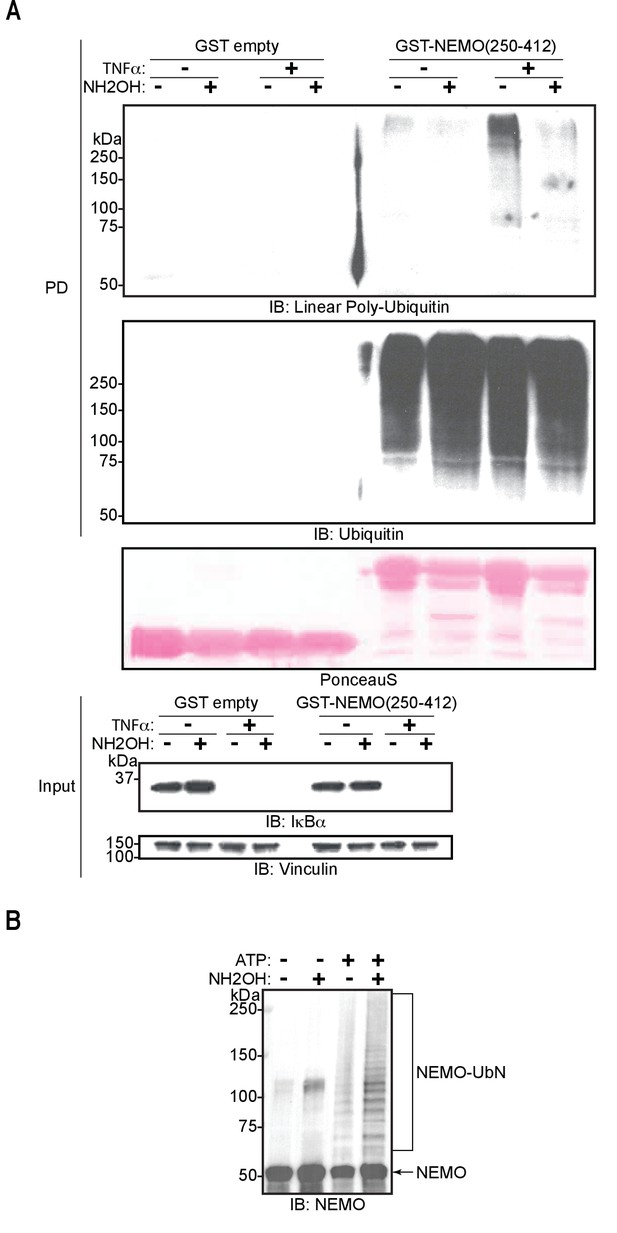
M1-linked/linear ubiquitin chains generated in cells and conjugated to NEMO in vitro are sensitive to hydroxylamine treatment.
(A) Hydroxylamine treatment of M1 linkage-containing poly-ubiquitin chains assembled in response to TNF in wild-type (WT) mouse embryonic fibroblasts (MEFs). MEFs were treated with TNF for 15 min and lysed, lysates were subjected to GST PD using GST or GST-NEMO(250-412), beads were treated with buffer or hydroxylamine for 30 min, bound ubiquitin species were then analysed by immunoblotting. (B) Treatment of linear ubiquitin chain assembly complex (LUBAC)-dependent NEMO in vitro ubiquitination reactions with hydroxylamine. All experiments were performed in triplicate representative results are shown.
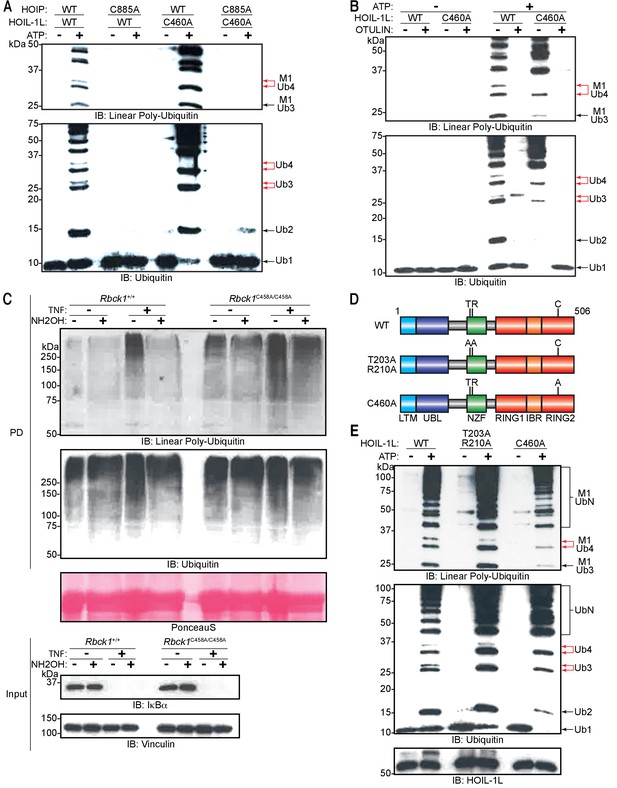
Heme-oxidized IRP2 ubiquitin ligase 1 (HOIL-1L) generates ester linkages on heterotypic chains but requires HOIL-1-interacting protein (HOIP) catalytic activity to polymerize ubiquitin.
