Temperature-dependent fasciation mutants provide a link between mitochondrial RNA processing and lateral root morphogenesis
Figures
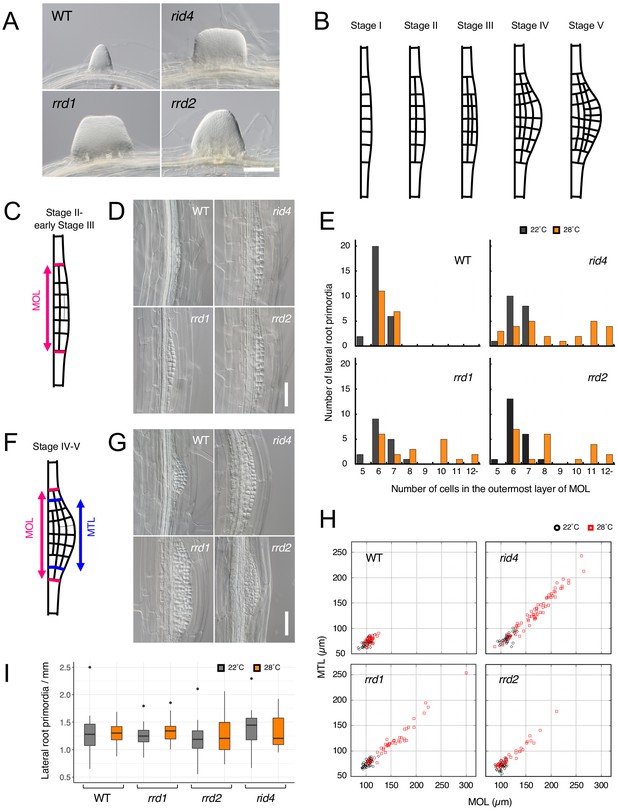
Effects of the temperature-dependent fasciation (TDF) mutations on the early stages of lateral root (LR) development.
(A) Fasciated LRs formed at 28°C in the TDF mutant explants vs. a normal root on the wild-type (WT) explant after 6 days of culture. (B) Schematic representation of LR development (stages I–V). (C) Schematic image of a primordium at stage II. The area consisting of more than one cell layer (MOL) is delimited by red lines. (D) Stage II primordia formed at 28°C in WT and TDF mutant explants. (E) Effects of the TDF mutations on the number of cells in the outermost layer of the MOL area of stage II primordia at 22°C (black) and 28°C (orange). N = 17–28. (F) Schematic image of a primordium at the transition from stage IV to stage V. The areas consisting of MOL and more than two cell layers (MTL) are delimited by red lines and blue lines, respectively. (G) Stage IV–V primordia formed at 28°C in WT explants and TDF mutant explants. (H) Scatterplot of the effect of the TDF mutations on the width of the MTL vs. the width of the MOL areas at 22°C (black) and 28°C (red). N = 31–66. (I) LR densities in the WT explants and TDF mutant explants cultured at 22°C or 28°C (including all developmental stages; median, 25–75% quantile, N = 21–29, p>0.3, Kruskal-Wallis test). Scale bars, 100 μm (A), 50 μm (D, G).
-
Figure 1—source data 1
Raw data and supplement to transparent reporting form for Figure 1I.
- https://cdn.elifesciences.org/articles/61611/elife-61611-fig1-data1-v2.xlsx
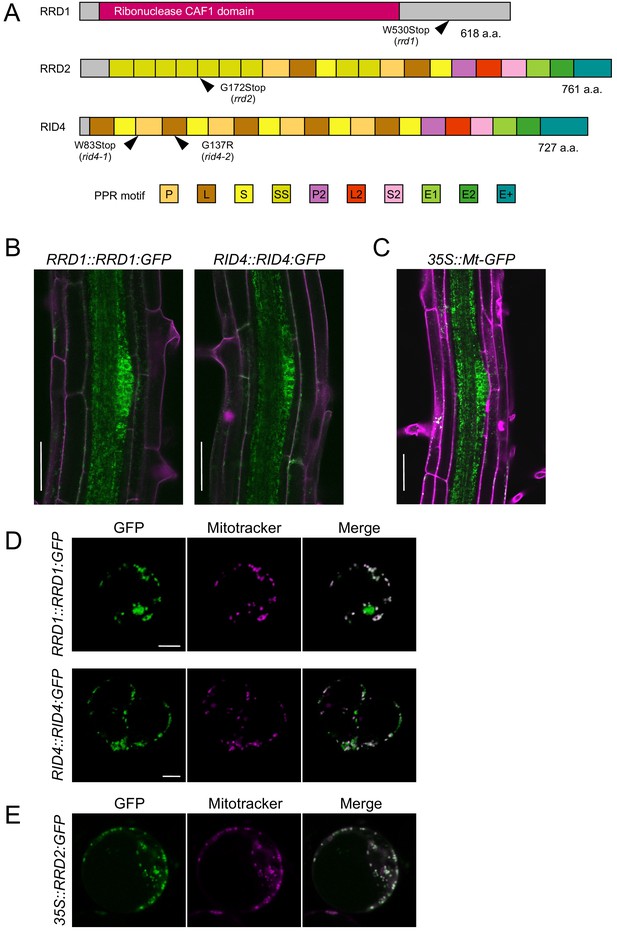
Tissue-specific expression and subcellular localization of the temperature-dependent fasciation (TDF) proteins.
(A) Structures of the RRD1, RRD2, and RID4 proteins. (B and C) Expression of RRD1::RRD1:GFP (B, left), RID4::RID4:GFP (B, right), and 35S::Mt-GFP (C) at stage II of lateral root (LR) primordium development. Propidium iodide was used as a red counterstain. (D and E) Expression of RRD1::RRD1:GFP (D, upper panels), RID4::RID4:GFP (D, lower panels), and 35S::RRD2:GFP (E) in callus-derived protoplasts. Mitochondria were labeled with MitoTracker Orange. Scale bars, 50 μm (B and C) and 5 μm (D and E).
