Behavioral role of PACAP signaling reflects its selective distribution in glutamatergic and GABAergic neuronal subpopulations
Figures
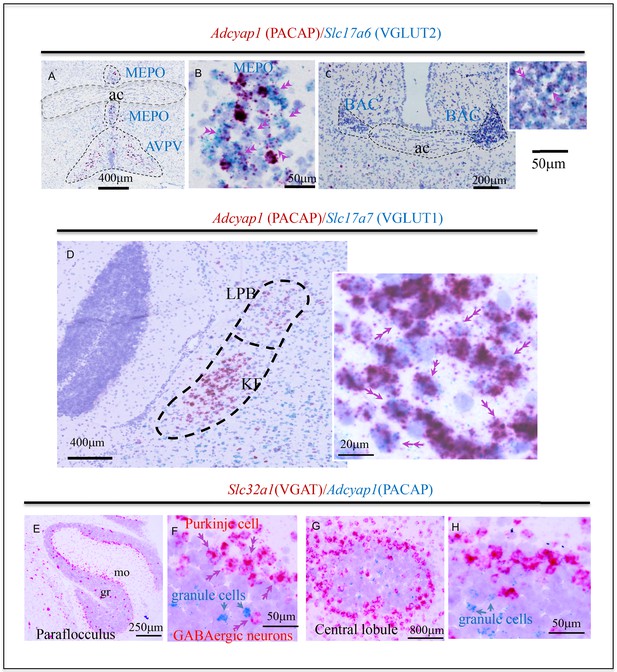
Examples of histological samples using the sensitive dual in situ hybridization (DISH) method that can label unambiguously the co-expression of two RNAs at single-cell level for light microscope examination.
(A and C) Adcyap1 (RNA coding for PACAP) co-expression with Slc17a6 (RNA coding for VGLUT2) in MEPO and BAC respectively. (B) High magnification of the MEPO region of (A). Double arrows indicate cells that co-express both mRNAs. This feature is better appreciated on the cells in which Adcyap1 is weakly expressed (weak staining), so the Slc17a6 staining can be clearly seen as independent dots. (D) Brain stem Koelliker Fuse (KF) nucleus of the parabrachial complex is another main PACAP-expressing nucleus. Adcyap1 was co-expressed intensely with Slc17a7 (RNA coding for VGLUT1). Double arrows indicate cells co-expressing both mRNAs. Panels E-H show two cerebellar regions, paraflocculus (E and F) and central (G and H) lobules, under low and high magnification, respectively, where the Adcyap1 expression was higher than rest of regions. Purkinje cells are the main GABAergic (expressing Slc32a1, RNA encoding VGAT) PACAP containing neurons, distributed in all regions of cerebellar cortex. Some GABAergic cells in granule cell layer of paraflocculus and central regions also co-expressed Adcyap1 and Slc32a1 (indicated with double pink arrowheads, see also SI-Fig.O1). In these two cerebellar regions, some granule cells also expressed Adcyap1 (indicated with single blue arrows). Nissl staining was used for counterstaining. Note: this figure contains excerpts from the more comprehensive figure supplement.
-
Figure 1—source data 1
Comprehensive DISH mapping of PACAP co-expression with VGLUT1, VGLUT2, and VGAT throughout mouse brain reveals an extensive distribution and diversity of cell types.
Adcyap1, PACAP-mRNA mapping within glutamatergic and GABAergic subpopulations of mouse brain. Slc17a7, Slc17a6, and Slc32a1: mRNAs encoding the VGLUT1, VGLUT2, and VGAT with respective color coding for the chromogens labeling the corresponding mRNAs. Double arrows indicate examples of colocalization of two probe mRNAs and single arrows indicate no-co-expression in given cells. Panels A–O: coronal and sagittal sections taken from the indicated Bregma or medio-lateral coordinates. The combination of mRNA probes is indicated with respective colors corresponding to the DISH method end products. The high-magnification panels are examples with relevant molecular features, of the regions labeled within low-magnification photomicrographs (see the abbreviations vide infra). Note that the high-magnifications photos are not from the same experiment of the low magnification. 3 v: third ventricle; ACA: anterior cingulate area; ac: anterior commissure; AHN: anterior hypothalamic nucleus; AI: agranular insular area; AONpv: anterior olfactory nucleus, postero-ventral; AUD: auditory areas; AVPV: antero-ventral periventricular nucleus; BAC: bed nucleus of anterior commissure; BSTpl: Bed nucleus of the stria terminalis, posterolateral; CA3v: ventral hippocampal formation, CA3; CBpj: purkinje layer of the cerebellum; CENT: central lobule of the cerebellum; COA: cortical amygdala; CSm: superior central nucleus raphe, medial part; CSl: superior central nucleus raphe, lateral part; DCO: dorsal cochlear nucleus; DR: Dorsal raphe nucleus; DMHa: dorsomedial nucleus of the hypothalamus, anterior part; ECT: ectorrinal area; ENT: entorrinal area; FL: flocculus; NLOT: nucleus of the lateral olfactory tract; gcl: granule cell layer; GU: gustatory area; IC: inferior colliculus; ICe: inferior colliculus external nucleus; IF: interfascicular nucleus of the raphe; IL: infralimbic area; IPN: interpeduncular nucleus; ISN: inferior salivatory nucleus; KF: Koelliker-Fuse subnucleus; LH: lateral habenula; ISN: inferior salivatory nucleus; LPO: lateral preoptic area; MD: mediodorsal nucleus of the thalamus; MEA: medial amygdala; MEPO: median preoptic nucleus; MGm: medial geniculate complex, medial part; MH: medial habenula; MM: medial mammillary nucleus; MOB: main olfactory bulb; MOp: primary motor area; MOs: supplemental motor area; MPO: medial preoptic area; MRN: midbrain reticular nucleus; NTS: nucleus of the tractus solitarius; opt: optic tract; ORB: orbital area; OV: vascular organ of lamina terminalis; PA: posterior amygdalar nucleus; PAG: periaqueductal gray; PB: parabrachial nucleus; PCG: contine central gray; PG: pontine gray; PH: posterior hypothalamic nucleus; PIR: piriform area; PL: prelimbic area; POL: posterior limiting nucleus of the thalamus; PP: peripeduncular nucleus; PPN: pedunculo pontine nucleus; PRN: pontine reticular nucleus; PSTN: parasubthalamic nucleus; PSV: principal sensory nucleus of the trigeminal nerve; RR: midbrain reticular nucleus, retrorubral area; RSPd: retrosplenial area dorsal; RSPv: retrosplenial area dorsal; PVH: periventricular hypothalamic nucleus; PVi: periventricular hypothalamic nucleus, intermediate part; RM: raphe magnus; SC: superior colliculus; SCm: superior colliculus, motor related; SFO: subfornical organ; SPF: subparafascicular nucleus; STN: subthalamic nucleus; SUBd: subiculum dorsal part; SGN: suprageniculate nucleus; SOC: superior olivary complex; SUM: supramammillary nucleus; TEa: Temporal association area; TM: tuberomammillary nucleus; VCO: ventral coclear nucleus; VISam: anteromedial visual área; VISp: primary visual area; VISal: anterolateral visual area; VISl: lateral visual area; VISC: visceral area; vHi: ventral hippocampus; VMH: ventromedial hypothalamic nucleus; VMHc: ventromedial hypothalamic nucleus, central part; VMHdm: ventromedial hypothalamic nucleus, dorsomedial part; VMHvl: ventromedial hypothalamic nucleus, ventrolateral part; VTA: ventral tegmental area; ZI: zona incerta.
- https://cdn.elifesciences.org/articles/61718/elife-61718-fig1-data1-v2.pdf
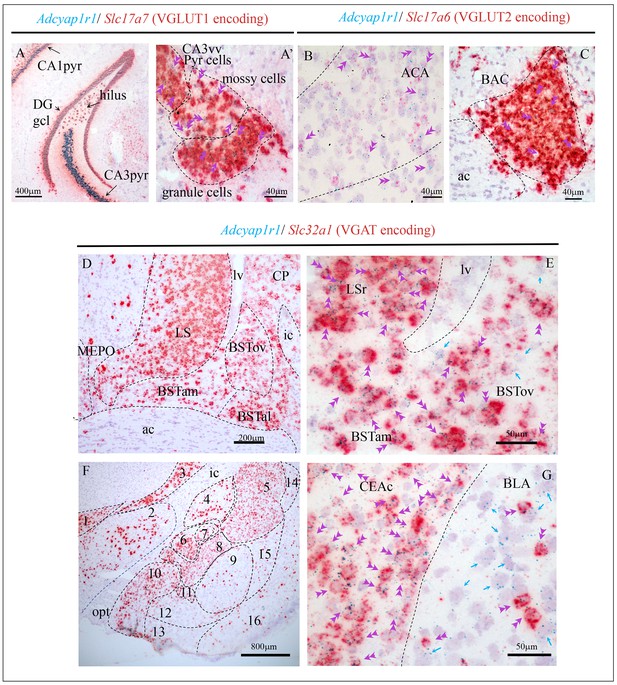
Examples illustrating Adcyap1r1 (RNA encoding PAC1) co-expression with glutamatergic (Slc17a7-VGLUT1 and Slc17a6-VGLUT2 expressing) and GABAergic (Slc32a1-expressing) neurons in cortical and subcortical regions.