(A) Comparison of linear ubiquitin chain assembly complex (LUBAC) in vitro chain assembly by complexes containing different catalytically inert mutants of HOIP and HOIL-1L. (B) OTULIN restriction of poly-ubiquitin chains assembled by LUBAC containing wild-type (WT) or catalytically inert HOIL-1L. (C) Hydroxylamine treatment of M1 linkage-containing poly-ubiquitin chains assembled in response to TNF in WT and Rbck1C458A/C458A mouse embryonic fibroblast (MEF) cells. Cells were treated with TNF (50 ng/ml) for 15 min and lysed, lysates were subjected to GST PD using GST-NEMO (250-412), beads were treated with buffer or hydroxylamine for 30 min, bound ubiquitin species were then analysed by immunoblotting. (D) Schematic representations of different HOIL-1L mutants. (E) Comparison of LUBAC in vitro chain assembly by complexes containing different HOIL-1L mutants. All experiments were performed in triplicate representative results are shown.
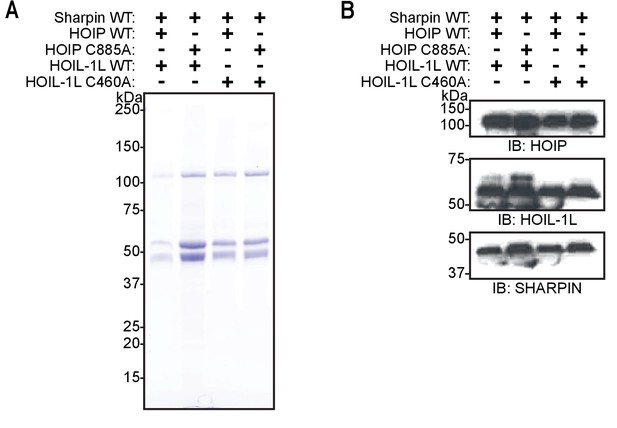
Purification of linear ubiquitin chain assembly complex (LUBAC) containing catalytically inert HOIL-1-interacting protein (HOIP) and heme-oxidized IRP2 ubiquitin ligase 1 (HOIL-1L) proteins.
(A) SDS-PAGE analysis of different purified LUBAC. (B) Immunoblot analysis of different purified LUBAC.
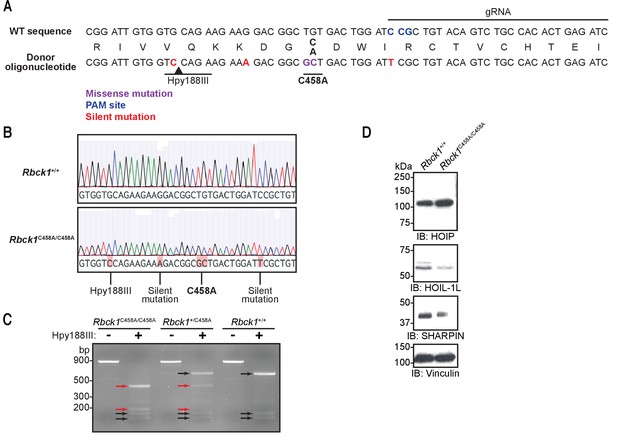
Generation of Hoil-1lC458A/C458A mice.
(A) Sequences of genomic DNA around C458 codon, gRNAs, and donor oligonucleotide used to target heme-oxidized IRP2 ubiquitin ligase 1 (HOIL-1L) C458A mutation. (B) Sanger sequencing confirming correct mutations at target sites. (C) Genotyping results of Hoil-1l+/+, Hoil-1l+/C458A, and Hoil-1lC458A/C458A mice. Hpy188III digest of a PCR fragment confirming correct targeting where a silent mutation is inserted. (D) Immunoblot analysis of linear ubiquitin chain assembly complex (LUBAC) component expression in mouse embryonic fibroblasts (MEFs) derived from Hoil-1l+/+ and Hoil-1lC458A/C458A mice.
