Erythrocyte CD55 mediates the internalization of Plasmodium falciparum parasites
Figures
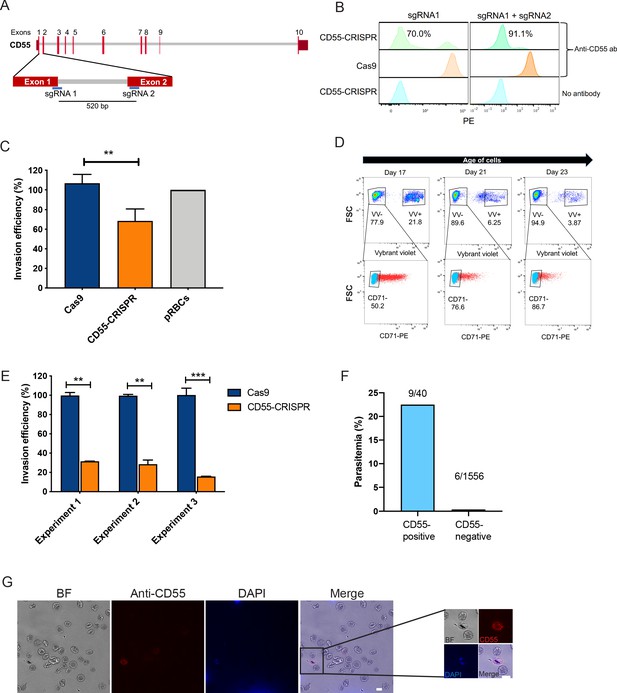
CD55 is required for P. falciparum invasion.
(A) Schematic of CD55 gene structure showing targeting sites of two single guide RNAs (sgRNAs). Vertical red lines indicate the positions of CD55 exons. (B) Expression of CD55 on mutant (CD55-CRISPR) cRBCs generated with one sgRNA (left) or two sgRNAs (right), as compared to control (Cas9) cRBCs. (C) Invasion efficiency of P. falciparum 3D7 in Day 18 CD55-CRISPR cRBCs compared to isogenic controls (Cas9), relative to the invasion efficiency in peripheral blood erythrocytes (N = 4 biological replicates; n = 3 technical replicates; error bars indicate SEM; **p<0.01). CD55-null cRBCs were generated using dual sgRNAs: sgRNA1, CD55-Cr1 and sgRNA2, CD55-Cr8. (D) Time course of expression of CD71 on cRBCs harvested on different days of differentiation. Enucleated versus nucleated cells were gated using a nuclear dye. (E) Invasion efficiency of P. falciparum strain 3D7 in CD71-negative, CD55-CRISPR cRBCs compared to CD71-negative isogenic controls (Cas9). Invasion efficiency is presented relative to the mean of Cas9 control. Three independent biological replicates are shown; error bars represent SEM (n = 2 or three technical replicates; **p<0.01, ***p<0.005). CD55-null cRBCs were generated using dual sgRNAs. (F) Parasitemia in population of CD55-CRISPR cRBCs, in which ~ 90% of the cells lack CD55 (CD55-negative), and the remaining are CD55-positive, as quantified by immunofluorescence assays. (G) Representative images of CD55-CRISPR cRBCs, including one P. falciparum-infected cell (magnified). Images are brightfield with fluorescence overlaid. Blue, dapi. Red, anti-CD55-PE. Scale bars indicate 10 µm.
-
Figure 1—source data 1
Source data for invasion assays.
- https://cdn.elifesciences.org/articles/61516/elife-61516-fig1-data1-v2.xlsx
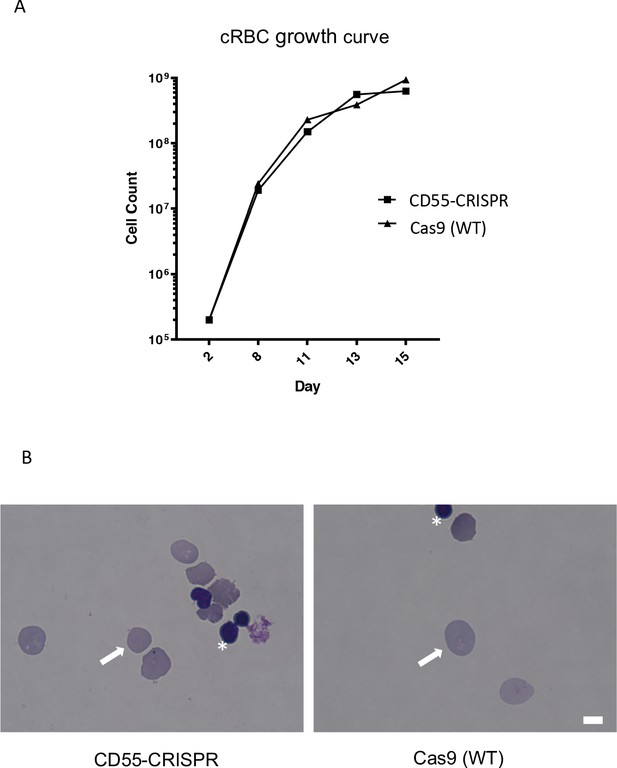
Deletion of CD55 does not impact growth or maturation of cRBCs derived from CD34 +primary human HSPCs.