(A and A') Temporal hippocampal formation where the Adcyap1r1 was strongly expressed in the principal neurons (pyr: pyramidal layer and DGgcl: dentate gyrus granule cell layer) as well as the VGLUT1+ mossy cells in the hilar region. Double arrows show single cells co-expressing Adcyap1r1 and Slc17a7. (B) Single-cell Adcyap1r1 co-expression with Slc17a6 (double arrowheads) was observed in ACA and (C) in the BAC. Regarding the GABAergic neurons expressing Adcyap1r1, the structures in the striatum and pallidum hosted very intensely expressing structures. Panel (D) shows the LS and BST in its three anterior divisions, anteromedial (BSTam), antero-lateral (BSTal) and oval (BSToval), as well as caudate-putamen (CP) with strong Adcyap1r1 expression. Panel (E) shows high-magnification photomicrograph where green dots (Adcyap1r1, PAC1 labeling), are mostly overlapped with red staining (Slc32a1, VGAT expression). Double pink arrowheads indicate co-expression within a single cell and single green arrows indicate cells only expressing Adcyap1r1. Panel (F) shows the amygdaloid complex and neighboring regions where Adcyap1r1 was strongly expressed in the GABAergic cell populations; (G) High-magnification photomicrograph showing that the Adcyap1r1 is exclusively expressed in Slc32a1 (VGAT) expressing neurons in the CEAc, while in the BLA it was expressed in the sparsely distributed GABAergic neurons as in most of the non-VGAT expressing neurons. 1. zona incerta of hypothalamus; 2. lateral hypothalamic area; 3: reticular nucleus of the thalamus; 4. globus pallidus; 5. caudate-putamen; 6: central amygdalar nucleus, medial part (CEAm); 7: lateral part (CEAl); 8: capsular part (CEAc); 9: basolateral amygdalar nucleus (BLA) 10: medial amigdalar nucleus; 11: intercalated nucleus of the amygdala; 12: basomedial nucleus of the amygdala; 13: cortical amygdalar area; 14: dorsal endopiriform; 15: ventral endopiriform; 16: piriform area. Fiber tracts: sm: stria medullaris; ac: anterior commissure; ic: internal capsule; opt: optic tract. lv: lateral ventricle.
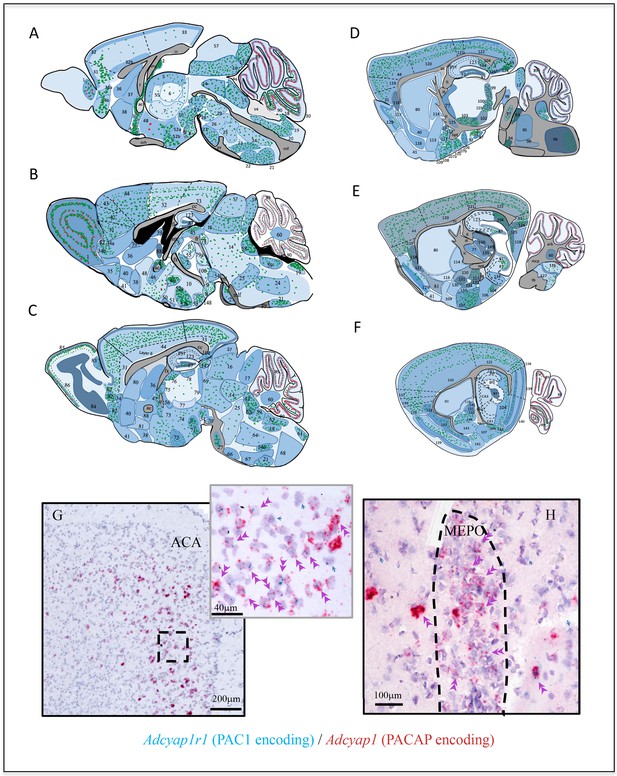
Adcyap1r1 expression assessment in relation to Adcyap1 expression suggests that the PACAP-PAC1 system uses autocrine, paracrine, and neuroendocrine modes for signal transduction.
(A–F) Mapping of Adcyap1r1 (RNA encoding Pac1) expression (symbolized by the intensity of blue shading) in six septo-temporal planes in relation to main PACAP containing brain nuclei and subfields, based on microscopic observations. Green and red dots represent Adcyap1 expressing neurons of glutamatergic (VGLUTs mRNA expressing) or GABAergic (VGAT mRNA expression) nature, respectively. Shaded regions with different blue intensity symbolize the strength of Adcyap1r1. For abbreviations see the corresponding table for abbreviations in Appendix 1. 1. MOB; 2. SFO; 3. MEPO; 4. OV; 5. PVT; 6. MD; 7. RE; 8. MBO; 9. SUM; 10. PH; 11. IF; 12. EW; 13. RL; 14. PAG; 15. DR.; 16 SCm; 17. IC; 18. AP; 19. NTS; 20. XII; 21. IO; 22. RPA; 23. RM; 24. GRN; 25. PRNr; 26. tegmental reticular n.; 27. PG; 28. IPN; 29. CLI 30. UVU; 31. ORB; 32. ACAd1; 32b. ACAd6; 33. RSP; 34 DP; 35. TTv; 36. LSr; 37. MS; 38. NDB; 39. LSc; 40. ACB; 41. OT; 42. ILA; 43. PL; 44. MOs; 45. MH; 46. BST.; 47. BAC; 48. MPO; 49. PVH; 50. SCH; 51. SO; 52a. DMH; 52b. VMH; 53. PVp; 54. VTA; 55. AM; 56. PT; 57. SCs; 58. PCG; 59. MV; 60. FN.; 61. CU; 62. SPIV.; 63. PB; 64. IRN; 65. LC; 66. SOC; 67. MARN; 68. MDRN; 69. MRN; 70. SN; 71. LM; 72. VLH; 73. LPO; 74. PHA; 75. AV; 76. AD; 77. RT; 78. PRC; 79. PF; 80. CP; 81. SI; 82. AON.; 83. AOB; 84. MOBgr; 85. MOBgl; 86: MOBml; 87.DG-gcl; 88. BSTov; 89. CBpj; 90. CBgcl; 91. CA3vv; 92. CA3v, 93. vhil; 94. MoV; 95. sV; 96. spV; 97. NLL; 98. PBG; 99. MG; 100. SPF; 101. PP; 102: ZI; 103. STN; 104. SUB; 105. MEApv; 106. PA; 107a. COAa; 107b. COAp; 108. NLOT; 109. AAA; 110. CEAm; 111. MEAad; 112. MEpd; 113. SI; 114. GPe; 115. GPi (entopeduncular nucleus); 116. FS; 117. EP; 118. CLA; 119. AI; 120. SS; 121. PTLp; 122. VIS; 123. DH; 124. POST; 125. PRE; 126. DCO; 127. VCO; 128. PAR; 129. PIR; 130. TT; 131. CEAc; 132. IA, 133. BMA; 134. CA2v; 135. LGv; 136. GU; 137. VISC; 138. ECT; 139. ENTl; 140. ENTm; 141. PAA; 142. TR; 143. BLA; 144. LA 145. CEAl 146. AMB; 147. OP; 148: PM.; 149. LGd; 150. IGL; 151. FL; 152. AN. Aq: aqueduct; och: optic chiasm; v4: forth ventricle; mlf: medial longitutinal fasciculus; cc: corpus callosum; vhc: ventral hippocampus commissure; fi/fx: fimbria/fornix; pyr: pyramidal layer; lot: lateral olfactory tract, mcp: middle cerebellar penducle; st: stria terminalis; opt: optic tract; ic: internal capsule; tb: trapezoid body; arb: arbor vidae. (G and H) Examples illustrating autocrine and paracrine features of PACAP-PAC1 signaling that Adcyap1r1 was expressed in PACAP containing (Adcyap1 expressing) neurons. (G) ACA in prefrontal cortex and (H) MEPO. Double arrowheads indicate co-expression and blue arrows indicate the Adcyap1r1 expressing neurons which are not Adcyap1 expressing but were adjacent to them.