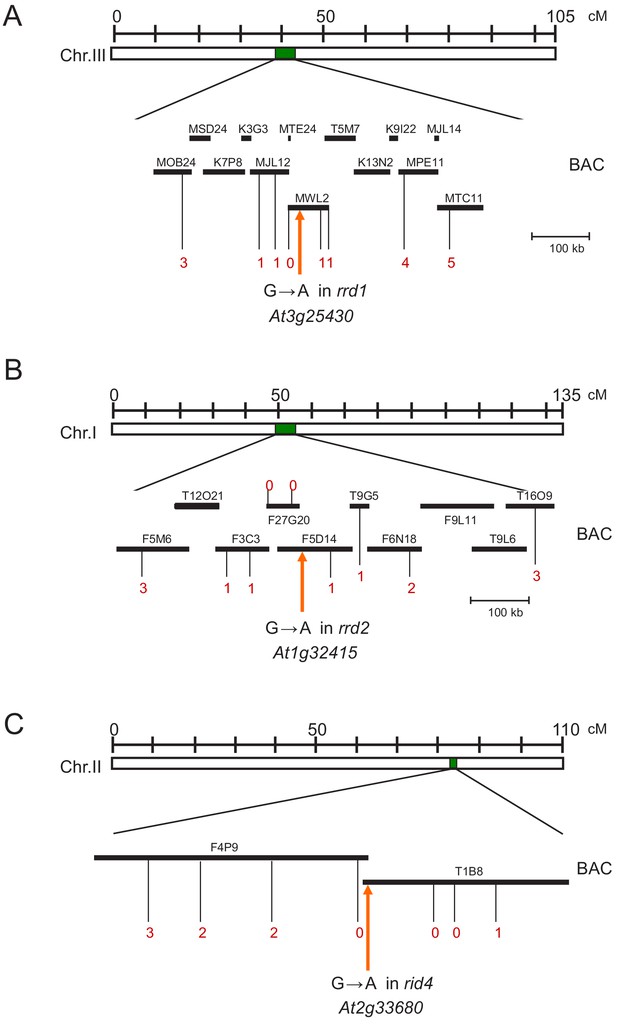
Chromosome mapping of the temperature-dependent fasciation (TDF) mutations, rrd1, rrd2, and rid4-1.
(A) Chromosome mapping of the rrd1 mutation. The black rectangles represent annotation units around the RRD1 locus on chromosome 3, and the red numerals correspond to the number of recombination events between DNA polymorphism markers and the RRD1 locus. The rrd1 mutation was mapped to the region covered by the annotation units MJL12, MTE24, and MWL2. Sequencing of this region, followed by complementation analysis, identified the rrd1 mutation as a G-to-A transition in At3g25430 (orange arrow). (B) Chromosome mapping of the rrd2 mutation. The black rectangles represent annotation units around the RRD2 locus on chromosome 1, and the red numerals correspond to the number of recombination events between DNA polymorphism markers and the RRD2 locus. The rrd2 mutation was mapped to the region covered by the annotation units F3C3, F27G20, and F5D14. Sequencing of this region, followed by allelism analysis, identified the rrd2 mutation as a G-to-A transition in At1g32415 (orange arrow). (C) Chromosome mapping of the rid4-1 mutation. The black rectangles represent the annotation units around the RID4 locus on chromosome 2, and the red numerals correspond to the number of recombination events between DNA polymorphism markers and the RID4 locus. The rid4 mutation was mapped to the region covered by the annotation units F4P9 and T1B8. Sequencing of this region, followed by complementation analysis, identified the rid4 mutation as a G-to-A transition in At2g33680 (orange arrow).
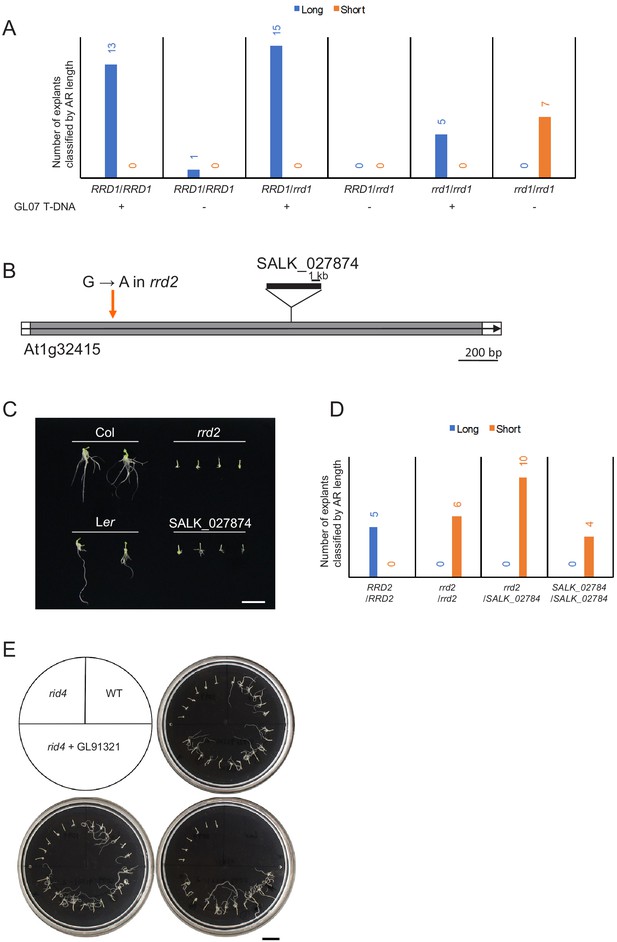
Complementation analysis and allelism test for the identification of the temperature-dependent fasciation (TDF) genes RRD1, RRD2, and RID4.