(A) Growth rate of CD55-null erythroid progenitors from CD34 +HSPCs over 15 days of differentiation, compared to isogenic wild-type cells. Results are from one representative experiment. (B) Day 20 cRBCs stained with May-Grünwald and Giemsa, and visualized by light microscopy. Enucleation rate of ~90.0% was observed in both CD55-null and Cas9 (WT) cRBCs. White arrows indicate enucleated cells. Asterisks indicate free nuclei. Scale bar 5 µM.
-
Figure 1—figure supplement 1—source data 1
Source data for growth curves.
- https://cdn.elifesciences.org/articles/61516/elife-61516-fig1-figsupp1-data1-v2.xlsx
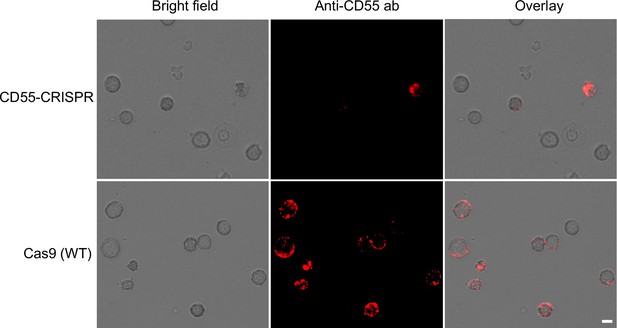
Expression of CD55 on cRBCs.
Immunofluorescence assays showing the expression of CD55 on the surface of CD55-CRISPR and Cas9 (WT) cRBCs using anti-CD55-PE antibody. Scale bar 5 µM.

Reticulocyte staining of day 17 cRBCs.
Reticulocytes are indicated by the purple reticular staining pattern. Scale bar 5 µM.
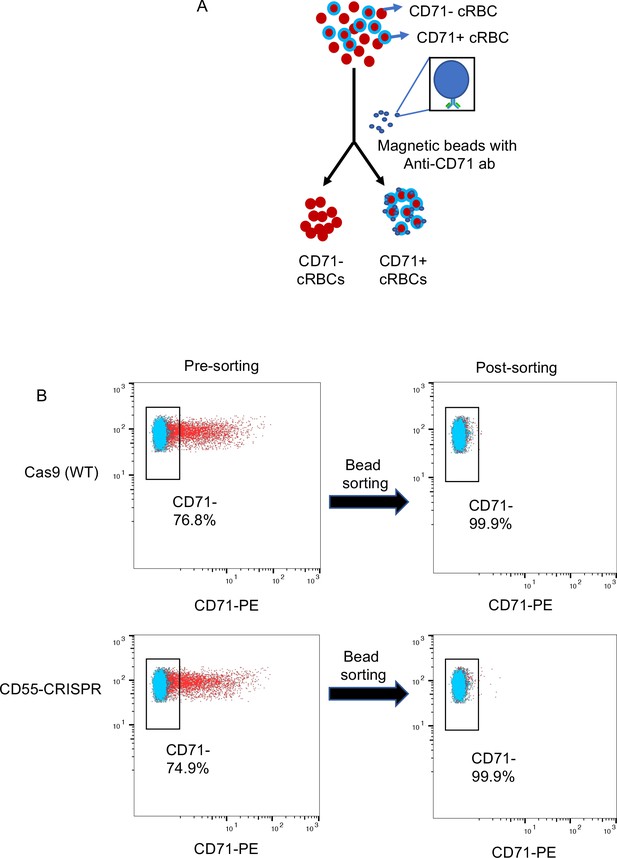
Enrichment of CD71- cRBCs by immunolabeled magnetic beads.
