Induction of the IL-1RII decoy receptor by NFAT/FOXP3 blocks IL-1β-dependent response of Th17 cells
Figures
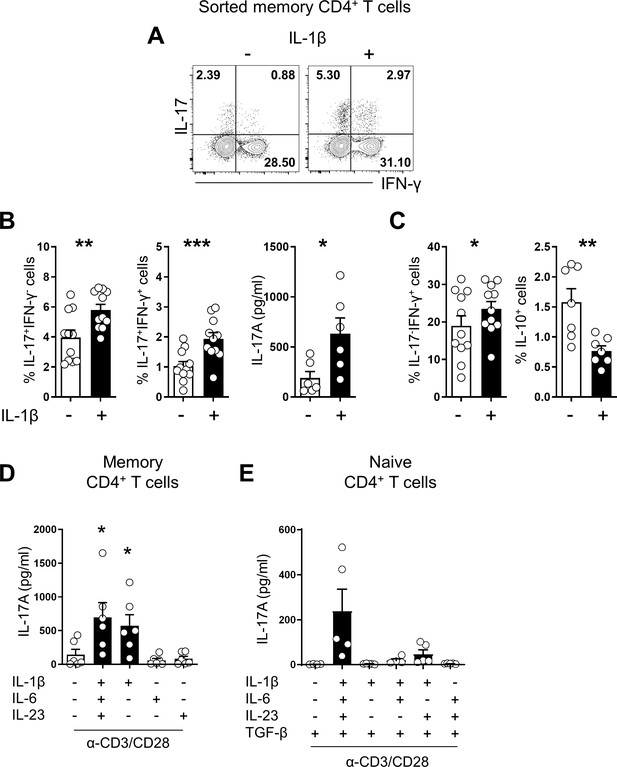
IL-1β is crucial for promoting human Th17 responses.
(A) Representative flow cytometric plot of IL-17 and/or IFN-γ production by human memory CD4+ T cells obtained from HCs and stimulated for 7 days with anti-CD3/28-coated microbeads with or without rhIL-1β (5 ng/ml). (B) Frequency (%) of IL-17-producing cells and IL-17/IFN-γ double-producing cells (flow cytometry) and the amount of IL-17 in the culture supernatant of (A) (ELISA). (C) Frequency (%) of IFN-γ-producing cells and IL-10-producing cells (flow cytometry). (D) The amount of IL-17 in the culture supernatant under the indicated cytokine conditions (ELISA): rhIL-6 (25 ng/ml), rhIL-23 (25 ng/ml), and rhL-1β (5 ng/ml). (E) Purifed naive CD4+ T cells were stimulated for 7 days with anti-CD3/28-coated microbeads in serum-free X-VIVO 10 medium under the indicated cytokine conditions. IL-17 in the culture supernatant (ELISA): rhIL-6 (25 ng/ml), rhIL-23 (25 ng/ml), rhL-1β (5 ng/ml), and rhTGF-β (10 ng/ml). Bar graphs show the mean ± SEM of six (A–D), and five (E) independent experiments. * = p<0.05, ** = p<0.01, and *** = p<0.001 by two-tailed paired t-test.
-
Figure 1—source data 1
Figure 1B IL-1β significantly enhance IL-17 & IFN-γ producing memory CD4+ T cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Figure 1C IL-1β regulated IL-10 producing memory CD4+ T cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Figure 1D Th17-polarizing cytokines intensify the Th17 response of memory CD4+ T cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig1-data3-v2.xlsx
-
Figure 1—source data 4
Figure 1E IL-1β is important for promoting Th17 differentiation form naïve CD4+ T cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig1-data4-v2.xlsx
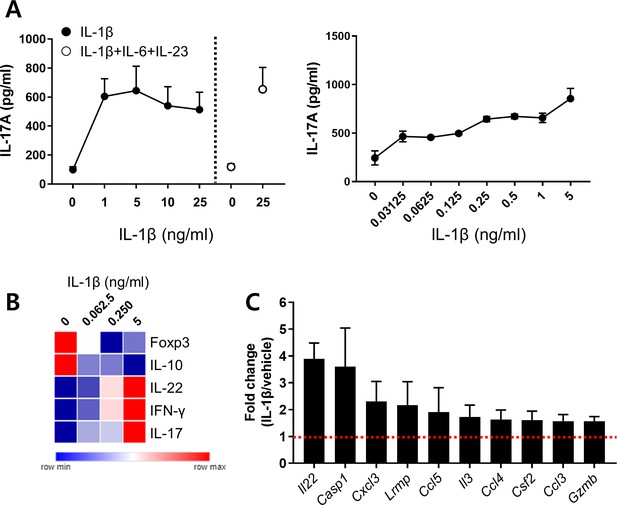
IL-1β is a crucial cytokine for promoting Th17 responses in humans.