-
Figure 3—source data 1
DISH mapping of Vipr1 co-expression with VGAT in selective brain regions.
Each panel from A to J show a low-magnification image of the coronal section analyzed, indicating with arrows the regions where the corresponding high-magnification photomicrographs were taken. The red signal corresponds to SLC32a1 (the mRNA for VGAT) and the green signal correspond to Vipr1 (the mRNA for VPAC1). The abbreviatures correspond to the Allen Brain Map and are indicated in the Figure 3—source data 5, where a comparison with the expression observed in the Vipr1 ISH experiments from Allen (73927619 and 77924538) and a semiquantitative analysis of the co-expression with VGAT mRNA was done. Scale bar: 2 mm for low-amplification and 50 µm for high-amplification photomicrographs.
- https://cdn.elifesciences.org/articles/61718/elife-61718-fig3-data1-v2.pdf
-
Figure 3—source data 2
DISH mapping of Vipr2 co-expression with VGAT in selective brain regions.
Each panel from A to J show a low-magnification image of the coronal section analyzed, indicating with arrows the regions where the corresponding high-magnification photomicrographs were taken. The red signal corresponds to Slc32a1 (the mRNA for VGAT) and the green signal correspond to Vipr2 (the mRNA for VPAC2). The abbreviations correspond to the Allen Brain Map and are indicated in the Figure 3—source data 5, where a comparison with the expression observed in the Vipr2 ISH experiments from Allen (1104 and 1105) and a semiquantitative analysis of the co-expression with VGAT mRNA was done. Scale bar: 2 mm for low-amplification and 50 µm for high-amplification photomicrographs.
- https://cdn.elifesciences.org/articles/61718/elife-61718-fig3-data2-v2.pdf
-
Figure 3—source data 3
Density of VipR1 mRNA expressing cells in the mouse brain: analysis of VGAT mRNA co-expression and comparison with data from Allen Brain Atlas.
- https://cdn.elifesciences.org/articles/61718/elife-61718-fig3-data3-v2.docx
-
Figure 3—source data 4
Density of VipR2 mRNA expressing cells in the mouse brain: analysis of VGAT mRNA co-expression and comparison with data from Allen Brain Atlas.
- https://cdn.elifesciences.org/articles/61718/elife-61718-fig3-data4-v2.docx
-
Figure 3—source data 5
Density distribution of PAC1 expressing cells in selective cortical regions.
- https://cdn.elifesciences.org/articles/61718/elife-61718-fig3-data5-v2.docx
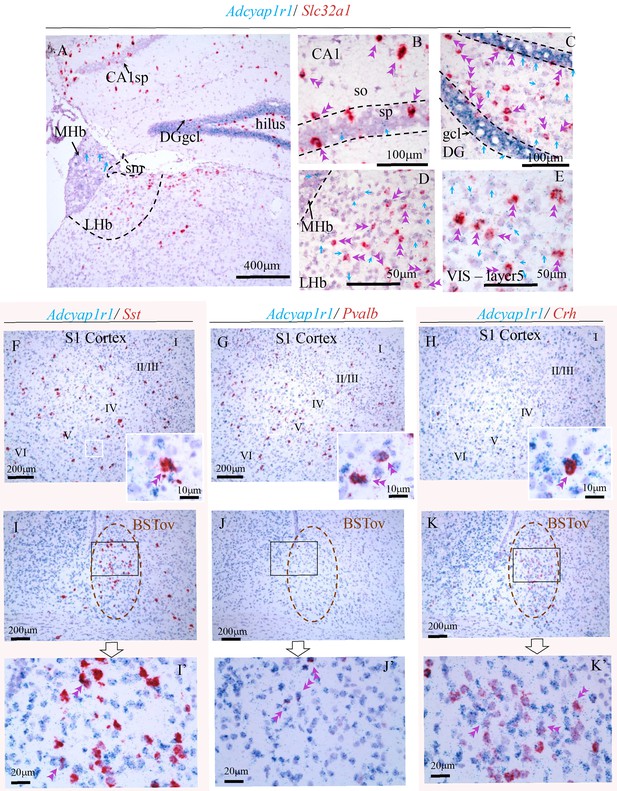
Examples illustrating PAC1 mRNA (Adcyap1r1) was highly co-expressed in cortical and subcortical GABAergic neurons.
(A) Low magnification showing epithalamic habenula and hippocampus with Slc32a1 (VGAT mRNA) expressing cells (red) sparsely distributed. B and C: in hippocampus CA1 (B), both stratum oriens (so) and stratum pyramidale (sp), and dentate gyrus (C), hilus of dentate gyrus (DG) and granule cell layer (gcl), PAC1 mRNA (Adcyap1r1, light blue) was co-expressed with all VGAT mRNA (Slc32a1, red) expressing neurons (double arrowheads). Some principal cells in sp also expressed PAC1 (single blue arrowheads) while in gcl, all principal cells co-expressed Adcyap1r1. (D) In lateral habenula, all the Slc32a1 cells co-expressed PAC1 mRNA (Adcyap1r1) as well as some no-VGAT expressing cells. (E) in primary visual cortex, layer 5, both Slc32a1 expressing and no-expressing cells express Adcyap1r1. F, G and H show the co-expression of Adcyap1r1 (PAC1) with main types of cortical GABAergic interneurons containing somatostatin, parvalbumin, and corticotropin releasing hormone (Sst, Pvalb and Crh respectively), in the primary somatosensory cortex (S1). I-VI indicate cortical layers. I, J, and K, the same mRNAs co-expression (of F, G, H) in the bed nucleus of stria terminalis, the oval subnucleus (BNSTov). I', J' and K' are amplifications of the corresponding black-squared fields. Double arrow heads indicate some co-expressed cells.
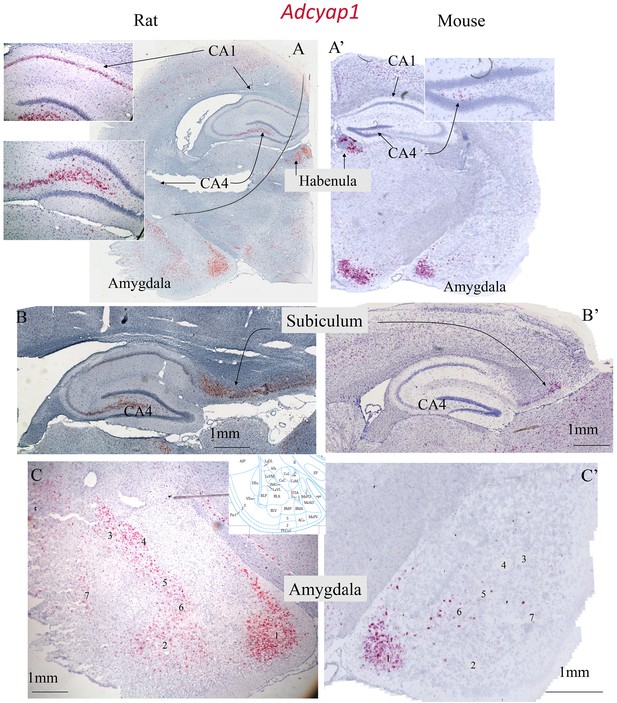
Neurochemical and anatomical divergence between the distribution of PACAP containing circuits in phylogenetically old and new brain areas were compared, revealing possible evolutionary divergence of PACAP function in mouse and rat.
PACAP containing cell distribution within the subcortical regions such as habenula and hypothalamus are relatively well conserved between the two rodent species. However, we observed more abundant Adcyap1 expression, in phylogenetically new regions in rats. Here are two limbic regions examined with RNAscope in situ hybridization using Adcyap1 probe (see the Materials and methods in the next section) as examples. A and A’: coronal sections from young adult male rat and mouse showing the conserved patterns of expression in subcortical regions of hypothalamus and habenula, but enhanced expression in rat in hippocampal formation and amygdala complex. Top inset of A shows hippocampus CA1 has high expression of Adcyap1, while in mouse this feature is not seen. Lower inset of A shows abundant expression of CA4/hilus region in rat while few of Adcyap1 expressed cells were observed in dorsal hilus region in mouse. B and B’, sagittal sections from rat and mouse showing the increased expression of Adcyap1 in rat subiculum. C and C’, comparison in amygdala complex while in rat, six regions with abundant expression of Adcypa1 were observed: 1. corticomedial amygdala CoM; 2. basal amygdala; 3. central amygdala, capsular; 4. central amygdala, lateral; 5. intercalated cells complex; 6. bed nucelus of stria terminalis, intra-amygdalar division. In the same regions, in mouse, some scattered Adcyap1-expressed cells can also be seen.