(A) Schematic showing the separation of CD71 +cRBCs from CD71- cRBCs using anti-CD71 antibody-immobilized magnetic beads. (B) Flow cytometric analysis of CD71 expression on cRBCs pre- and post-sorting using anti-CD71 antibody immobilized magnetic beads.
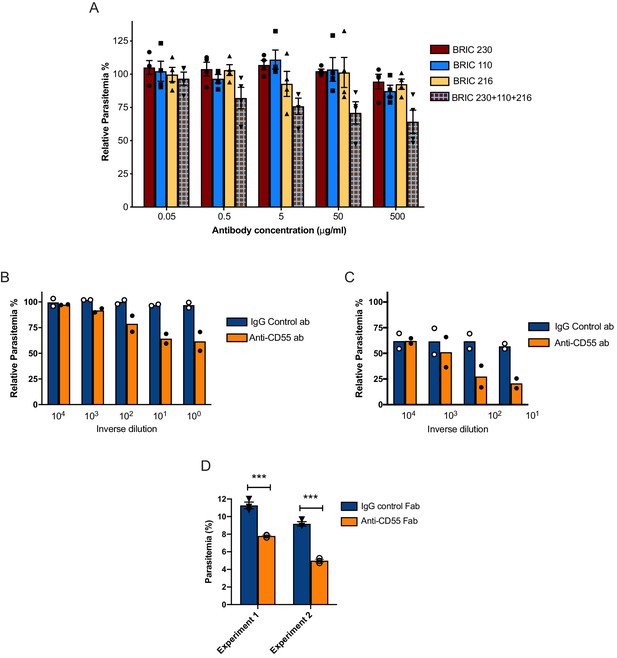
Blocking CD55 with antibody inhibits growth of P. falciparum.
(A) P. falciparum strain 3D7 parasitemia after 72 hr of growth in RBCS in the presence of increasing concentrations of anti-CD55 monoclonal antibodies, relative to isotype control antibody (BRIC 170) at the same concentration. For the pooled antibodies, the indicated concentration was the total combined value, and there were equimolar amounts of each antibody. (N = 3 biological replicates; n = 2 technical replicates). Error bars indicate SEM. (B) Parasitemia of P. falciparum strain 3D7 after 72 hr growth in non-enzyme-treated RBCs with increasing concentrations of polyclonal anti-CD55 IgG antibody, relative to that in isotype control antibody at same concentration. (N = 2 biological replicates; n = 2 technical replicates). The highest antibody concentration (100) was 400 µg/ml. (C) As in B, but with neuraminidase-treated RBCs. The highest antibody concentration (101) was 40 µg/ml. (D) P. falciparum strain 3D7 parasitemia after 72 hr growth in 400 µg/ml Fab fragments generated from anti-CD55 polyclonal antibody or isogenic control. Error bars indicate SEM. ***p<0.005. The starting parasitemias were 0.3% (Experiment 1) or 0.5% (Experiment 2).
-
Figure 2—source data 1
Source data for antibody inhibition assays.
- https://cdn.elifesciences.org/articles/61516/elife-61516-fig2-data1-v2.xlsx
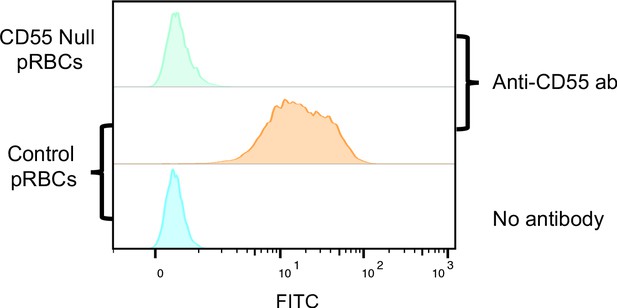
Specificity of rabbit polyclonal anti-CD55 antibody.
Flow cytometric analysis of control and CD55-null RBCs incubated with rabbit polyclonal anti-CD55 antibody. The anti-CD55 polyclonal antibody detects CD55 on wild-type pRBCs but not CD55-null RBCs.