(A) Complementation analysis for the identification of the RRD1 gene. The genomic fragment GL07 encompassing At3g25430, where an rrd1 phenotype-linked mutation was found, was introduced into the wild-type (WT) plant, which was then crossed with rrd1. Each individual of the F2 progeny was genotyped for the rrd1 allele and the GL07 transgene. Hypocotyl explants of the F2 progeny were cultured on RIM at 28°C for 14 days and examined for adventitious rooting. The explants were categorized according to the length of the ARs (Short, shorter than 5 mm; Long, longer than 5 mm) and counted. The results showed that the development of ARs, which was highly temperature sensitive in the rrd1 mutant, was clearly rescued by the introduction of GL07. Therefore, we concluded that the RRD1 gene corresponds to At3g25430. (B and C) Defect of AR formation in a T-DNA insertion mutant of At1g32415. SALK_027874 carries a T-DNA insertion in the middle of At1g32415 (B). The transcribed region and open reading frame of At1g32415 are indicated by the open arrow and gray box, respectively. Hypocotyl explants of Col, Ler, rrd2, and SALK_027874 were cultured on RIM at 28°C for 27 days and examined for adventitious rooting (C). The results indicated that SALK_027874 and rrd2 are defective in AR formation at this temperature. As AR formation was not significantly affected at 22°C in both rrd2 and SALK_027874 (data not shown), SALK_027874 was shown to be temperature sensitive for root development, as was rrd2. Bar, 1 cm. (D) Allelism test for the identification of the RRD2 gene. The rrd2 mutant was crossed with SALK_027874 carrying a T-DNA insertion in At1g32415, in which an rrd2 phenotype-linked mutation was found. Each individual of the F2 progeny was genotyped for the rrd2 and the T-DNA insertion alleles. Hypocotyl explants of the F2 progeny were cultured on RIM at 28°C for 14 days and examined for adventitious rooting. The explants were categorized according to the length of the ARs (Short, shorter than 5 mm; Long, longer than 5 mm) and counted. The results indicated clearly that rrd2 and SALK_027874 are allelic. Therefore, we concluded that the RRD2 gene corresponds to At1g32415. (E) Complementation analysis for the identification of the RID4 gene. The genomic fragment GL91321 encompassing At2g33680, where we found an rid4 phenotype-linked mutation, was introduced into rid4, and the resultant transgenic rid4 mutant harboring GL91321 (rid4/GL91321) was used for complementation analysis. Hypocotyl explants of the WT plant, rid4, and rid4/GL91321-2 were cultured on RIM at 28°C for 19 days and examined for AR formation. The development of ARs, which was highly temperature sensitive in the rid4 mutant, was clearly rescued by the introduction of GL91321. Therefore, we concluded that the RID4 gene corresponds to At2g33680. Scale bar, 1 cm.
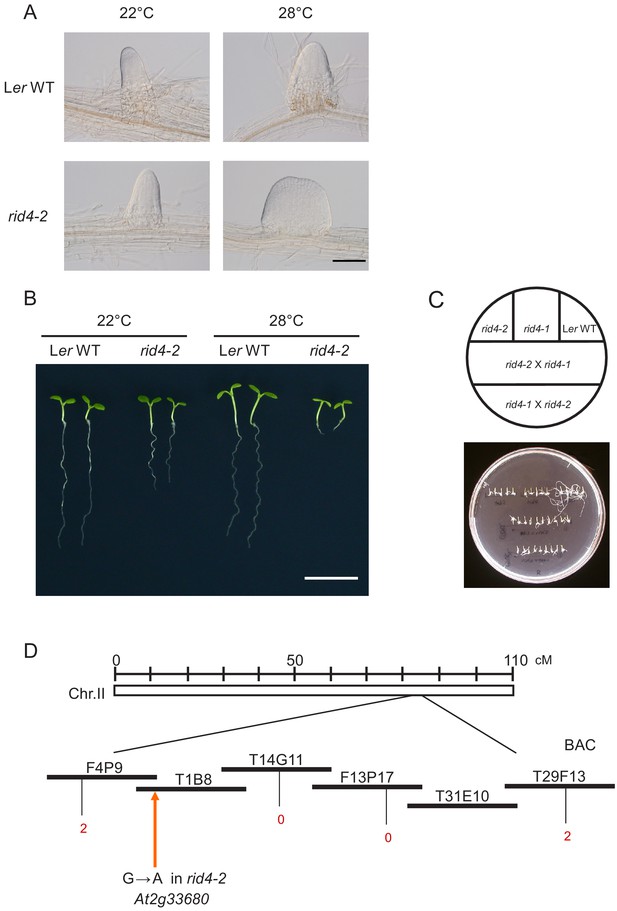
Identification and characterization of the rid4-2 mutant.
(A) Representative images of lateral roots (LRs) formed at 22°C or 28°C in the explants of the wild-type plant or the rid4-2 mutant after 6 days of culture. Fasciated LRs were observed in rid4-2 explants at 28°C. (B) Phenotypes of seedlings that were grown for 7 days on vertical agar plates. Seedlings were grown either at 22°C or 28°C. (C) Allelism test between rid4-1 and rid4-2. F1 plants derived from a reciprocal crossing between rid4-1 and rid4-2 were subjected to phenotypic analysis regarding AR formation. Hypocotyl explants of rid4-1, rid4-2, Ler WT, and F1 plants were cultured on RIM for 24 days at 28°C. (D) Chromosome mapping of the rid4-2 mutation. The black rectangles represent annotation units around the RID4 locus on chromosome 2, and the red numerals correspond to the number of recombination events between DNA polymorphism markers and the RID4 locus. rid4-2 was mapped to the region covered by the annotation units F4P9 and T29F13. Sequencing of this region, followed by complementation analysis, identified the rid4-2 mutation as a G-to-A transition in At2g33680 (orange arrow). Scale bars, 100 μm (A) and 1 cm (B).