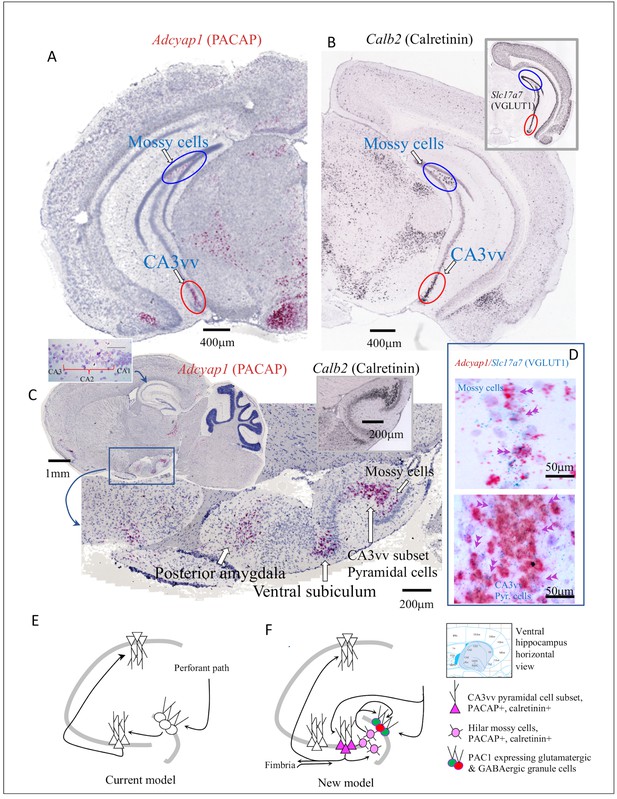
Ventral (temporal pole) hippocampus CA3 (CA3vv) contained a newly identified subset of pyramidal neurons distinguished by its molecular signatures of VGLUT1, PACAP and calretinin mRNA expression.
Low-magnification bright field coronal (A) and sagittal (C) whole-brain sections with ISH (RNAscope 2.5 High Definition (HD) Red Assay), showing the selective expression of mRNA of PACAP (Adcyap1) in a subset of CA3 of the temporo-ventral pole of hippocampus (red ovals circumscribed regions in A and C). Hilar mossy cells in the dorsal (A) and ventral (C) hippocampus, also Adcyap1 expressing, are circumscribed by blue ovals. (B) and inset show the corresponding coronal sections in low magnification of Calb2 (calretinin) and slc17a7 (VGLUT1) taken from Allen Brain Atlas (Ng et al., 2009) where CA3vv subset and hilar mossy cells are indicated with red and blue ovals, respectively). Right inset of C corresponds to calretinin mRNA expression in the same hippocampus sagittal squared region of C. Both the subset of CA3vv pyramidal neurons and the mossy cells co-expressed VGLUT1 mRNA (Slc17a7) and Adcyap1 (D). Left inset of C shows dorsal CA2 pyramidal layer expressed Adcyap1. The ‘trisynaptic-centric’ (E) vs ‘CA3-centric’ (F) view of hippocampal information processing, where the newly identified CA3vv subset of Adcyap1 and Calb2 containing glutamatergic neurons are presented in dark pink triangles and the mossy cells are in light pink circles. The Adcyap1r1 expressing granule cells (green) and interneurons (red) in the granule cell layer are symbolized with pink circle. Chartings were based on ventral pole of hippocampus (shaded region of atlas segment (Paxinos mouse brain), where this chemically distinct subset of CA3c pyramidal neurons was identified. Circuits were modified from Scharfman, 2007, with adaptation to the new finding from this study.
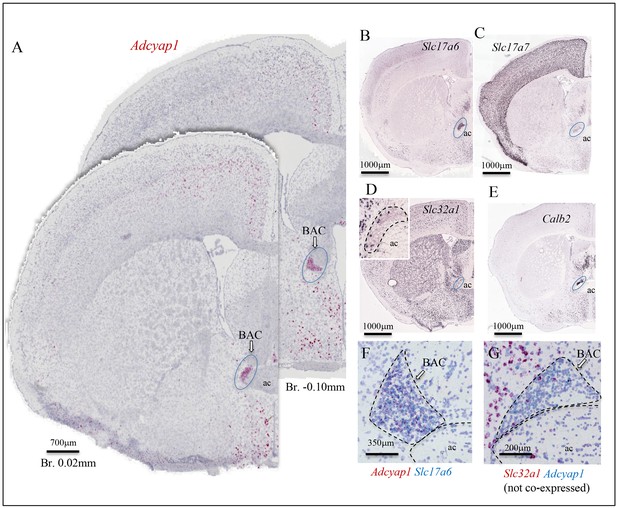
Bed nucleus of anterior commissure (BAC): a prominent PACAP containing glutamatergic nucleus chemo-anatomically identified.
(A) Two coronal sections at Bregma 0.02 mm and −0.10 mm of mouse brain showing Adcyap1 ISH (RNAscope 2.5 High Definition (HD) Red Assay) expressing BAC (ac: anterior commissure). Panels B–E are low-magnification photomicrographs taken from Allen Brain Atlas (Ng et al., 2009) showing the Slc17a6 (VGLUT2, (B)) Slc17a7 (VGLUT1, (C)) Slc32a1 (VGAT, (D) and inset), Calb2 (calretinin, (E)) expressed in BAC. The Adcyap1 expressing neurons were densely packed and co-expressed Slc17a6 (F) and we did not observe co-expression within the Slc32a1-expressing cells (G).
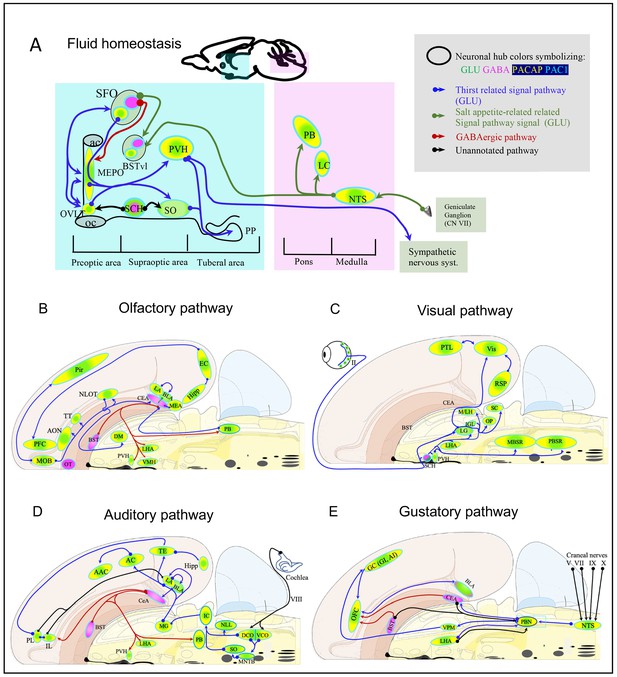
Mapping the spatial distribution of PACAP-PAC1 hubs within glutamate/GABA context in relevant sensory circuits in mice.
For abbreviations also see the corresponding table in Appendix 1. (A) Thirst and salt appetite-related pathways with PACAP-PAC1 glutamatergic / GABAergic signaling noted. The main figure is the enlargement of color-shaded areas of the box in the inset at the upper right, projected against a midsagittal section of mouse brain. Blue shaded area symbolizes the hypothalamus and pink shaded area the hindbrain. (B) Olfactory pathway. The projection neurons from the OB send their axons to the different structures of the olfactory cortex, among them AON, TT, OT, PIR, the amygdaline complex (LA, BLA, CEA, BST), ENT, and nLOT). (C) Visual pathway and circadian circuit for brain states. II: optic nerve; PVN: paraventricular nucleus; LHA: lateral hypothalamic area; LG: lateral geniculate nuclei; IGL: intergeniculate leaflet; M/LHb: medial and lateral habenula; OP: olivary pretectal nucleus, SC: superior colliculus; Vis: visual area; PTL: parietal association area; RSP: retrosplenial area; MBSR: midbrain behavioral state related (pedunculopontine nucleus, substantia nigra, midbrain raphe nuclei; PBSR: pons behavioral state related (locus coeruleus, superior central nucleus of raphe, pontine reticular nucleus). (D) Auditory pathway. VIII: cochlear nerve; DCO/VCO: dorsal and ventral cochlear nuclei; MNTB: medial nucleus of the trapezoid body; SO: superior olivary complex; NLL: Nucleus of the lateral lemniscus; IC: Inferior colliculus; MG: medial geniculate complex; AC: Auditory cortex; AAC: associate auditory cortex; TE: temporal association area; Hipp: hippocampus; LHA: lateral hypothalamic area; PVN: hypothalamic paraventricular nucleus; IL: infralimbic cortex; PL: prelimbic cortex. (E) Gustative pathway. V, VII, IX, X: represent trigeminal, facial, glossopharyngeal and vagus nerves respectively. NTS: Nucleus of the solitary tract (nucleus of tractus solitarius); NA: nucleus ambiguus; PBN: Parabrachial nucleus; VPM: ventropostero medial nucleus of the thalamus; BLA: Basolateral amygdala; CeA: central Amygdala; BST: bed nucleus of the stria terminalis; LHA: lateral hypothalamic area; RF: reticular formation.