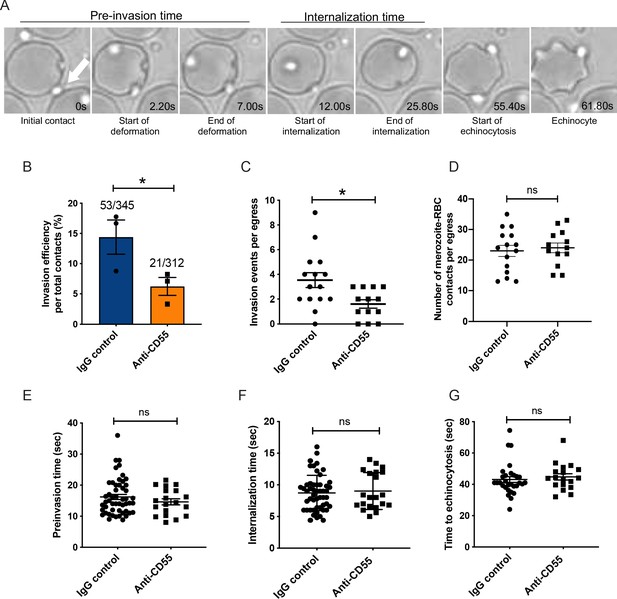
Blocking CD55 with antibody inhibits invasion but not pre-invasion kinetics.
(A) Time-lapse images of invasion after initial merozoite contact. Arrowhead indicates invading merozoite. Time in seconds. (B) Percentage of merozoites that invaded an RBC after initial contact in presence of polyclonal anti-CD55 antibody (anti-CD55) or isotype control (IgG control). Bottom number is the total number of merozoites followed that made contact with the RBC, and top number is the subset that invaded. The data were acquired in three independent experiments, and the dots indicate the mean invasion efficiency from each experiment; *p=0.03. (C) Number of successful invasion events per schizont rupture (egress) in presence of anti-CD55 antibody or isotype control; *p=0.01. The data were acquired in three independent experiments and the dots indicate individual egress events. (D) Number of merozoites that made contact with an RBC (>0.5 s) in the presence of anti-CD55 antibody or isotype control, per egress event. The data were acquired in three independent experiments. (E) Pre-invasion time (in s) for merozoites that contact an RBC in the presence of anti-CD55 antibody or isotype control. (F) Internalization time (in s) for merozoites in the presence of anti-CD55 antibody or isotype control antibody. (G) Time to echinocytosis from the end of merozoite internalization in the presence of anti-CD55 antibody or isotype control. Error bars indicate SEM; ns, not significant (B– G).
-
Figure 3—source data 1
Source data for live microscopy studies.
- https://cdn.elifesciences.org/articles/61516/elife-61516-fig3-data1-v2.xlsx
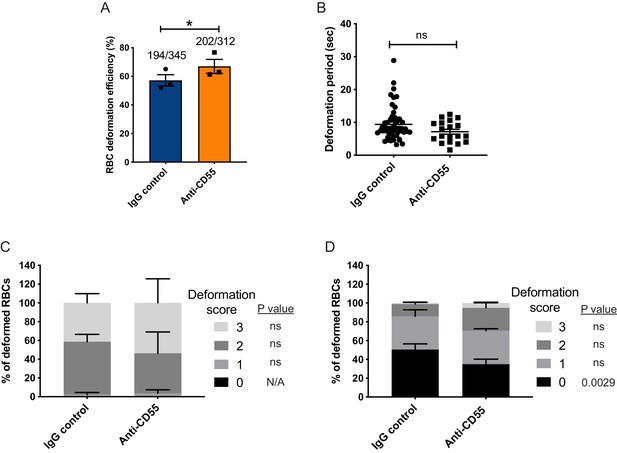
Merozoite-induced erythrocyte deformation is not affected by blocking CD55.
(A) Efficiency of RBC deformation by merozoites that make contact in presence of anti-CD55 or IgG control. The fractions indicate the number of deformed RBCs out of the total merozoite-RBC contacts observed. The data were acquired in three independent experiments, and the dots represent the mean for each experiment.; *p=0.02. (B) Duration of erythrocyte deformation induced by attached merozoites that ultimately invaded in presence of polyclonal anti-CD55 antibody (anti-CD55) or isotype control (IgG control). Time in seconds. (C–D) Strength of merozoite-induced deformation in presence of anti-CD55 or isotype control for cases where invasion was successful (C) or not successful (D); N/A, not applicable; ns, not significant, *p=0.0029. Error bars indicate SEM (A–D).
-
Figure 4—source data 1
Source data for merozoite-induced deformation experiments.