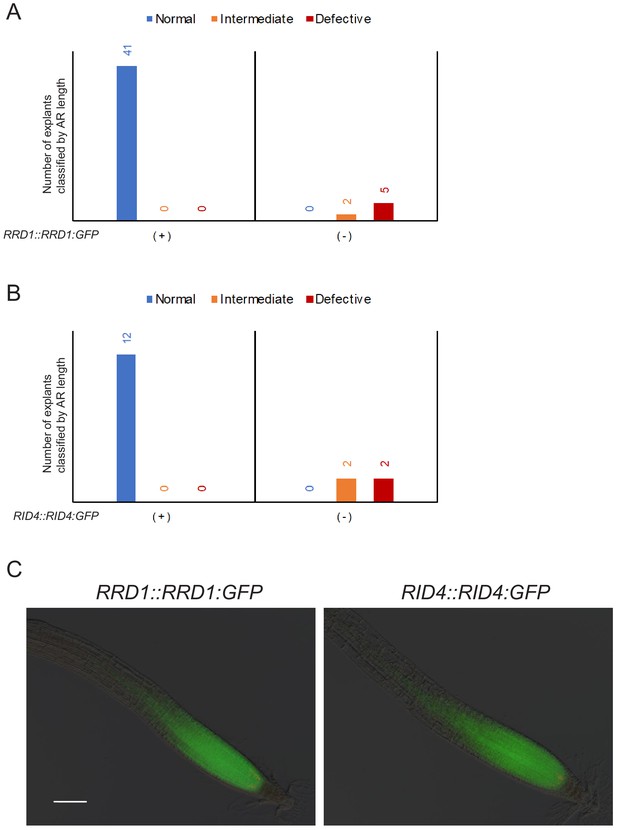
Functionality and expression of RRD1::RRD1:GFP and RID4::RID4:GFP.
(A) Phenotypic complementation of rrd1 by the introduction of the GFP reporter gene RRD1::RRD1:GFP. F3 plants homozygous for rrd1 derived from the cross between rrd1 and Col carrying RRD1::RRD1:GFP were phenotyped for AR formation from hypocotyl explants at 28°C and genotyped for the presence of RRD1::RRD1:GFP (+, present; –, absent). (B) Phenotypic complementation of rid4 by the introduction of the GFP reporter gene RID4::RID4:GFP. rid4 homozygotes in which the genetic background had been partially replaced by crossing with the Col strain were transformed with RID4::RID4:GFP. Plants of the resultant T2 line were phenotyped for AR formation from hypocotyl explants at 28°C and genotyped for the presence of RID4::RID4:GFP (+, present; –, absent). (C) Expression patterns of RRD1 and RID4 in the root apical region. GFP signals in the primary roots of the transgenic plants harboring RRD1::RRD1:GFP and RID4::RID4:GFP revealed strong expression of RRD1 and RID4 in the root apical meristem. Scale bar, 100 μm.
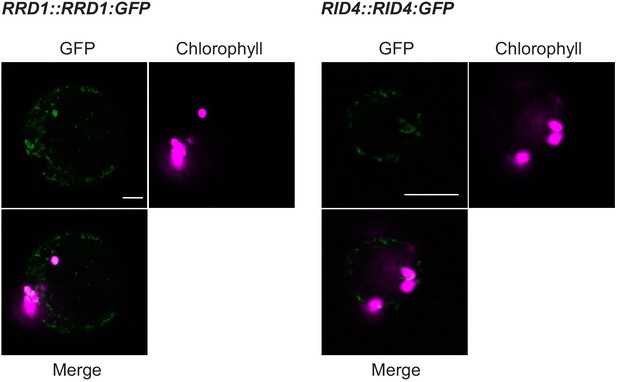
Colocalization of chlorophyll autofluorescence and RRD1::RRD1:GFP and RID4::RID4:GFP.
Greening-induced calli were produced by culturing hypocotyl segments on CIM(G), which consisted of B5 medium supplemented with 2.0% (w/v) glucose, 0.1 mg l−1 2,4-dichlorophenoxyacetic acid (1/5 of CIM) and 0.1 mg l−1 kinetin and solidified with 0.25% (w/v) gellan gum. Protoplasts were prepared from the green calli and the signals of GFP and chlorophyll were observed using the LSM710 system (Carl Zeiss). Scale bar, 10 µm.
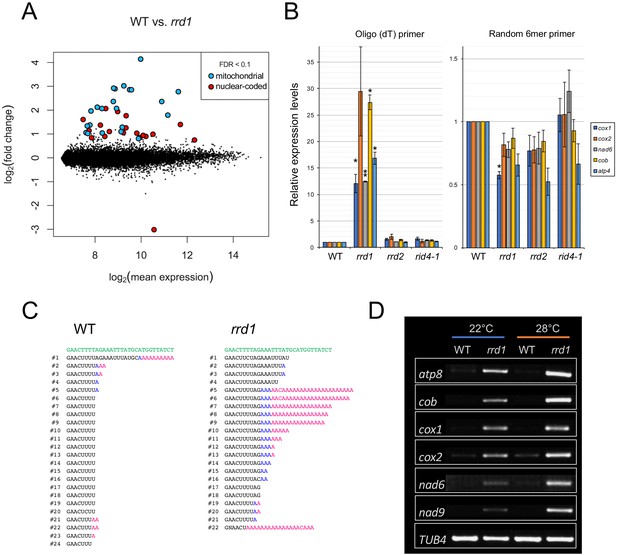
Accumulation of polyadenylated mitochondrial transcripts in rrd1.
(A) MA plot for the microarray analysis of poly(A)+ transcripts of rrd1 vs. wild-type (WT) explants in which lateral roots (LRs) were induced at 28°C for 12 hr. (B) qRT–PCR analysis of explants in which LRs were induced at 28°C for 12 hr. The total and polyadenylated transcript levels are shown for cytochrome oxidase subunit 1 (cox1), cox2, NADH dehydrogenase subunit 6 (nad6), apocytochrome B (cob), and ATP synthase subunit 4 (atp4) (mean ± s.d., N = 3, *p<0.05, **p<0.01, one sample t test with Benjamini-Hochberg correction). (C) Analysis of the 3´ end of the cox1 mRNA by CR–RT PCR. mRNAs were prepared from WT and rrd1 seedlings that were first grown at 22°C for 7 days, and then at 28°C for 2 days. The genomic sequence of cox1 is shown in green. (D) RACE-PAT assay showing the accumulation of polyadenylated transcripts of atp8, cob, cox1, cox2, nad6, nad9, and TUB4. mRNAs were prepared from explants in which LRs were induced at 22°C or 28°C for 12 hr.