Purified human memory CD4+ T cells were stimulated for 7 days with anti-CD3/28-coated microbeads under the indicated cytokine conditions: rhIL-6 (25 ng/ml) and rhIL-23 (25 ng/ml). (A) Amount of IL-17A in culture supernatants was quantified by conventional ELISA. (B) mRNA expression of the indicated genes was analyzed by qRT-PCR on day 7 post-stimulation. The relative gene expression under different concentrations of IL-1β is displayed using a heatmap. (C) Expression of pathogenic Th17 cell-associated genes in IL-1β (5 ng/ml)-treated CD4+ T cells was presented relative to their expression in PBS-treated CD4 T cells.
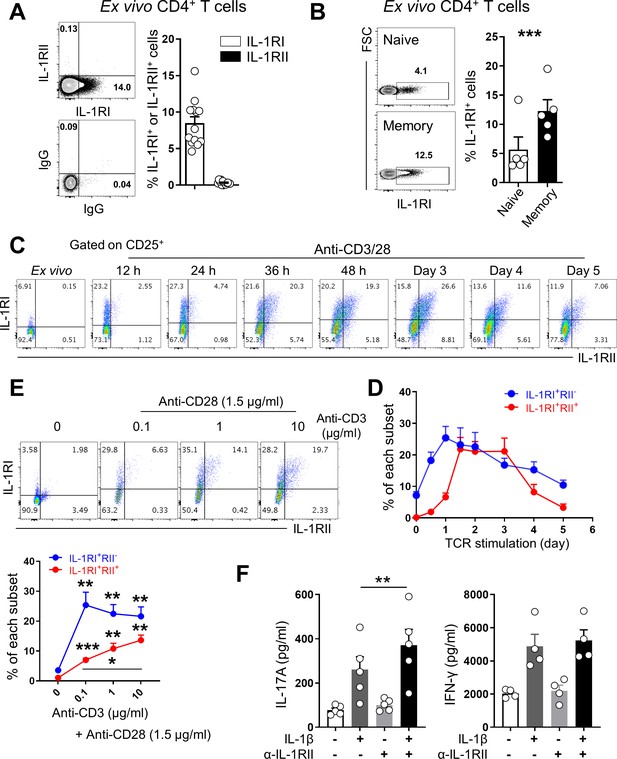
Dynamic regulation of IL-1β receptors by memory CD4+ T cells upon TCR stimulation.
(A) Representative flow cytometric plot and the frequency (%) of functional IL-1RI- and decoy IL-1RII-expressing CD4+ T cells of HCs (n = 13). (B) Representative flow cytometric plot and the frequency (%) of IL-1RI+ cells in naive (CD45RA+CCR7+) or memory (CD45RA-) CD4+ T-cell subset of HCs (n = 13). (C) Representative flow cytometric plot of change in IL-1RI and IL-1RII expression by TCR-stimulated memory CD4+ T cells (n = 7). (D) Time kinetics of IL-1RI and IL-1RII expression on TCR-stimulated memory CD4+ T cells (n = 5). (E) Representative flow cytometric plot and the frequency (%) of IL-1RI and IL-1RII expression by memory CD4 + T cells in response to stimulation with different concentrations of anti-CD3 and 1.5 μg/ml of anti-CD28 Ab for 48 hr (n = 5). (F) The amounts of IL-17A (left) and IFN-γ (right) in culture supernatants of memory CD4+ T cells stimulated with anti-CD3/28-coated microbeads for 7 days with or without IL-1β (n = 5). Anti-IL-1RII neutralizing Ab or control isotype Ab was added into the culture at day 2 post-stimulation. Bar graphs and line graphs show the mean ± SEM. * = p<0.05, ** = p<0.01, and *** = p<0.001 by two-tailed paired t-test.
-
Figure 2—source data 1
Figure 2A Ex vivo expression of IL-1RI & IL-1RII on CD4+ T cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Figure 2B Ex vivo expression of IL-1RI between naïve and memory CD4+ T cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Figure 2B Ex vivo expression of IL-1RI between naïve and memory CD4+ T cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig2-data3-v2.xlsx
-
Figure 2—source data 4
Figure 2D Time kinetics of IL-1RI & IL-1RII.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig2-data4-v2.xlsx
-
Figure 2—source data 5
Figure 2E Effect of TCR signaling strength on expression of IL-1RI & IL-1RII.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig2-data5-v2.xlsx
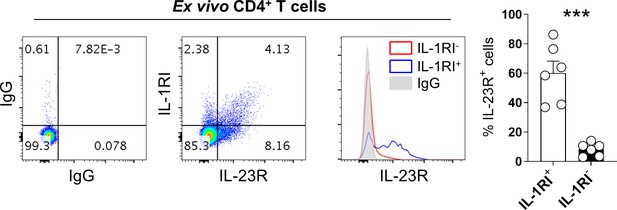
The differential expression of IL-23R on IL-1RI+ and IL-1RI- memory CD4+ T cells in humans.