- https://cdn.elifesciences.org/articles/61516/elife-61516-fig4-data1-v2.xlsx
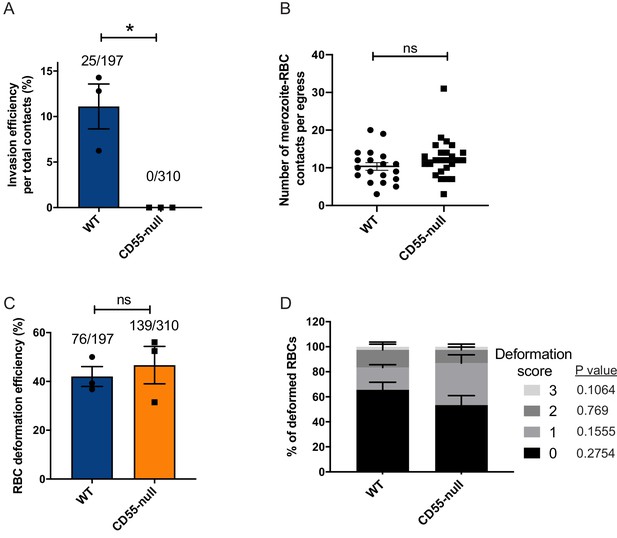
Absence of CD55 prevents invasion but does not impact deformation.
(A) Percentage of merozoites that invaded wild-type (WT) or CD55-null RBCs after initial contact. Bottom number is the total number of merozoites followed that made contact with the RBC, and top number is the subset that invaded. The data were acquired in three independent experiments, and the dots indicate the mean invasion efficiency from each experiment; *p=0.04. (B) Number of merozoites per egress that made contact (>0.5 s) with WT or CD55-null RBCs. (C) Efficiency of WT or CD55-null RBC deformation upon merozoite contact. Bottom number is the total number of RBCs contacted by a merozoite, and top number is the subset that were deformed upon contact. The data were acquired in three independent experiments, and the dots indicate the mean deformation efficiency for each experiment; ns, not significant. (D) Strength of merozoite-induced deformation of WT or CD55-null RBCs among non-invading merozoites; ns, not significant. Error bars indicate SEM (A– D).
-
Figure 5—source data 1
Source data for live imaging experiments with CD55-null erythrocytes.
- https://cdn.elifesciences.org/articles/61516/elife-61516-fig5-data1-v2.xlsx
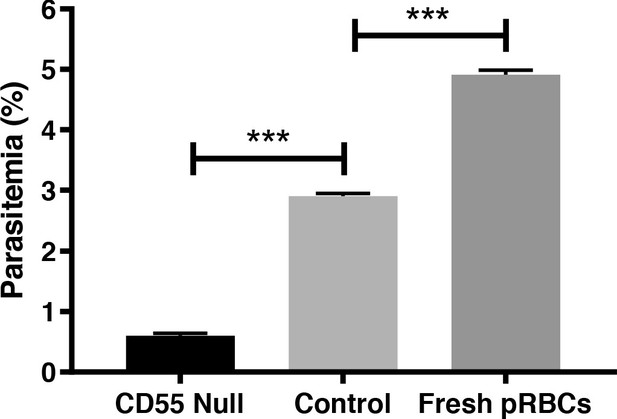
CD55-null pRBCs are refractory to invasion by P. falciparum.
Invasion efficiency of P. falciparum strain 3D7 in previously cryopreserved CD55-null pRBCs from rare Inab donor, control wild-type pRBCs that had been similarly cryopreserved, or fresh pRBCs. n = 3 technical replicates, error bars indicate SD; ***, p<0.05.
-
Figure 5—figure supplement 1—source data 1
Source data for CD55-null invasion assays.
- https://cdn.elifesciences.org/articles/61516/elife-61516-fig5-figsupp1-data1-v2.xlsx
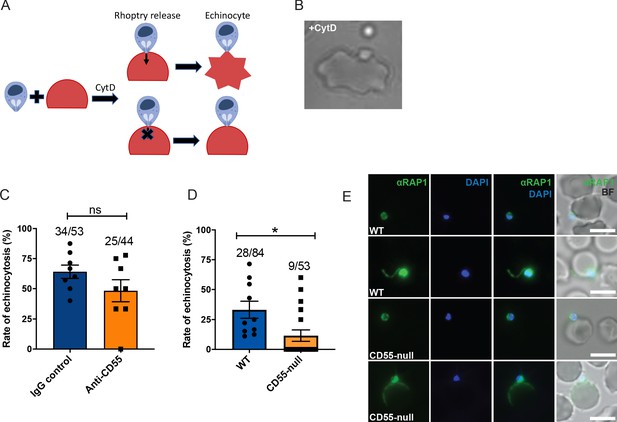
Blocking CD55 with antibody does not impact rhoptry discharge.