-
Figure 3—source data 1
Raw data and supplement to transparent reporting form for Figure 3B.
- https://cdn.elifesciences.org/articles/61611/elife-61611-fig3-data1-v2.xlsx
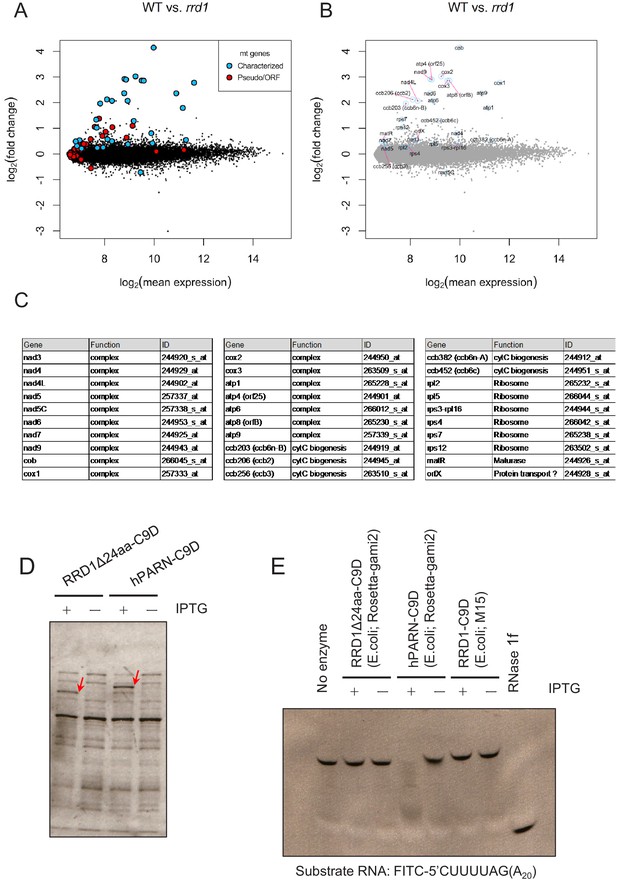
Characterization of RRD1 function.
(A–C) Microarray analysis of mitochondrial genes in rrd1. MA plot for the microarray analysis of poly(A)+ transcripts of rrd1 vs. wild-type (WT) explants in which lateral roots (LRs) were induced at 28°C for 12 hr. The characterized mitochondrial genes are shown in blue, while the uncharacterized mitochondrial ORFs or pseudogenes are shown in red (A). The names of the characterized genes are indicated in (B). (C) The function and array element ID of the characterized mitochondrial genes in the GeneChip Arabidopsis Genome ATH1 Array. (D and E) PARN activity assay of RRD1. TALON-purified fraction of the total cell lysate with or without IPTG induction (D). PARN activity assay using the TALON-purified fraction (E).
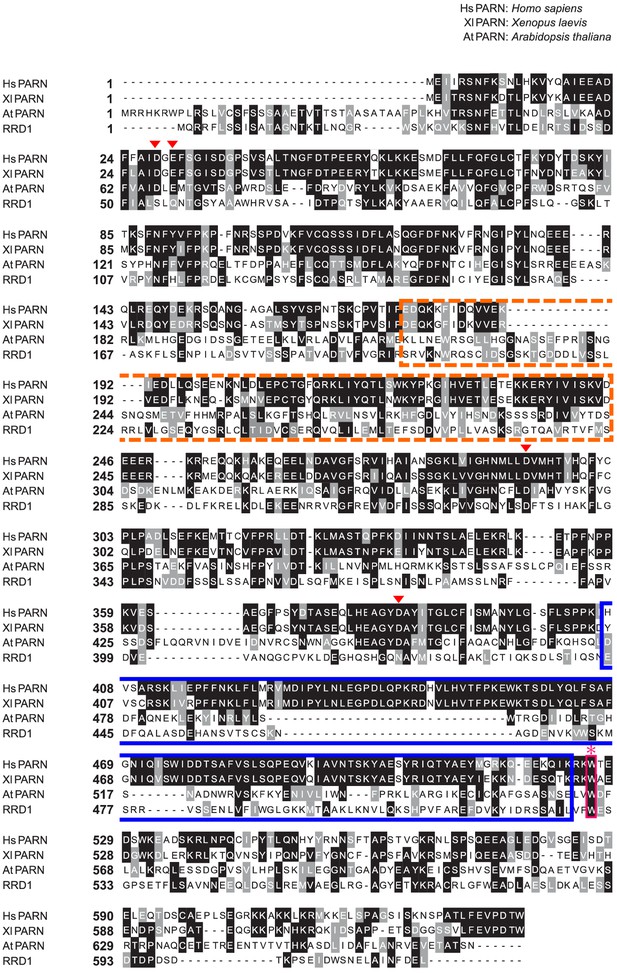
Sequence alignment between RRD1 and PARNs from various organisms.
An alignment of amino acid sequences was generated between RRD1 and PARNs from humans, Xenopus laevis, and Arabidopsis using the ClustalW program and processed with BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html). Identical and similar amino acid residues are highlighted on black and gray backgrounds, respectively. The R3H domain is marked by the dotted orange box, and the RNA recognition motif (RRM) is marked by the solid blue box. These domains are conserved in the animal PARNs (human and Xenopus laevis), but are not clearly present in the Arabidopsis PARN and RRD1. The pink asterisk represents the tryptophan codon that was changed to a stop codon by the rrd1 mutation. The red arrowheads represent the four residues that are important for PARN activity (29).