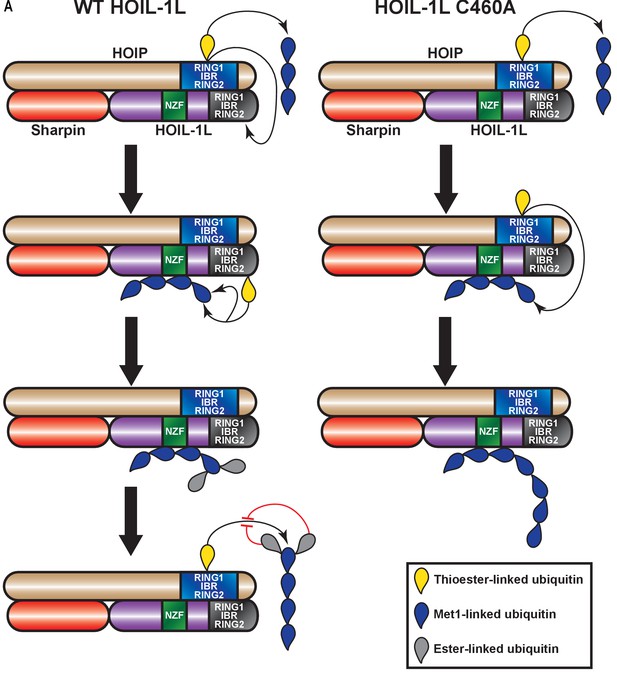
Proposed model for concerted action between HOIL-1-interacting protein (HOIP) and heme-oxidized IRP2 ubiquitin ligase 1 (HOIL-1L) in heterotypic chain assembly.
HOIP and HOIL-1L assemble heterotypic chains through a Cys relay mechanism. HOIP forms a thioester bond to ubiquitin, which can be either transferred to a thioester bond on HOIL-1L or added to a nascent linear ubiquitin chain. HOIL-1L subsequently binds the linear ubiquitin chain through its Npl zinc finger (NZF) domain and branches it with ester linkages. The resulting heterotypic poly-ubiquitin chains contain predominantly linear linkages with ester-linked branches.
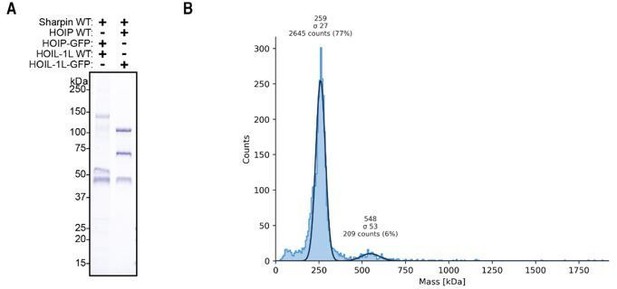
GFP-tagged LUBAC purification.
(A) Purification of LUBAC containing GFP-tagged HOIP or HOIL-1L. (B) Mass photometry measurement of GFP-tagged LUBAC showing 27 kDa shift in the monomeric (250kDa) and dimeric (500kDa) mass of the complex.
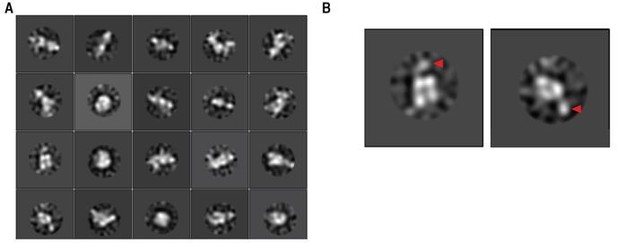
GFP-tagged LUBAC class averages.
(A) Class averages of GFP-tagged LUBAC. (B) Selected class averages that may show GFP tag.
Additional files
-
Supplementary file 1
Population masses determined by mass photometry.
- https://cdn.elifesciences.org/articles/60660/elife-60660-supp1-v2.xlsx
-
Supplementary file 2
Unique inter-protein cross-links detected by linear ubiquitin chain assembly complex (LUBAC) 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium tetrafluoroborate (DMTMM ) XL-MS analysis.
- https://cdn.elifesciences.org/articles/60660/elife-60660-supp2-v2.xlsx
-
Supplementary file 3
Unique intra-protein cross-links detected by linear ubiquitin chain assembly complex (LUBAC) 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium tetrafluoroborate (DMTMM) XL-MS analysis.
- https://cdn.elifesciences.org/articles/60660/elife-60660-supp3-v2.xlsx
-
Supplementary file 4
Primers.
- https://cdn.elifesciences.org/articles/60660/elife-60660-supp4-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60660/elife-60660-transrepform-v2.docx