(A) Cartoon illustrating echinocytosis elicited by an attached merozoite that has discharged its rhoptry contents in the presence of cyt-D, which prevents internalization. Reagents that block rhoptry discharge prevent echinocytosis. (B) Image showing an echinocyte with an attached merozoite. (C) Echinocytosis efficiency in the presence of cyt-D and anti-CD55 antibody or isotype control. Bottom number is the total number of merozoite-RBC pairs observed to make sustained contact (>30 s), and top number is the subset in which echinocytosis occurred. Each data point represents the average rate of echinocytosis from a single egress event and error bars represent SEM, obtained in three independent experiments. ns, not significant. (D) Echinocytosis efficiency for WT versus CD55-null RBCs with attached merozoites in the presence of cyt-D. Bottom number is the total number of merozoite-RBC pairs observed to make sustained contact (>30 s), and top number is the subset in which echinocytosis occurred. Each data point represents the average rate of echinocytosis from a single egress event and error bars represent SEM, obtained in five independent experiments. *, p<0.05. (E) Representative images from immunofluorescence assays showing PfRAP1 localization in WT or CD55-null RBCs with attached merozoites in the presence of cyt-D. Assays were performed twice and ~50 attached merozoites were assessed for each RBC genetic background.
-
Figure 6—source data 1
Source data for echinocytosis assays.
- https://cdn.elifesciences.org/articles/61516/elife-61516-fig6-data1-v2.xlsx
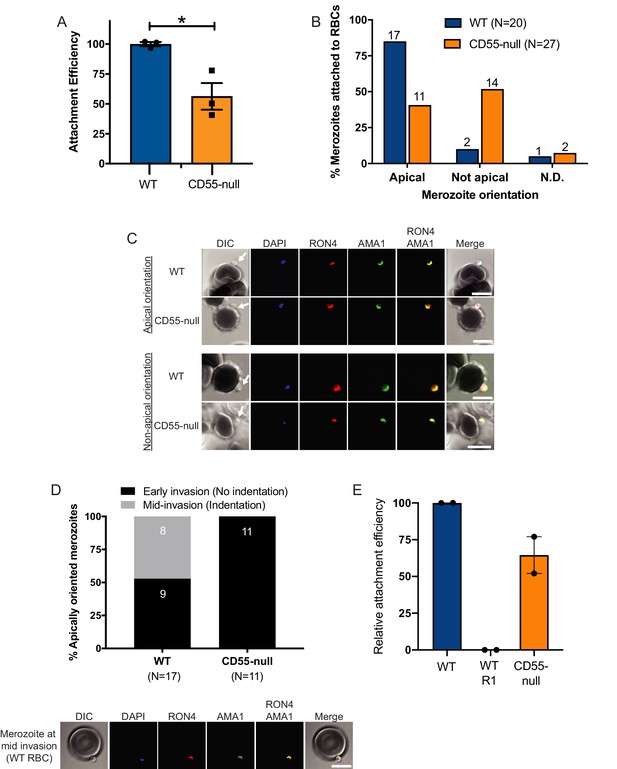
Invasion fails to progress in the absence of CD55.
(A) Attachment efficiency of P. falciparum merozoites to CD55-null versus control RBCs in the presence of cyt-D, as measured by flow cytometry 90 min after the addition of synchronized schizonts (N = 3, n = 3). Error bars indicate SEM; *p<0.018. (B) Orientation of merozoites attached to WT versus CD55-null RBCs, indicated by the localization of AMA1, RON4, and a density at the merozoite’s apical end as measured by confocal microscopy. Apical, AMA1 and RON4 co-localized at the cellular interface and merozoite density abutting RBC; Not apical, AMA1 and RON4 not co-localized at the cellular interface and merozoite density not abutting RBC; N.D., indeterminate. The numbers above each bar indicate the subset of attached merozoites in that orientation. The data were acquired in three biological replicates and were scored in a blinded manner. (C) Representative confocal images showing merozoites attached to RBCs with apical or non-apical orientation. Merozoite density at the apical end of the merozoite is indicated by an arrow. (D) Progression of invasion for merozoites apically attached to CD55-null or WT RBCs in the presence of cyt-D. Panel below shows representative confocal images of a merozoite at the mid-invasion stage. (E) Attachment efficiency of P. falciparum merozoites to WT RBCs, WT RBCs in the presence of the moving junction-blocking R1 peptide (1 mg/ml), or CD55-null RBCs, all in the presence of cyt-D. Attachment was measured by flow cytometry 90 min after the addition of synchronized schizonts (N = 2, n = 2). Data are presented relative to the efficiency of attachment to WT cells. Error bars represent SEM.