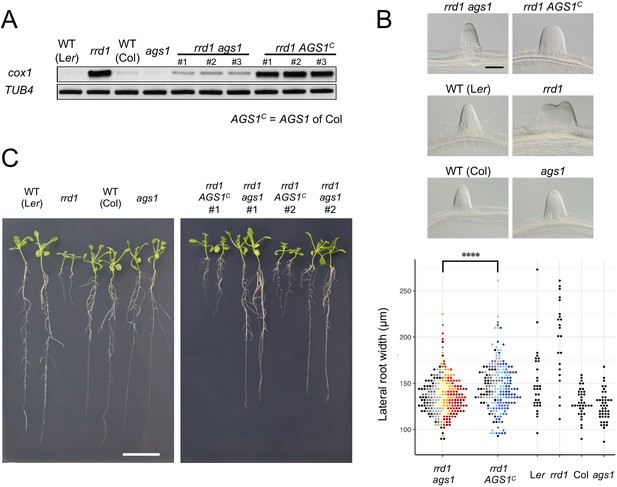
Effects of ags1 on the phenotypes of rrd1.
(A) RACE-PAT assay showing the accumulation of polyadenylated transcripts of cox1 and TUB4. rrd1 mutant strains harboring either ags1 or AGS1c (AGS1 of Col background) were obtained by rrd1 (Ler background) × ags1 (Col background) and rrd1 × Col crosses, respectively. mRNAs were prepared from seedlings that were first grown at 22°C for 5 days, and then at 28°C for 3 days. (B) Representative images of lateral roots (LRs) formed at 28°C after 6 days of culture (upper panels). The basal width of the LRs that were formed in this way was scored (lower panel, N = 115–116 for rrd1 ags1, and rrd1 AGS1c, N = 22–43 for others, ****p<10–4, Mann–Whitney–Wilcoxon test with Bonferroni correction). For rrd1 ags1 and rrd1 AGS1c, data were gathered from seven strains, which are shown by different colors. (C) Seedlings grown at 28°C for 13 days on gellan gum plates. Scale bars, 100 μm (B) and 2 cm (C).
-
Figure 4—source data 1
Supplement to transparent reporting form for Figure 4B.
- https://cdn.elifesciences.org/articles/61611/elife-61611-fig4-data1-v2.xlsx
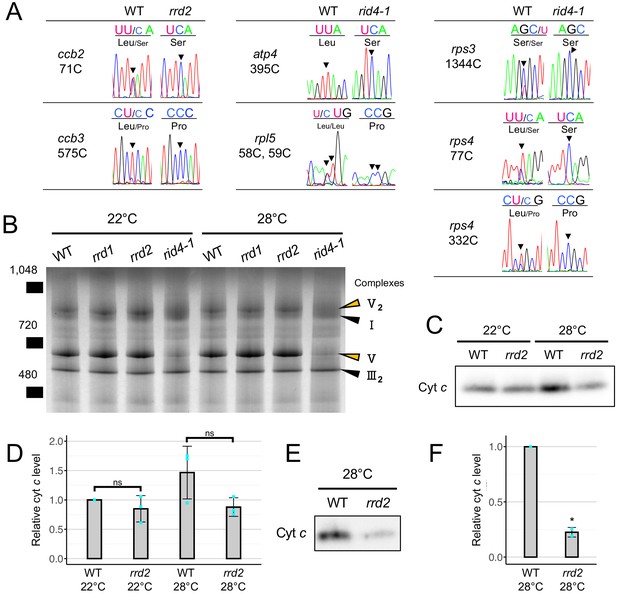
Effects of rrd2 and rid4 on mitochondrial mRNA editing and protein synthesis.
(A) Sequencing analysis of mitochondrial mRNA editing in explants in which LRs were induced at 28°C for 12 hr. Arrowheads indicate the RNA editing sites. (B) BN-PAGE analysis of mitochondrial protein complexes. Mitochondria were extracted from seed-derived liquid-cultured callus that were first incubated at 22°C for 20 days, and then at 22°C or 28°C for an additional 3 days. Arrowheads indicate the mitochondrial complexes. (C and D) Immunoblot analysis of cyt c. Mitochondria were extracted in the same conditions as in (B). The signal intensities determined by densitometry are shown as values relative to that of the wild type at 22°C in (D) (N = 3, mean ± s.d., Welch’s t test). (E and F) Immunoblot analysis of cyt c using mitochondria extracted from callus that were cultured first at 22°C for 14 days, and then at 28°C for 7 days. The signal intensities determined by densitometry are shown as values relative to that of the wild type in (F) (N = 2, mean ± s.d., **p<0.01, Welch’s t test).
-
Figure 5—source data 1
Raw data and supplement to transparent reporting form for Figure 5C.
- https://cdn.elifesciences.org/articles/61611/elife-61611-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Raw data and supplement to transparent reporting form for Figure 5E.
- https://cdn.elifesciences.org/articles/61611/elife-61611-fig5-data2-v2.xlsx
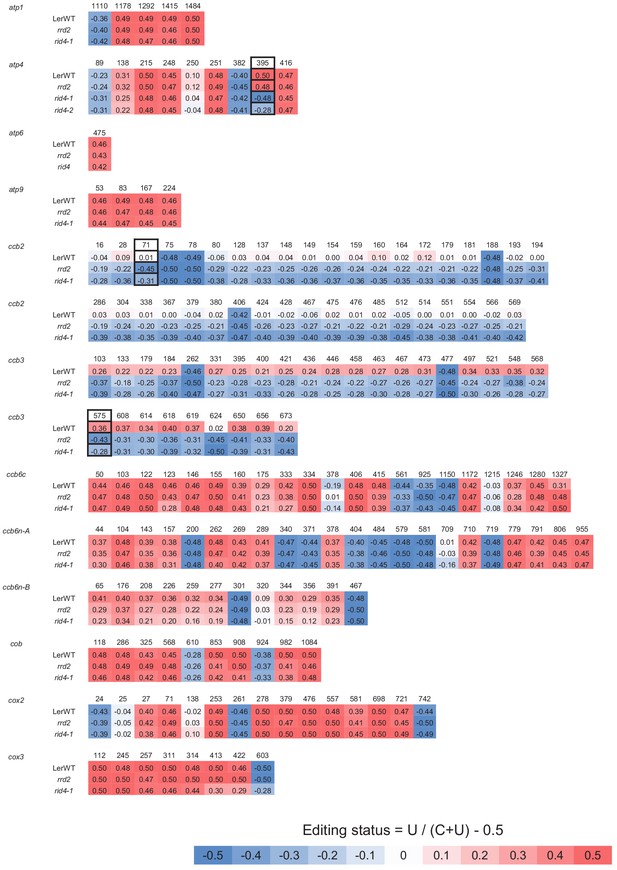
Comprehensive analysis of mitochondrial mRNA editing in rrd2 and rid4-1 (atp1 to cox3).