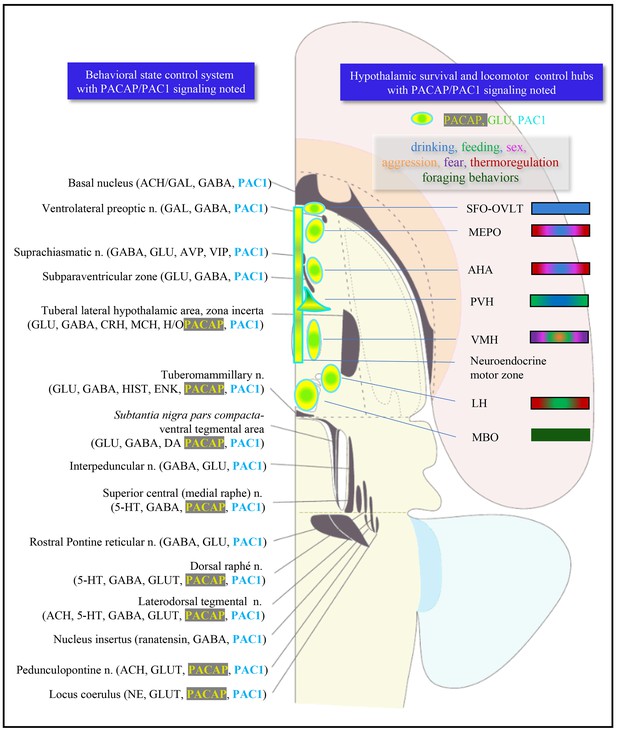
Presence of PACAP-PAC1 in major cells groups associated with behavioral state (left) and behavioral control system and hypothalamic instinctive survival system (right column).
Left column: critical nodes for behavioral state symbolized by dark gray shaded objects, modified from Swanson, 2012. In the longitudinal cell group-column of brain stem the key neurotransmitters are annotated. ACH, acetylcholine; CRH, corticotropin-releasing hormone; DA, dopamine; ENK, encephalin: GABA, gamma-amino butyric acid; GAL, galanin; GLUT, glutamate; H/O, hypocretin/orexin; HIST, histamine; MCH, melanin-concentrating hormone; NE, norepinephrine; 5HT, serotonin. Right column: hypothalamic survival circuit that consisted of discrete hypothalamic regions contain interoceptors for a variety of substances and have neuronal afferences from primary sensory systems to control the secretory and instinctive motor outputs. The rectangle in the midline represents the neuroendocrine motor zone for secretion of hypophysiotropic hormones, which include thyrotropin-releasing hormone, corticotropin-releasing hormone, growth hormone-releasing hormone, somatostatin, gonadotropin-releasing hormone, dopamine, neurotensin. SFO, subfornical organ; OVLT, organum vasculosum of lamina terminalis; MnPO, preoptic nucleus; AHN, anterior hypothalamic area; PVH, paraventricular hypothalamic nucleus; VMH, ventromedial hypothalamic nucleus; LH, lateral hypothalamic area; MBO, mammillary body (for general reference see Swanson, 2012).
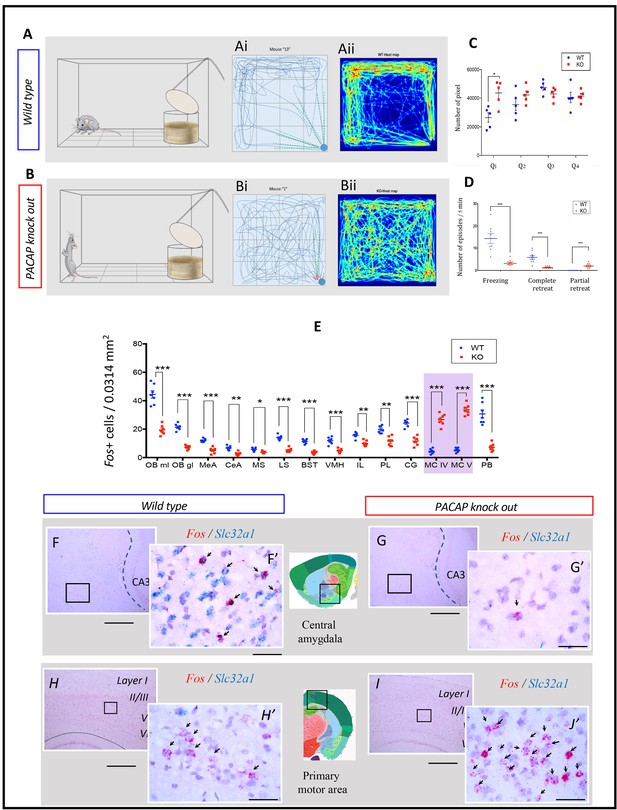
PACAP-deficient mice exhibited aberrant predator odor response and defensive behavior.
(A and B) Schematics of behavioral procedure using a modified open field test (OFT) to study defensive behavior during predator odor exposure, with typical behaviors of wild type (A: freezing) and PACAP KO (B, wandering, hyperactive and jumping) symbolized. (Ai and Bi) representative 2D movement tracking in the OFT of WT (Ai) and OK (Bi) mice, with dashed lines symbolizing complete (green lines) /incomplete (red lines) approach/sniff > retreat cycles. (Aii and Bii) 2D movement heat-map, WT, n = 9 mice; KO, n = 9 mice. (C) Pixel analysis reflecting total distance traveled in each of the four quadrants, being the Q1 the container located quadrant (clockwise numbering). The pixel numbers of per quadrant (Q) per mouse were compared that revealed a significant increase only in Q1 (WT vs KO, 26500 ± 3288 vs. 43640 ± 3921, *p<0.05. (D) Numbers of freezing, complete sniff>retreat or partial sniff>retreat episodes were assessed using Student t-test. For freezing (n = 8) and complete sniff>retreat (n = 9) behaviors, KO mice showed significant reduction WT:14.29 ± 2.18 vs KO: 3.18 ± 0.64; and WT: 5.89 ± 0.84 vs KO: 1.33 ± 0.17; for partial sniff>retreat behavior (n = 9) KO mice showed significant increase (WT: 0 vs KO: 2 ± 0.41). (E) Number of cells expressing fos mRNA 45 min after the predator odor test were quantified in a 0.0314 mm2 area and statistic significant differences between KO and WT were determined using a multiple t-test and the Bonferroni-Dunn method. There was a significant decrease in the expression of fos in the following regions of KO mice: olfactory bulb mitral layer ‘OB ml’ (WT: 44.4 ± 2.60 vs KO: 19.43 ± 1.25); olfactory bulb granule layer ‘OB gl’(WT: 21.43 ± 0.84 vs KO: 7.29 ± 0.52); medial amygdala ‘MeA’ (WT: 12 ± 0.44 vs KO: 5 ± 0.76); central amygdala ‘CeA’ (WT: 6.71 ± 0.57 vs KO: 2.71 ± 0.47); medial septum ‘MS’ (WT: 5.71 ± 0.42 vs KO: 3.71 ± 0.29); lateral septum ‘LS’ (WT: 14 ± 0.62 vs KO: 5.14 ± 0.56); bed nucleus of stria terminalis ‘BST’ (WT: 11.43 ± 0.57 vs KO: 3.57 ± 0.37); ventromedial hypothalamus ‘VMH’ (WT: 11.86 ± 0.91 vs KO: 4.86 ± 0.50); infralimbic cortex ‘IL’ (WT: 15.71 ± 0.714 vs KO: 9.86 ± 0.74); prelimbic cortex ‘PL’ (WT: 19.71 ± 0.97 vs KO: 11.43 ± 1.23); cingulate cortex ‘CG’ (WT: 24.28 ± 0.92 vs KO: 11.57 ± 1.15) and parabrachial nucleus ‘PB’ (WT: 30.71 ± 2.82 vs KO: 7.29 ± 1.02) and granule cell layer of cerebellum ‘CBgcl’ (WT: 13.14 ± 0.96 vs KO: 8 ± 0.76). In contrast, the number of fos nuclei was augmented in two motor related areas in KO animals related to WT: motor cortex, layer IV ‘MC IV’ (WT: 4.43 ± 0.65 vs KO: 26.71 ± 1.54) and motor cortex, layer V (WT: 5.28 ± 0.57 vs KO: 33.57 ± 1.43). (F and G) example of reduced fos expression pattern in central amygdala in KO mice and (H and I) augmented fos expression in primary motor cortex of KO mice. Scale bars: 200 µm for low-magnification and 20 µm for high-magnification insets. Statistic significant differences are depicted as ***p<0.001, **p<0.01, *p<0.05. Note that during the freezing analysis, two outliers, one in each experimental group were discarded because being 45 and 24 (counts), with 5 times and 11 times higher than the standard deviations for the corresponding groups, that is WT:14.29 ± 6.17 (mean ± SD) vs KO: 3.18 ± 1.80 (mean ± SD). This exclusion was based on criteria explained in NIH Rigor and Reproducibility Training course, https://www.nigms.nih.gov/training/documents/module4-sample-size-outliers-exclusion-criteria.pdf and http://www.itl.nist.gov/div898/handbook/prc/section1/prc16.htm.