-
Figure 7—source data 1
Source data for attachment assays.
- https://cdn.elifesciences.org/articles/61516/elife-61516-fig7-data1-v2.xlsx
Videos
P. falciparum 3D7 invasion in the presence of isotype control antibody.
P. falciparum 3D7 invasion in the presence of anti-CD55 antibody.
P. falciparum 3D7 merozoite-induced deformation with deformation score 1.
P. falciparum 3D7 merozoite-induced deformation with deformation score 2.
P. falciparum 3D7 merozoite-induced deformation with deformation score 3.
An example of a failed invasion despite successful merozoite-induced deformation in CD55-null pRBC.
An example of a successful invasion of a wild-type pRBC.
Echinocytosis induced by attached P. falciparum merozoite to pRBCs in the presence of cytochalasin D (cyt-D).
Black arrows: Attachment resulting echinocyte formation. White arrows: Attachment failing to result in echinocytosis.
Echinocytosis induced by attached P. falciparum merozoites to previously cryopreserved WT pRBCs in the presence of cyt-D.
Black arrows: Attachment resulting in echinocyte formation. White arrows: Attachment failing to result in echinocytosis.
Echinocytosis induced by attached P. falciparum merozoites to previously cryopreserved CD55-null pRBCs in the presence of cyt-D.
Black arrows: Attachment resulting in echinocyte formation. White arrows: Attachment failing to result in echinocytosis.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Plasmodium falciparum) | 3D7 | Walter and Eliza Hall Institute, Melbourne, Australia | ||
| Cell line (Mus musculus) | Murine stromal cell layer; MS-5 | PMID:1375698 | Early passage obtained from Luc Douay Lab, Mycoplasma-negative. | |
| Biological sample (Homo sapiens) | Bone marrow-derived primary human CD34 + HSPCs (de-identified) | Stem Cell Technologies | Catalog numbers 70002.1 or 70002.2 | |
| Biological sample (Homo sapiens) | Erythrocytes; RBCs (de-identified) | Stanford Blood Center | ||
| Biological sample (Homo sapiens) | CD55-null pRBCs (CROM7) | doi:10.3925/jjtc.54.359; obtained via MTA from Japanese Red Cross Kinki Block Blood Center | Cryopreserved | |
| Biological sample (Homo sapiens) | control WT pRBCs | Obtained from Japanese Red Cross Osaka Blood Center | Cryopreserved | |
| Antibody | anti-CD55 (Rabbit polyclonal) | This paper | Generated at New England Peptide using human CD55 cDNA immunogen (Asp35-Ser353). Negative affinity purified via 8x HIS column and affinity purified by protein A column. WB, FC (75 µg/ml). Growth assay (serial dilutions). Reacts with WT RBCs and not CD55-null RBCs. | |
| Antibody | anti-CD55; BRIC 216 (Mouse monoclonal IgG1) | International Blood Group Reference Laboratory, UK | Cat# 9404 | FC (1:3000) Growth assay (serial dilutions) |
| Antibody | anti-CD55; BRIC 230 (Mouse monoclonal IgG1) | International Blood Group Reference Laboratory, UK | Cat# 9428 | FC (1:3000) Growth assay (serial dilutions) |
| Antibody | anti-CD55; BRIC 110 (Mouse monoclonal IgG1); | International Blood Group Reference Laboratory, UK | Cat# 9402 | Growth assay (serial dilutions) |
| Antibody | Anti-Band3 cytoplasmic domain; BRIC 170 (Mouse monoclonal IgG1) | International Blood Group Reference Laboratory, UK | Cat# 9450 | Growth assay (serial dilutions) |
| Antibody | Isotype Control Rabbit IgG antibody (Rabbit polyclonal) | Novus Biologicals | Cat# NB810-56910 | Growth assay (serial dilutions) |