Bar graphs show the mean ± SEM. *** = p<0.001 by two-tailed paired t-test.
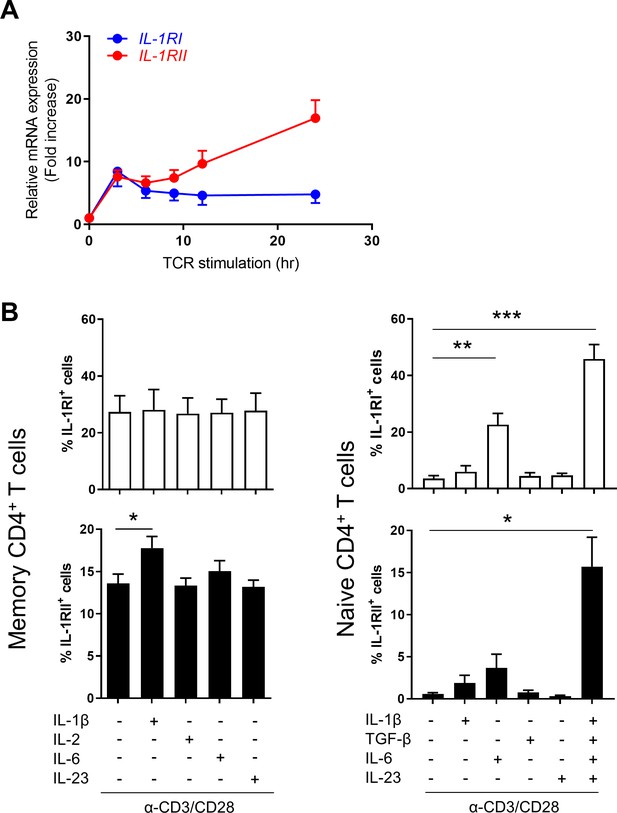
The expression of receptors for IL-1β is dynamically changed by various stimulations of CD4+ T cells.
(A) Purified human memory CD4+ T cells were stimulated with anti-CD3/28-coated microbeads. mRNA expression of IL-1RI and IL-1RII was analyzed by qRT-PCR at the indicated time-points (B) Purified human naive and memory CD4+ T cells were stimulated for 48 hr with anti-CD3/28-coated microbeads under the indicated cytokine conditions. The expression of IL-1RI and II was analyzed by flow cytometry (n = 6 for memory, n = 5 for naive CD4+ T cells). Bar graphs and line graphs show the mean ± SEM. * = p<0.05, ** = p<0.01, and *** = p<0.001 by two-tailed paired t-test.
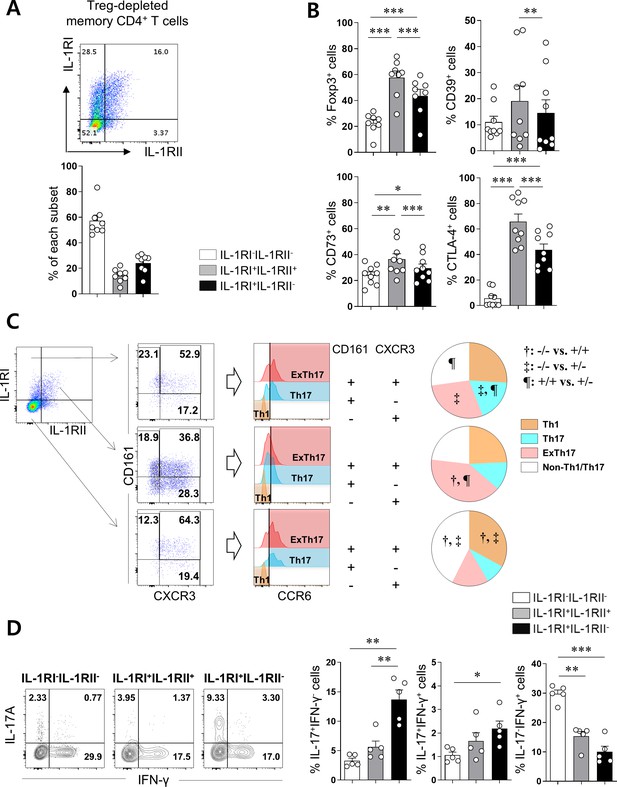
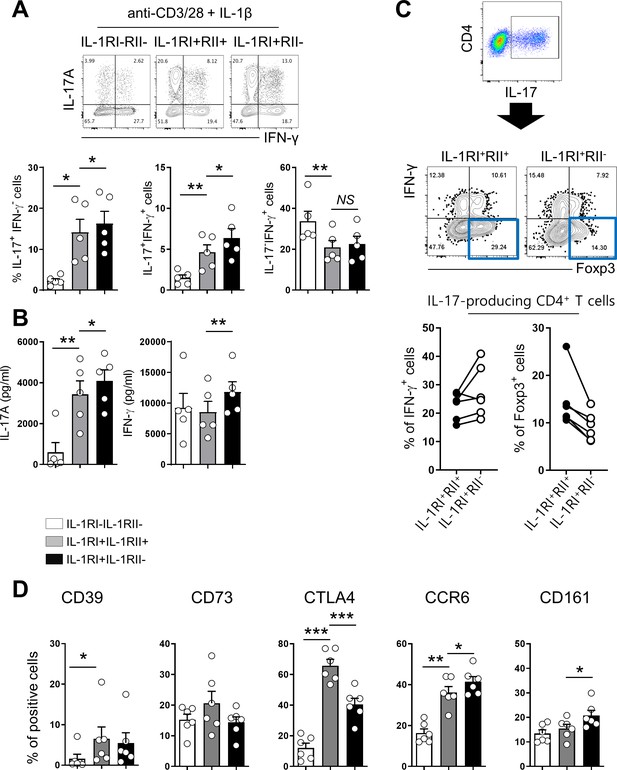
Differential expression pattern of IL-1Rs defines immunological features of CD4+ T cells.