-
Figure 8—source data 1
MatLab script for motion heatmap analysis.
- https://cdn.elifesciences.org/articles/61718/elife-61718-fig8-data1-v2.docx
-
Figure 8—source data 2
Raw data for panels C, D and E.
- https://cdn.elifesciences.org/articles/61718/elife-61718-fig8-data2-v2.docx
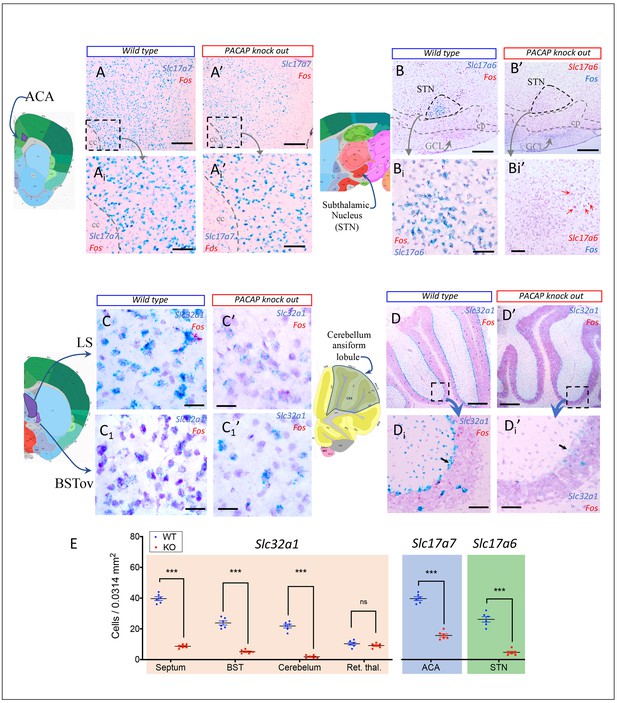
PACAP-deficient (KO) mice showed significant down-regulation of vesicular transporters for glutamate and GABA in regions where Adcyap1 or Adcyap1r1 were strongly expressed in WT mice.
(A–D) Examples of in situ hybridization using RNAscope method showing down-regulation in KO mice (A’, B’, C’, D’) of Slc17a7 (VGLUT1 mRNA) in anterior cingulate area (ACA) (panels As), Slc17a6 (VGLUT2 mRNA) in hypothalamic subtalamic nucleus (STN) (Panels B) and of Slc32a1 (VGAT mRNA) in lateral septum (LS) and bed nucleus of stria terminalis, oval subnucleus (BSTov) (panels Cs), and cerebellar cortex, the ansiform lobule's, Purkinje’s cells (panels Ds). Note that the feature of reduced expression of Slc32a1 at both single-cell and cell density levels, can be clearly observed (arrows) in the Purkinje cells. (E) Number of cells expressing Slc32a1 (orange shading), Slc17a7 (blue shading) and Slc17a6 (green shading) were quantified in a 0.0314 mm2 area and statistically significant reductions of KO compared to WT were detected septum (WT: 39.67 ± 1.23 vs KO: 8.67 ± 0.49); BST (WT = 23.83 ± 1.35 vs KO = 5.17 ± 0.48); ansiform lobule of cerebellum (WT: 21.83 ± 1.19 vs KO: 1.83 ± 0.4); anterior cingulate area (WT: 39.67 ± 1.23 vs KO: 15.67 ± 1.05) and subthalamic nucleus (WT: 26.17 ± 1.78 vs KO: 4.67 ± 0.76), while in the reticular thalamic nucleus, a region that did not contain Adcyap1 expressing cells, no significant difference was detected (negative control). Statistic differences are depicted as ***p<0.001 and ns: not significant. Scale bars: A and A’: 300 µm; Ai and Ai’: 100 µm; Bs and Cs: 20 µm; D and D’: 500 µm; D1 and D1’: 100 µm.
-
Figure 9—source data 1
Raw data for panel E.
- https://cdn.elifesciences.org/articles/61718/elife-61718-fig9-data1-v2.docx
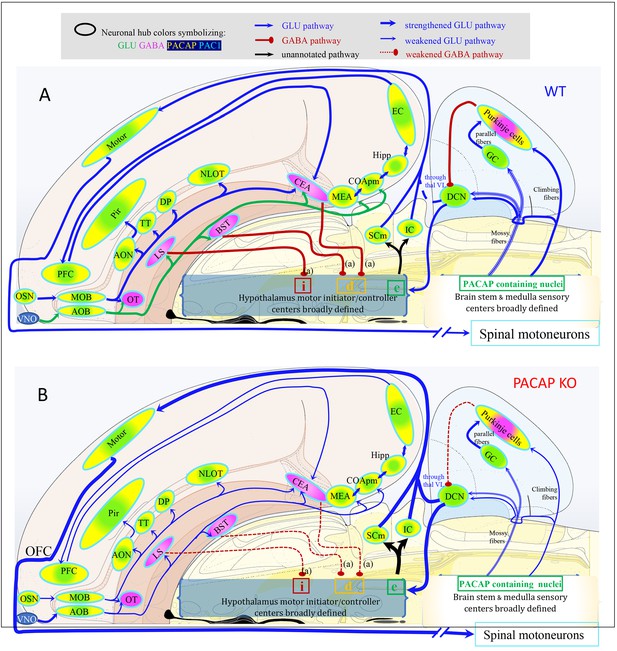
Proposed model to interpret how PACAP deficiency influence the olfactory information salience processing to impact motor output, bases on the analysis of this study.
(A) Schematic representations of the mouse peripheral and central olfactory pathways to cognitive centers and motor higher control centers in brain stem under normal condition. PACAP-Pac1 glutamatergic/GABAergic signaling is symbolized by colors. (B) PACAP deficiency weakens the corresponding neuronal hubs inducing ultimately unbalanced excitation/inhibition in sensory-cognitive and motor pattern initiator and controller centers, which may underlie the hyperactivity and attention deficit to salient olfactory stimulus during predator odor exposure test. OSN: olfactory sensorial neurons; VNO: vomeronasal organ; MOB: main olfactory bulb; AOB: accessory olfactory bulb; AON, anterior olfactory n.; TT: taenia tecta; OT: olfactory tubercle; DP: dorsal peduncular area; Pir: piriform cortex; NLOT: n. lateral olfactory tract; EC: entorhinal cortex; PFC: prefrontal cortex; COApm: corticoamygdalar, posteromedial; MEA and CEA: medial and central amygdala; LS: lateral septum; BNST: bed nucleus of stria terminalis; SCm: superior colliculus, motor related; IC: inferior colliculus; DCN: deep cerebellar nuclei; GC: granule cells; thal VL: thalamic ventrolateral n.; i: inhibition; d: disinhibition; e: excitation.