| Antibody | Anti-AMA1 1F9 (mouse monoclonal) | Gift from Alan Cowman PMID:11707616 | IFA (1:200) | |
| Antibody | Anti-RON4 (Rabbit polyclonal) | Gift from Dave Richard PMID:20228060 | IFA (1:200) Affinity purified | |
| Antibody | anti-RAP1/2 209.3 (Mouse monoclonal) | Gift from Tony Holder. PMID:16102004 | IFA (1:500) | |
| Antibody | Anti-CD55-PE; BRIC 216-PE (Mouse monoclonal) | International Blood Group Reference Laboratory, UK | Cat# 9404-PE | FC, IFA (1:50) |
| Antibody | Anti-CD71-PE (Mouse monoclonal) | Miltenyi | Cat # 130-091-728 | FC (1:20) |
| Biological sample (Homo sapiens) | Plasma (Octaplas AB pooled solvent/detergent treated human plasma solution) | Octapharma | ||
| Software, algorithm | Two sgRNAs (Guide RNA design) | Designed using GPP web portal Broad Institute | ||
| Software, algorithm | ImageJ | ImageJ 1.50i PMID:22930834 | Wayne Rasband, NIH | |
| Software, algorithm | Fiji | Version 2.0.0-rc-69/1.52i | ||
| Software, algorithm | GraphPad Prism | Version 8.0.2 (159) for macOS | ||
| Sequence-based reagent | CD55-Cr1; sgRNA1 (sgRNA sequence) | This paper | Chemically modified sgRNA | GGGCCCCUACUCACCCCACA; Targets exon 1 of human CD55; synthesized by Synthego |
| Sequence-based reagent | CD55-Cr8; sgRNA2 (sgRNA sequence) | This paper | Chemically modified sgRNA | CUGGGCAUUAGGUACAUCU; Targets exon 2 of human CD55; synthesized by Synthego |
| Peptide, recombinant protein | Cas9 protein; Cas9 (Cas9-NLS purified protein) | QB3 MacroLab; University of California, Berkeley | ||
| Peptide, recombinant protein | Neuraminidase; Neuraminidase from Vibrio cholerae | Sigma | Cat# N7885 | Confirmed to cleave RBC sialic acid through P. falciparum invasion assay using sialic-dependent strain W2mef |
| Peptide, recombinant protein | Recombinant human insulin | Sigma | Cat# 91077C | |
| Peptide, recombinant protein | R1 inhibitory peptide; R1 peptide | Gift from Manoj Duraisingh; PMID:29078358 | ||
| Commercial assay or kit | Pierce Fab Preparation kit | Thermo Fisher Scientific | Cat# 44685 | |
| Commercial assay, kit | 4D-Nucleofector X kit | Lonza | Cat# V4XP-3032 | |
| Peptide, recombinant protein | Holotransferrin | BBI solutions | Cat# T101-5 | >98% purity, from human serum |
| Chemical compound, drug | Heparin (Heparin sodium salt) | Affymetrix | Cat# 9041-08-1 | |
| Chemical compound, drug | Hydrocortisone | Sigma | Cat# H2270-100MG | |
| Peptide, recombinant protein | SCF (Recombinant human Stem Cell Factor/c-kit ligand) | R and D Systems | Cat# 255 SC/CF | |
| Peptide, recombinant protein | IL-3 (Recombinant human IL-3 Protein) | R and D Systems | Cat# 203-IL-050 | |
| Peptide, recombinant protein | Epo (Recombinant Epoetin alpha (Procrit)Epo) | Amgen | NDC# 59676-0312-04 | |
| Chemical compound, drug | E64 | Sigma | Cat# E3132-5MG | |
| Chemical compound, drug | Compound 2 | PMID:23675297 | Gift from Simon Osborne (MTA with LifeArc) | |
| Chemical compound, drug | Cyt-D; Cytochalasin-D | Sigma | Cat# C8273-1MG | |
| Chemical compound, drug | ML-10 | PMID:28874661 | Gift from Simon Osborne (MTA with LifeArc) | |
| Other | SYBR Green 1 Nucleic Acid Gel Stain | Invitrogen | Cat# S-7563 | FC: 1:2000 |
| Other | Vybrant DyeCycle Violet | Life Technologies | Cat# V35003 | FC: 1:10,000 |