A sequencing analysis of mitochondrial mRNA editing was performed using explants in which lateral roots (LRs) were induced at 28°C for 12 hr. The color code indicates the level of C-to-U RNA editing at each site (editing status: –0.5 = 100% C, 0.5 = 100% U). The presumptive specific editing sites of RRD2 and RID4 are marked by solid black boxes. RNA editing in rid4-2 was also analyzed for sites affected in rid4-1.
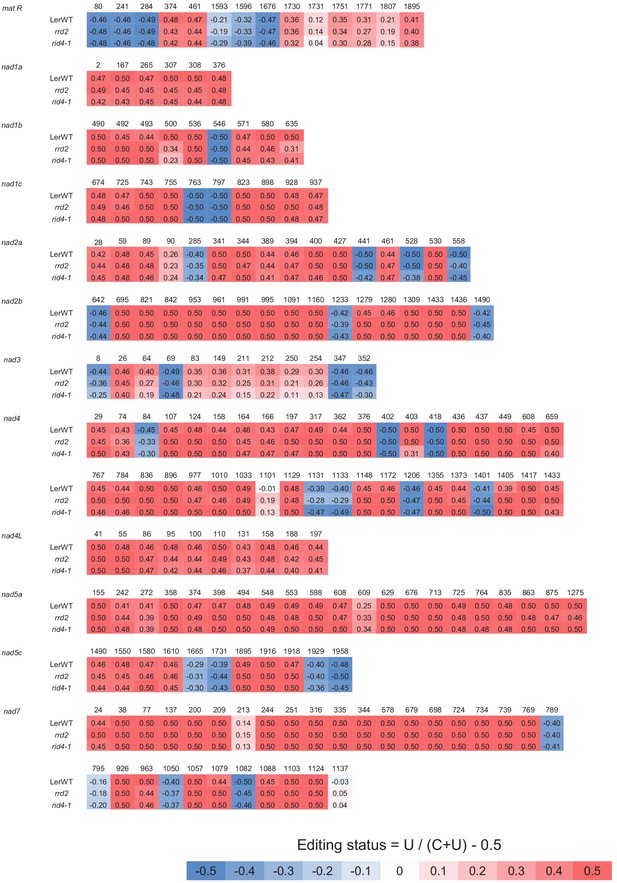
Comprehensive analysis of mitochondrial mRNA editing in rrd2 and rid4-1 (matR to nad7).
A sequencing analysis of mitochondrial mRNA editing was performed using explants in which lateral roots (LRs) were induced at 28°C for 12 hr. The color code indicates the level of C-to-U RNA editing at each site (editing status: –0.5 = 100% C, 0.5 = 100% U).

Comprehensive analysis of mitochondrial mRNA editing in rrd2 and rid4-1 (orf240A to rps14(ψ)).
A sequencing analysis of mitochondrial mRNA editing was performed using explants in which lateral roots (LRs) were induced at 28°C for 12 hr. The color code indicates the level of C-to-U RNA editing at each site (editing status: –0.5 = 100% C, 0.5 = 100% U). The presumptive specific editing sites of RRD2 and RID4 are marked by solid black boxes. RNA editing in rid4-2 was also analyzed for sites affected in rid4-1.
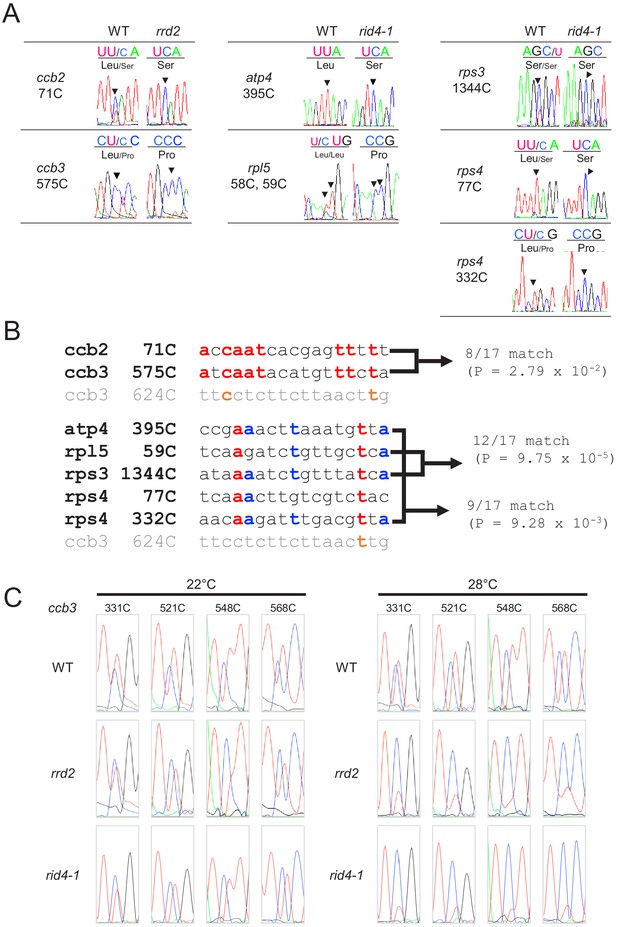
Analysis of mitochondrial mRNA editing in rrd2 and rid4-1.
(A) A sequencing analysis of mitochondrial mRNA editing was performed using explants in which lateral roots (LRs) were induced at 22°C for 12 hr. (B) Alignment of the estimated binding sequence of RRD2 and RID4 (31). (C) Analysis of the mRNA editing of ccb3 in explants that were cultured at 22°C or 28°C for 12 hr.