Tables
Distribution, cell types, and strength of main PACAPergic cell groups in mouse brain with comparison of rat brain reported by Hannibal, 2002*.
| Cell group / sub-field† | Hannibal, 2002 | Slc17a7 (VGLUT1) | Slc17a6 (VGLUT2) | Slc32a1 (VGAT) |
|---|---|---|---|---|
| Retina | ||||
| Ganglion cell layer‡ | + | - | +++ | - |
| Cerebrum: Cortical plate | ||||
| Olfactory area | ||||
| Main olfactory bulb | ||||
| Granular cell layer | - | - | - | - |
| Inner plexiform layer | - | - | - | - |
| Mitral cell layer | + | +++ | ++ | + |
| Outer plexiform layer | n.r. | +++ | +++ | + |
| Glomerular layer | - | + | + | + |
| Periglomerular cells | n.r. | + | + | + |
| Accessory olfactory bulb | ||||
| Mitral cell layer | + | ++++ | ++ | + |
| Glomerular layer | n.r. | + | + | + |
| Granular layer | n.r. | + | - | - |
| Other olfactory areas | ||||
| Ant olfactory n. lateral | ++ | ++++ | ++ | - |
| Ant olfactory n. medial | ++ | ++++ | ++ | - |
| Dorsal peduncular area | n.r. | +++ | ++ | ++ |
| Taenia Tecta | n.r. | +++ | - | - |
| Piriform area: Pir2 | n.r. | + | - | - |
| Piriform area: Pir3 | n.r. | ++++ | + | - |
| N. lat. olfactory tract (NLOT) | ++++ | ++++ | ++ | + |
| Cortical amygdalar area (CoA) | n.r. | + | ++++ | - |
| Hippocampal formation | ||||
| Hippocampal region | ||||
| Dorsal dentate gyrus | n.r. | - | - | - |
| Dorsal hippocampus CA1 | + | - | - | - |
| Dorsal hippocampus CA2 | n.r. | + | - | - |
| Dorsal hippocampus CA3 | + | - | - | - |
| Dorsal hilus | n.r. | ++ | - | - |
| Ventral dentate gyrus | n.r. | - | - | - |
| Ventral CA3vv | n.r. | +++++ | - | - |
| Ventral hilus | n.r. | ++ | - | - |
| Retrohippocampal regions | ||||
| Entorhinal area | n.r. | + | + | - |
| Parasubiculum | ++ | +++ | +++ | - |
| Postsubiculum | ++ | +++ | +++ | - |
| Presubiculum | n.r. | +++ | +++ | - |
| Subiculum | n.r. | +++ | +++ | - |
| Isocortex§ | ||||
| Layer I | + | n.a | n.a | n.a |
| Layer II-II | ++ | n.a | n.a | n.a |
| Layer IV | - | n.a | n.a | n.a |
| Layer V | ++ | n.a | n.a | n.a |
| Layer VI | + | n.a | n.a | n.a |
| Agranular insular cortex | n.r. | ++++ | ++ | - |
| Somatomotor areas | ||||
| 2ry motor area, layer 2–3 | n.r. | +++ | - | - |
| 2ry motor area layer 5 | n.r. | ++++ | ++ | + |
| 1ry motor area, layer 2–3 | n.r. | +++ | - | - |
| 1ry motor area, layer 5 | n.r. | ++++ | ++ | + |
| Orbital frontal cortex (OFC) | ||||
| OFC 1 | n.r. | ++ | ++ | - |
| OFC 2/3 | n.r. | +++ | + | - |
| OFC 5 | n.r. | +++ | - | - |
| Prefrontal cortex (PFC) | ||||
| Ant cingulate cortex (ACC): ACC 2/3: ACC 5: | n.r. n.r. | ++++ +++ | + + | - - |
| Prelimbic (PL) PL 2/3 PL 5 | n.r. n.r. | ++++ +++ | + + | - - |
| Infralimbic (IL) IL 2/3 IL 5 | n.r. n.r. | ++++ +++ | + + | - - |
| Cell group / sub-field† | Rat Hannibal JCN, 2002 | Slc17a7 (VGLUT1) | Slc17a6 (VGLUT2) | Slc32a1 (VGAT) |
| Prim somatosensory a. SSp, SSp 1 SSp 2/3 SSp 4 (mouth) SSp 5 SSp 6a | - - n.r. - - | + +++ ++ ++ ++ | - - - - - | - - - - - |
| Gustatory areas | n.r. | ++++ | - | - |
| Auditory area | n.r. | +++ | - | - |
| Visual area | n.r. | +++ | - | - |
| Visceral area | n.r. | +++ | - | - |
| Temporal association area | n.r. | +++ | - | - |
| Ectorhinal area | n.r. | +++ | - | - |
| Perirhinal area | n.r. | ++ | - | - |
| Retrosplenial area | n.r. | ++++ | - | - |
| Post parietal association area | n.r. | ++++ | - | - |
| Cortical subplate | ||||
| Claustrum | n.r. | + | + | - |
| Endopiriform nucleus | n.r. | + | + | - |
| Lateral amygdalar nucleus | n.r. | ++++ | ++ | - |
| Post amygdalar nucleus (PA) | n.r. | ++++ | + | - |
| Basomedial amygdala | - | + | + | - |
| Basolateral amygdala | + | + | + | - |
| Cerebral nuclei | ||||
| Striatum | ||||
| Lateral septal nucleus | n.r. | ++ | ++ | - |
| Anterior amygdala area | n.r. | + | + | - |
| Central amygdalar nucleus | + | - | - | + |
| Medial amygdalar nucleus | ++ | +++ | +++ | + |
| Pallidum | ||||
| Bed nucleus of Stria Terminalis (BST) | + | n.a | n.a | n.a |
| BST oval | n.r. | - | + | + |
| BST am | n.r. | - | ++ | ++ |
| BST dm | n.r. | - | + | + |
| BST pr | n.r. | + | ++ | ++ |
| Bed nucleus of anterior commissure | n.r. | +++++ | +++++ | - |
| Brain stem, inter-brain | ||||
| Thalamus | ||||
| Somato-motor related | ||||
| Subparafacicular nucleus, magnocellular part | n.r. | + | ++ | - |
| Subparafacicular area | n.r. | - | +++ | - |
| Peripeduncular nucleus | n.r. | - | +++ | - |
| Medial geniculate complex | n.r. | - | +++ | - |
| Polymodal association cortex related | ||||
| Lat. Posterior n. thal | n.r. | + | ++ | - |
| Post. Limiting nucleus | n.r. | + | ++ | - |
| Suprageniculate n. | n.r. | + | ++ | - |
| Anterodorsal n. | - | +++ | + | - |
| Anteromedial n. | n.r. | ++ | ++ | - |
| Parataenial n. | n.r. | ++ | ++ | - |
| Intermedial n. | n.r. | + | + | - |
| Laterodorsal n. | n.r. | + | ++ | - |
| Centrolateral n | n.r. | - | ++ | - |
| Intermediodorsal n. | n.r. | + | ++ | - |
| Mediodorsal n. | n.r. | +++ | + | - |
| Pariventricular n. | - | + | +++ | - |
| Parateanial n. | n.r. | + | ++ | - |
| N. of reuniens | - | + | +++ | - |
| Posterior pretectal n. | +++ | - | +++ | - |
| Precommissural n. | +++ | - | + | - |
| Cell group / sub-field† | Rat Hannibal JCN, 2002 | Slc17a7 (VGLUT1) | Slc17a6 (VGLUT2) | Slc32a1 (VGAT) |
| Epithalamus | ||||
| Medial habenula¶ | ++++ | ++++ | ++++ | - |
| Lateral habenula | ++++ | - | ++++ | - |
| Hypothalamus | ||||
| Paraventricular n | + | - | + | - |
| Periventricular n | + | - | ++ | - |
| Anterodorsal preoptic n. | n.r. | - | + | - |
| Anteroventral | n.r. | - | +++ | - |
| Dorsomedial n. | +++ | - | +++ | - |
| Median preoptic n. | +++ | - | ++++ | - |
| Medial preoptic area | +++ | - | ++ | - |
| Vascular organ of lamina terminalis | +++ | - | ++++ | - |
| Posterodorsal preoptic n. | n.r. | - | + | - |
| Subfornical organ | ++++ | - | ++++ | - |
| Lateral preoptic area | n.r. | - | ++ | - |
| Anterior hyp. area | ++ | - | ++ | - |
| Premammillary n. | n.r. | - | ++ | + |
| Lateral mammillary n. | ++++ | - | ++++ | - |
| Medial mammillary n. | - | - | +++ | - |
| Supramammillary n. | - | - | ++ | + |
| Median preoptic n. | ++ | - | ++ | - |
| Lateral hyp. area | ++ | - | ++ | - |
| Preparasubthalamic n. | n.r. | - | +++ | - |
| Parasubthalamic n. | n.r. | - | +++ | - |
| Subthalamic nucleus | - | - | +++++ | - |
| Retrochiasmatic area | n.r. | - | +++ | - |
| Tuberomammillary nucleus | - | - | ++ | + |
| Zona incerta | + | - | ++ | - |
| Ventromedial hyp. n | ++++ | - | +++++ | - |
| Post. hypothalamic n. | n.r. | - | +++ | - |
| Midbrain | ||||
| Sensorial related | ||||
| Inf. colliculus (IC), central and external n. | n.r. | - | ++ | - |