(A) Treg-depleted memory CD4+ T cells were stimulated with anti-CD3/28-coated microbeads for 48 hr. The expression of IL-1RI and IL-1RII were analyzed by flow cytometry (n = 12). (B) The frequency (%) of Treg-related marker-expressing cells in IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- memory CD4+ T cell populations at day 2 post-stimulation (n = 12). (C) Representative flow cytometric plot and the frequency (%) of Th1 (CCR6-CD161-CXCR3+), Th17 (CCR6+CD161+CXCR3-), and ex-Th17 (CCR6+CD161+CXCR3+) cells in IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- memory CD4+ T cell populations (n = 9). (D) Intracellular cytokine staining (ICS) of IL-17- and IFN-γ in IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- memory CD4+ T cell populations (n = 4). Bar graphs and pie charts show the mean ± SEM. * = p<0.05, ** = p<0.01, and *** = p<0.001 by two-tailed paired t-test. †, ‡, and ¶ = p<0.05: compared between indicated subsets by two-tailed paired t-test (C).
-
Figure 3—source data 1
Figure 3A Frequency of IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- subset of Treg-depleted memory CD4+ T cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Figure 3B Expression of Treg related markers on L-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- subsets.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Figure 3C Frequency of ex-Th17, Th17, and Th1 of L-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- subsets.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig3-data3-v2.xlsx
-
Figure 3—source data 4
Figure 3D Expression of IL-17 & IFN-γ in L-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- subsets.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig3-data4-v2.xlsx
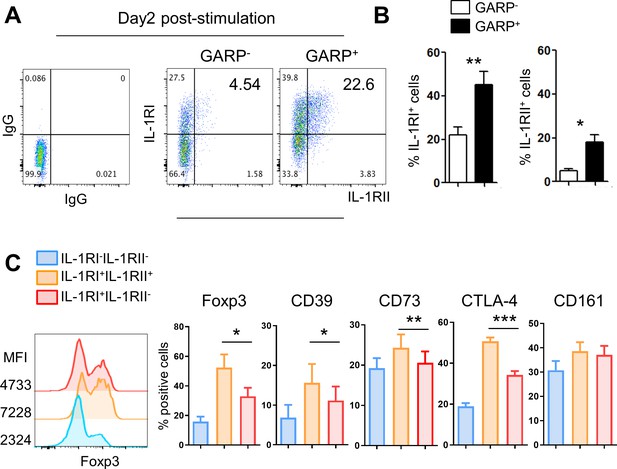
CD4+ T cells expressing GARP, a marker of activated Tregs, preferentially, but not exclusively, increase IL-1RII expression.
Purifed memory CD4+ T cells were stimulated with anti-CD3/28-coated microbeads. (A) Representative flow cytometric plot (A) and the frequencies (B) of IL-1RI and IL-1RII expression on GARP+ or GARP- memory CD4+ T cells at day 2 post-stimulation (n = 4). (C) The frequencies (%) of Treg- or Th17-related marker-expressing cells on IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- memory CD4+ T cells at day 2 post-stimulation (n = 12). Bar graphs show the mean ± SEM. * = p<0.05, ** = p<0.01, and *** = p<0.001 by two-tailed paired t-test.