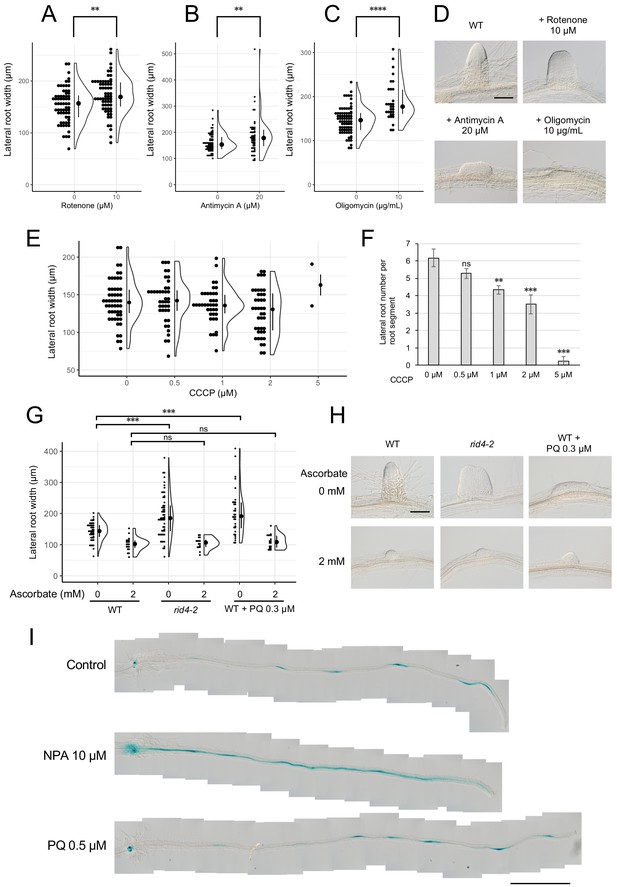
Formation of fasciated lateral roots (LRs) after treatment with chemicals that inhibit mitochondrial respiration or induce ROS.
(A–D) LRs were induced at 28°C from the wild-type (WT) plant in the presence of rotenone (A), antimycin A (B), or oligomycin (C), and the basal width of the LRs that were formed was scored after 6 days in culture (median, 25–75% quantile, N = 30–76, **p<0.01, ****p<0.0001, Mann–Whitney–Wilcoxon test). Typical LRs that were formed in each treatment are shown in (D). (E and F) LRs were induced from the WT plant in the presence of CCCP. The basal width of LRs (E, median, 25–75% quantile, N = 2–53, p>0.1, Kruskal-Wallis test) and the number of LRs per segment (F, Number of segments = 12, **p<0.01, ***p<0.001, Dunnett's test) were scored on the 6th day. (G and H) The effects of the application of ascorbate on WT, paraquat (PQ)-treated, or rid4-2 segments during LR formation. The basal width of the LRs formed was measured on the 6th day of LR induction (G, median, 25–75% quantile, N = 16–58, ***p<0.001, Mann–Whitney–Wilcoxon test with Bonferroni correction). Representative images of LRs in each condition are shown in (H). (I) DR5::GUS expression at 12 hr after LR induction under treatment with naphthylphthalamic acid (NPA) or PQ. Scale bars, 100 μm (D and H) and 1 mm (I).
-
Figure 6—source data 1
Supplement to transparent reporting form for Figure 6, A–F.
- https://cdn.elifesciences.org/articles/61611/elife-61611-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Supplement to transparent reporting form for Figure 6G.
- https://cdn.elifesciences.org/articles/61611/elife-61611-fig6-data2-v2.xlsx
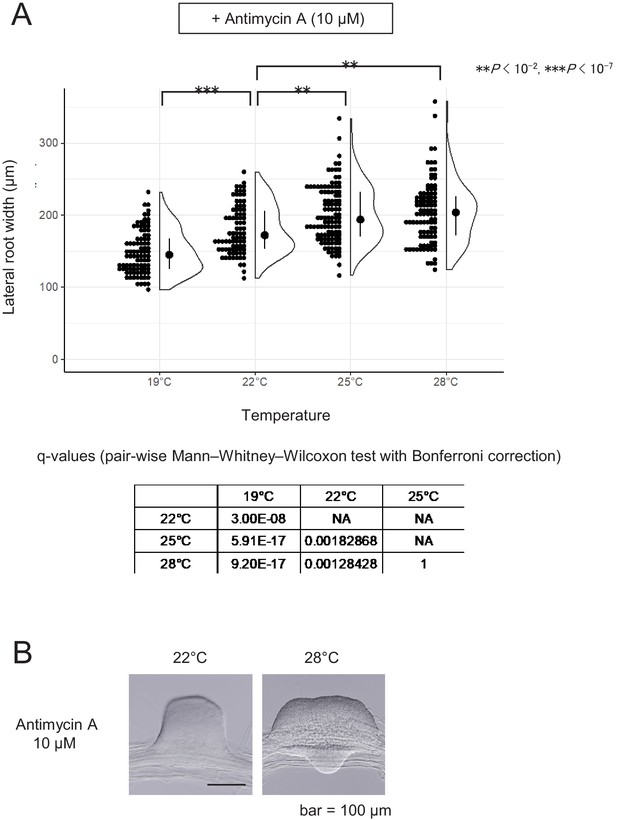
Effects of antimycin A on lateral root (LR) formation at different temperatures.
(A) LRs were induced at from the wild-type (WT) plant in the presence of antimycin A at the indicated temperatures and the basal width of the LRs that were formed was scored after 6 days in culture (median, 25–75% quantile, N = 83–109, **p<0.01, ***p<10−7, Mann–Whitney–Wilcoxon test). Typical LRs that were formed at 22°C and 28°C are shown in (B).

Effects of naphthylphthalamic acid (NPA) and paraquat (PQ) on lateral root (LR) formation.
Root explants 6 days after LR induction under treatment with NPA or PQ. Scale bar, 1 mm.
Additional files
-
Supplementary file 1
Primers used in this study.
- https://cdn.elifesciences.org/articles/61611/elife-61611-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61611/elife-61611-transrepform-v2.docx