| N. of the brachium of IC | n.r. | - | ++ | - |
| N. saculum | n.r. | - | + | - |
| Parabigeminal n. | n.r. | - | + | - |
| Midbrain trigeminal n. | n.r. | - | ++ | - |
| Motor related | ||||
| Ventral tegmental area | n.r. | - | ++ | - |
| Midbrain reticular n. | n.r. | - | + | - |
| Superior colliculus, motor related | n.r. | - | +++ | - |
| Periaqueductal gray | n.r. | - | +++ | - |
| Cuneiform n. | n.r. | - | ++ | - |
| Edinger-Westphal n. | n.r. | - | + | - |
| Interfascicular n. Raphe | n.r. | - | ++ | - |
| Behavior state related | ||||
| Midbrain raphe nuclei | n.r. | - | - | - |
| Pedunculopontine n. | n.r. | - | ++ | - |
| Dorsal n. raphe | n.r. | - | - | - |
| Central linear n. raphe | n.r. | - | ++ | - |
| Rostral linear n. raphe | n.r. | - | - | - |
| Olivary pretectal nucleus | n.r. | - | +++ | - |
| Cell group/sub-field† | Rat Hannibal JCN, 2002 | Slc17a7 (VGLUT1) | Slc17a6 (VGLUT2) | Slc32a1 (VGAT) |
| Hindbrain | ||||
| Pons | ||||
| Sensory related | ||||
| N. lateral lemniscus | n.r. | - | + | - |
| Principal sensorial nucleus of trigeminal nerve | - | - | + | - |
| Koelliker-Fuse subnucleus | n.r. | ++++ | - | - |
| Parabrachial n. lateral div. | n.r. | - | ++++ | - |
| Parabrachial n, rest subfields | n.r. | - | +++ | - |
| Superior olivary comp (lat) | n.r. | + | - | - |
| Motor related | ||||
| Tegmental reticular n. | n.r. | - | +++ | - |
| Barrington’s nucleus | n.r. | - | +++ | - |
| Dorsal tegmental n. | n.r. | - | + | - |
| Pontine gray | n.r. | - | +++ | - |
| Pontine central gray | n.r. | - | + | - |
| Supratrigeminal nucleus | n.r. | - | + | - |
| Behavior state related | ||||
| Locus Coerulus (state) | + | ++ | +++ | - |
| Laterodorsal tegmental n. | + | - | +++ | - |
| Pontine reticular n. | n.r. | - | + | - |
| Superior central n. raphe | n.r. | - | + | + |
| Medulla | ||||
| N. tractus solitarii medial | +++ | ++ | ++++ | - |
| N. tractus solitarii lateral | +++ | - | ++++ | - |
| Hypoglossal (XII) n. | - | ++ | - | |
| Dorsal motor n. of the vagus nerve (X) | +++ | +++ | - | - |
| Dorsal cochlear n. | +++ | ++ | ++ | - |
| Ventral cochlear n. | n.r. | ++ | ++ | - |
| Spinal n. trigeminal | n.r. | - | ++ | - |
| N. prepositus | n.r. | - | ++ | - |
| Inferior salivatory complex | n.r. | - | ++ | - |
| Facial motor n. (VII) | n.r. | - | ++ | - |
| N. ambiguus | +++ | - | ++ | - |
| Magnocellular reticular n. | n.r. | - | ++ | - |
| Parapyramidal n. | n.r. | - | ++ | - |
| Spinal vestibular n. | +++ | - | ++ | - |
| N. X | n.r. | - | + | - |
| N. raphe magnus (state related) | n.r. | - | ++ | - |
| N. raphe pallidus (state related) | n.r. | - | ++ | - |
| N. raphe obscurus (state rel.) | n.r. | - | ++ | - |
| Cuneate n. | - | ++ | ++ | - |
| Inferior olivary | n.r. | - | ++ | - |
| Cerebellar cortex | ||||
| Purkinje’s cells | ++ | - | - | +++++ |
| Golgi’s cells | n.r. | - | - | + |
| Granule cells¶ | - | ++ | - | - |
| Cerebellar nuclei | ||||
| Interposed n. | ++ | + | - | - |
| Dentate n. | n.r. | - | + | - |
-
n.a.: not applicable.
n.r.: not reported (blue color text refers to Hannibal, 2002) rat PACAPergic cell group and expression strength analysis.
-
*Similar semiquantitative annotations are used here the percentage of expressing cell/total Nissl stained nuclei: '-', not detectable; '+', weak (<20%); '++', low (40–20%); '+++', moderate (60–40%); '++++', intense (80–60%); '+++++', very intense (>80%).
† Functional neuroanatomy order and annotations are based on Allen Institute Mouse Reference Atlas.
-
‡ Circadian oscillating expression (Lindberg et al., 2019).
§ Isocortex expression was regionally evaluated.
-
¶ Dorsal half of the MHb which co-express Calb2 (RNA encoding calretinin).
** Prominent in lobules paraflocculus, central and uvula. Coincide with calretinin (Calb2) expression.
Distribution, cell types, and strength of main PAC1 expressing group in mouse cerebral nuclei (striatum and pallidum).
| Cell group / sub-field | Slc17a7 (VGLUT1) | Slc17a6 (VGLUT2) | Slc17a8 (VGLUT3) | Slc32a1 (VGAT) |
|---|---|---|---|---|
| Striatum | ||||
| Caudoputamen | ||||
| Nucleus accumbens | - | - | - | +++ |
| Fundus of striadum | - | - | - | ++ |
| Olfactory tubercle | - | - | - | + |
| Lateral septum complex | - | - | - | ++++ |
| Medial amygdala (MeA) | ||||
| MeAav | - | ++++ | - | + |
| MeApd | - | + | - | ++++ |
| Central amygdala (CeA) | ||||
| CeA medial | - | - | - | +++ |
| CeA lateral | - | - | - | +++ |
| CeA capsular | - | - | - | ++++ |
| Anterior amygdala area | - | - | - | +++ |
| Intercalated nucleus | - | - | - | ++++ |
| Cell group / sub-field | Slc17a7 (VGLUT1) | Slc17a6 (VGLUT2) | Slc17a8 (VGLUT3) | Slc32a1 (VGAT) |
| Pallidum | ||||
| Globus pallidum internal | - | - | - | ++ |
| Globus pallidum external | - | - | - | ++ |
| Globus pallidum ventral (VP) | ||||
| Substantia innominata | - | - | - | ++ |
| Magnocellular nucleus | - | - | - | +++ |
| Medial septal complex | - | - | - | +++ |
| Bed nuclei stria terminalis (BNST) | ||||
| BNSToval | - | - | - | ++++ |
| BNSTam | - | - | - | +++ |
| BNSTdm | - | - | - | +++ |
| BNSTpr | - | - | - | +++ |
| Nucleus of Diagonal Band | - | - | - | ++++ |
| Bed nucleus of anterior commissure | + | + | - | - |
-
Semiquantitative annotations are used here the percentage of expressing cell/total Nissl stained nuclei: ‘-”, not detectable; '+”' weak (<20%); '++', low (40–20%); '+++', moderate (60–40%); '++++', intense (80–60%); '+++++', very intense (>80).
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus), both sexes | Wildtype C57Bl/6N | In-house breeding program in accordance with NIH guidelines and standards and housed in cages containing3–5 male siblings. | C57Bl/6N | |
| Strain, strain background (Mus musculus), both sexes | PACAP KO mice C57Bl/6N | Hamelink et al Proc Natl Acad Sci, 2002, 99(1):461–6, PMID:11756684 NIH in-house breeding program | C57Bl/6N Adcyap1-/- | |
| Commercial assay or kit | RNAscope detection assay probes | 2020 Advanced Cell Diagnostics, Inc | 405911 409561 319171 416331 319191 316921 502231 465391 404631 421931 316091 | Adcyap1 Adcyap1r1 Slc17a6 Slc17a7 Slc32a1 Fos Vipr1 Vipr2 Sst Pvalb Crh |
| Commercial assay or kit | RNAscope 2.5 HD Duplex detection kit Detection reagent-red | 2020 Advanced Cell Diagnostics, Inc | ---------- 322500 322360 | |
| Software, algorithm | MATLAB scripts | This paper | Script | For motion heatmaps |
Additional files
-
Source code 1
Matlab script for locomotion analysis.
- https://cdn.elifesciences.org/articles/61718/elife-61718-code1-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61718/elife-61718-transrepform-v2.pdf