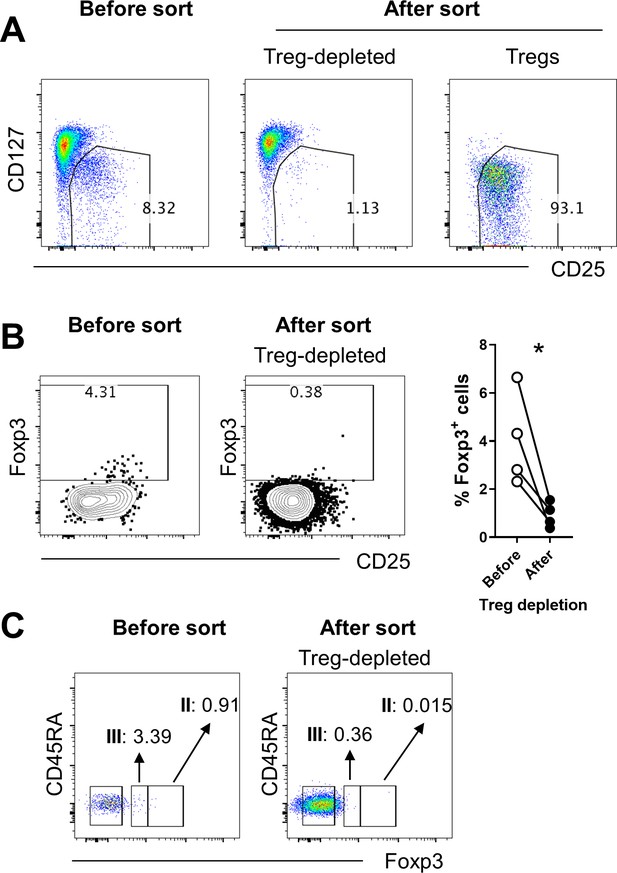
Efficiency of sorting-based Treg depletion from CD4+ T cells.
(A) Expression of CD127 and CD25 before and after sorting-based depletion of CD25highCD127dim/- Treg from memory CD4+ T cells. (B) Expression of Foxp3 before and after sorting-based depletion as performed in (A). (C) Expression of CD45RA and Foxp3 before and after sorting-based depletion as performed in (A) (n = 4). Faction II and III indicate activated Treg cells and non-suppressive cytokine-producing Foxp3low T cells, respectively, according to the definition of the Sakaguchi group.
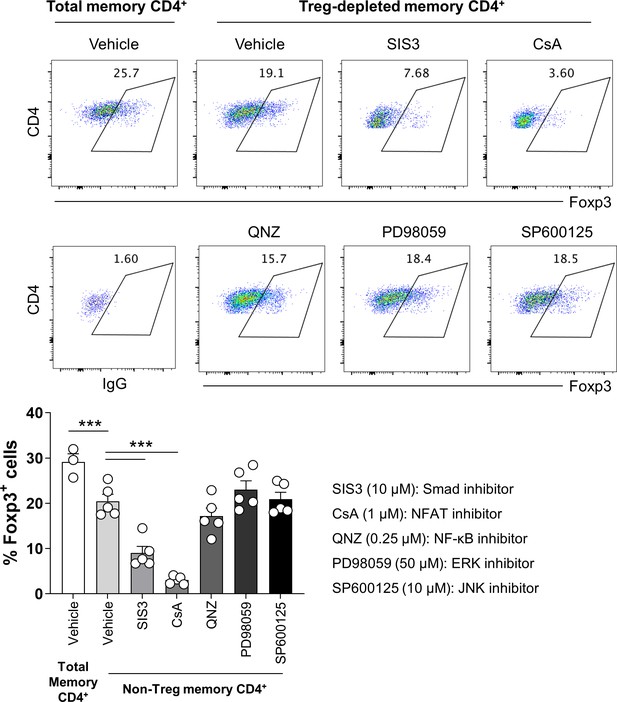
Critical roles of Smad3 and NFAT for Foxp3 expression in non-Treg memory CD4+ T cells.
Total or Treg-depleted memory CD4+ T cells were stimulated for 2 days with anti-CD3/28-coated microbeads in the presence of various signaling inhibitors. The expression of Foxp3 was analyzed by flow cytometry. Bar graphs show the mean ± SEM. *** = p<0.005 by two-tailed paired t-test.
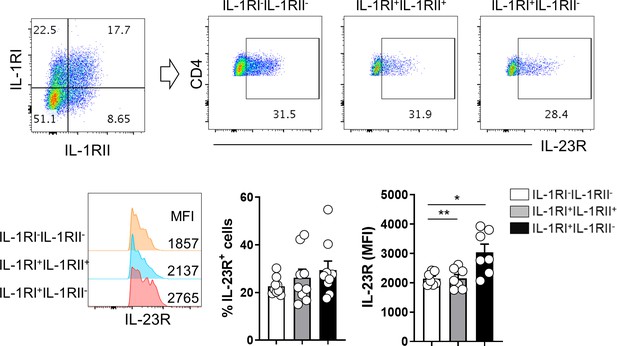
The correlation of IL-23R with IL-1RI+IL-1RII+ and IL-1RI+IL-1RII- subset of memory CD4+ T cells.
Purified memory CD4+ T cells were stimulated for 48 hr to induce the expression of IL-1RI and IL-1RII. The expression of IL-23R were compared among IL-1RI-IL-1RII-, IL-1RI+IL-1RII-, and IL-1RI+IL-1RII+ cells. MFI indicates mean fluorescent intensity. Bar graphs show the mean ± SEM. * = p<0.05 and ** = p<0.01 by two-tailed paired t-test.
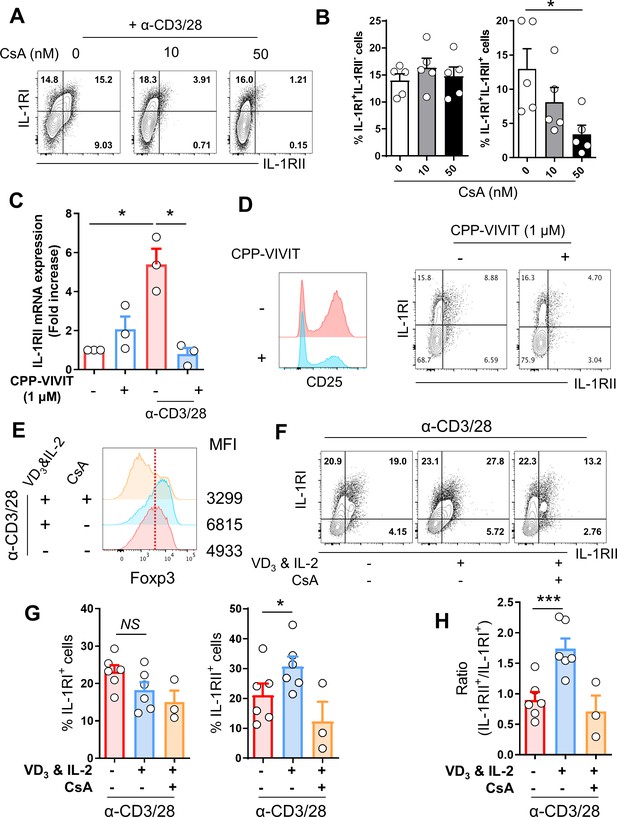
NFAT and Foxp3 are essential for IL-1RII expression in memory CD4+ T cells.
(A and B) Representative flow cytometric plot (A) and the frequency (B) of IL-1RI and IL-1RII expression on TCR-stimulated memory CD4+ T cells treated with the indicated concentration of CsA, a chemical inhibitor of NFAT at day 2 post-stimulation (n = 5). (C) The expression of IL-1RII mRNA in TCR-stimulated or unstimulated memory CD4+ T cells in the presence or absence of CPP-VIVIT (1 μM), NFAT-specific peptide inhibitor (n = 4). (D) The expression of CD25, IL-1RI, and IL-1RII on TCR-stimulated memory CD4 T cells in the presence or absence of CPP-VIVIT (n = 3). (E) Representative histogram plot of Foxp3 in TCR-stimulated memory CD4+ T cells in the presence or absence of 1,25-dihydroxyvitamin D3 (VD3; 10 μM) and rhIL-2 (250 IU/ml) with or without CsA. MFI indicates mean fluorescent intensity. (F) Flow cytometric analysis of IL-1RI and IL-1RII on memory CD4+ T cell under same conditions as in (E). (G and H) The frequencies (%) of IL-1RI+ and IL-1RII+ cells and the ratio of IL-1RII+ to IL-1RI+ cells under same conditions in (E) (n = 6). Bar graphs show the mean ± SEM. * = p<0.05, ** = p<0.01, and *** = p<0.001 by two-tailed paired t-test.
-
Figure 4—source data 1
Figure 4A NFAT inhibitor CsA selectivley repress the expression of IL-1RII.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Figure 4B NFAT inhibiton peptide VIVIT repress the expression of IL-1RII.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Figure 4G expression of IL-1RII significantly upregulated by treatment with 1,25(OH)2VD3 and IL-2 in memory CD4+ T cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig4-data3-v2.xlsx
-
Figure 4—source data 4
Figure 4H Ratio of IL-1RII+/IL-1RI+.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig4-data4-v2.xlsx
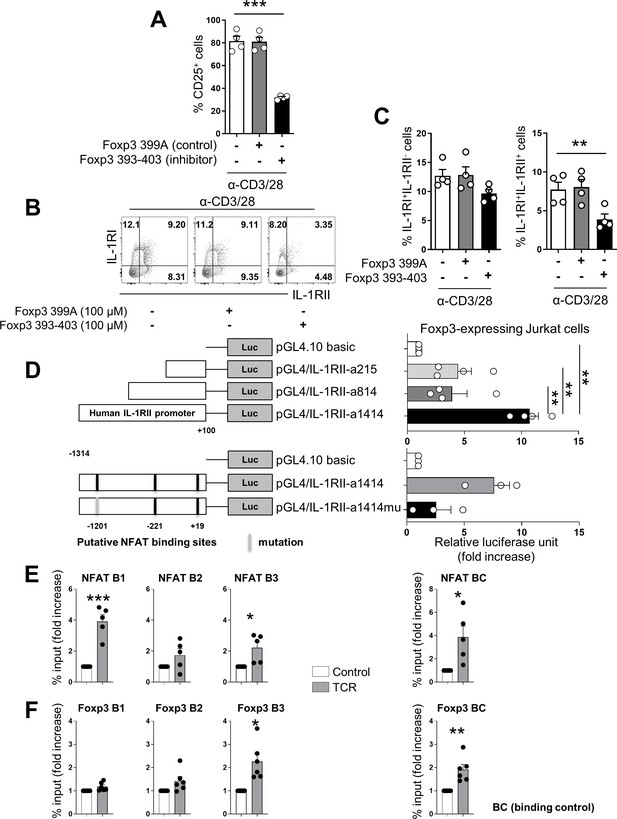
NFAT/FOXP3 interaction is responsible for expression of IL-1RII by human memory CD4+ T cells.
(A) The frequency (%) of CD25, a NFAT-dependent molecule, on TCR-stimulated memory CD4+ T cells in the presence of Foxp3 393–403 peptide (100 μM), NFAT/FOXP3 interaction inhibition peptide, or control Foxp3 399 peptide (100 μM) at day 2 post-stimulation (n = 4). (B and C) Representative flow cytometric plot (B) and the frequencies (C) of IL-1RI and IL-1RII expression on TCR-stimulated memory CD4+ T cells in the presence of Foxp3 393–403 peptide or control Foxp3 399 peptide at day 2 post-stimulation (n = 4). (D) Foxp3-expressing Jurkat cells were transfected with the indicated pGL4.10 luciferase vectors (pGL4/IL-1RII-a1414, pGL4/IL-1RII-a814, and pGL4/IL-1RII-a215) and pGL4.74 control renilla vector as internal control, followed by stimulation with PMA and ionomycin for 48 hr (upper panel: n = 4). Comparison of luciferase activity between pGL4/IL-1RII-a1414 and pGL4/IL-1RII-a1414mu (−1209 to −1188 NFAT-binding motif mutant) transfected Foxp3-expressing Jurkat cells (lower panel: n = 4). Luciferase activity was determinded using dual lucifease assay system. (E and F) Purified memory CD4+ T cells were stimulated with plate-bound anti-CD3/28 mAbs for 24 hr. ChIP qPCR was perfomed using anti-NFATc2 or anti-Foxp3 Ab at the IL-1RII promoter region. Enrichment of NFAT within four putative NFAT binding motifs (E) and enrichment of Foxp3 within three putative Foxp3-binding motifs (F) was analyzed by qPCR (n = 5 or 6). There are two (−1209 ~ −1188 of IL-1RII promoter), one (−649 ~ −639), and one (+10 ~ +18) binding sites for NFAT B1, B2 and B3, respectively. NFAT BC (binding control) has a binding site in human IL-2 promoter region. There are three (−4652 ~ −4611), one (−1236 ~ −1230), and one (−425 ~ −418) binding sites for Foxp3 B1, B2 and B3, respectively. Foxp3 BC (binding control) has a binding site in the human IL-2Rα promoter region (Zhang et al., 2013). Bar graphs show the mean ± SEM. * = p<0.05, ** = p<0.01, and *** = p<0.001 by two-tailed paired t-test.
-
Figure 5—source data 1
Figure 5A The inhibitory effect of FOXP3 393–403, a specific inhibitor of the NFAT/FOXP3 interaction.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Figure 5C NFAT/Foxp3 interaction inhibitor significantly repress the IL-1RII expression.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig5-data2-v2.xlsx
-
Figure 5—source data 3
Figure 5D IL-1RII promoer activity measured via luciferase assay.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig5-data3-v2.xlsx
-
Figure 5—source data 4
Figure 5E and F Result of NFAT & Foxp3 ChIP-qPCR via IL-1RII promter.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig5-data4-v2.xlsx
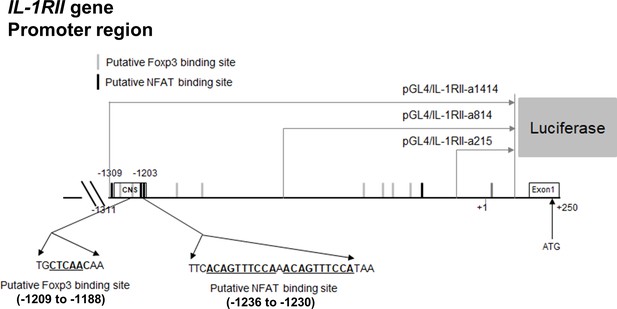
Analysis of potential binding sites of NFAT and Foxp3 in the minimal promoter of the human IL-1RII gene and schematic diagram of three luciferase reporter constructs.
CNS (conserved noncoding sequences).
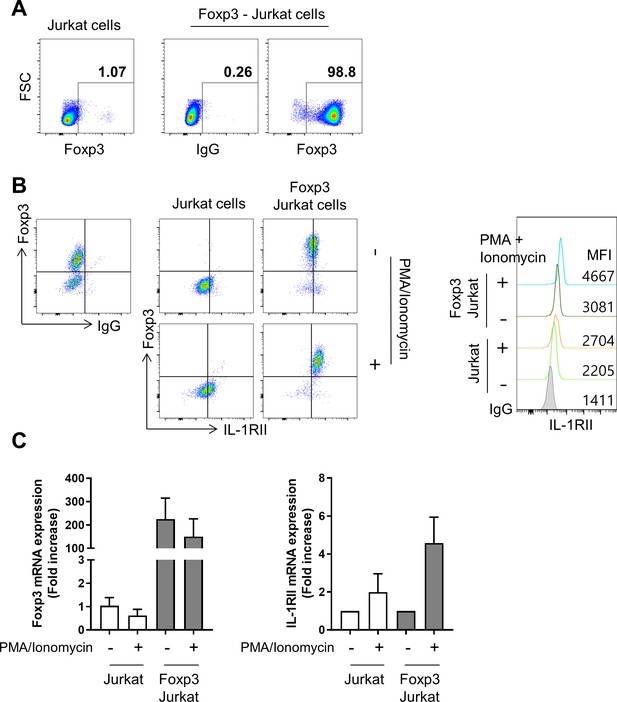
Overexpression of Foxp3 in Jurkat cells leads to an increase in the expression of IL-1RII.
(A) Flow cytometric analysis of Foxp3 expression in Puro-control Jurkat (mock transfected: hereafter Jurkat) or Foxp3-expressing Jurkat cells. The numbers indicate the frequecy of Foxp3+ cells (B) Representative flow cytometric plot of IL-1RII and Foxp3 expression in Jurkat or Foxp3-expressing Jurkat cells upon stimulation with PMA and ionomycin. Histogram plot shows MFIs of IL-1RII expression on Jurkat cells under the indicated condtions. (C) mRNA expression of Foxp3 and IL-1RII in PMA/Ionomycin-stimulated Jurkat cells or Foxp3-expressing Jurkat cells (n = 3).
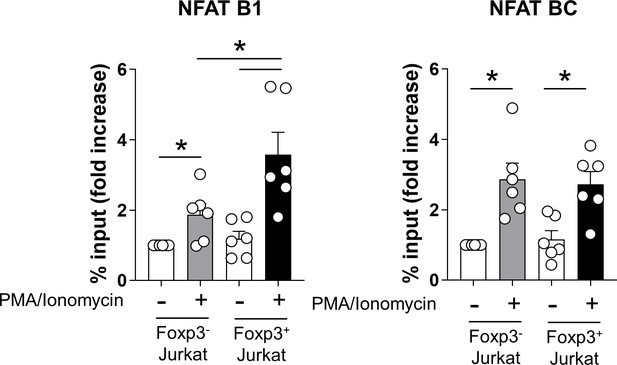
Cooperation of NFAT/Foxp3 via binding of NFAT on NFAT B1 site.
Control Jurkat or Foxp3-expressing Jurkat cells were stimulated with or without PMA (50 ng/ml)/ionomycin (1 μg/ml) for 2 hr. ChIP qPCR was performed using anti-NFATc2 at the IL-1RII promoter region. NFAT enrichment at NFAT B1 (−1209 ~ −1188 of IL-1RII promoter) in IL-1RII gene or NFAT BC in IL-2 gene was analyzed by qPCR. Bar graphs show the mean ± SEM. * = p<0.05 by two-tailed paired t-test.
Differential expression of IL-1RI and IL-1RII on activated CD4+ T cells defines unique immunological features of cells.
Treg-depleted memory CD4+ T cells were stimulated for 48 hr with anti-CD3/28-coated microbeads to induce IL-1RI and IL-1RII expression. Cells were sorted into IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- cells and cultured for another 5 days with rhIL-1β (5 ng/ml) and rhIL-2 (50 IU/ml). (A) Representative flow cytometric plot and the frequencies of IL-17- and IFN-γ-producing cells in sorted IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- memory CD4+ T cells at day 7 post-stimulation (n = 6). (B) The amount of IL-17 and IFN-γ in the culture supernatant (A) by ELISA (n = 6). (C) Representative flow cytometric plot and the frequency of IFN-γ+ cells and Foxp3+ cells in the IL-17 producing IL-1RI+IL-1RII- or IL-1RI+IL-1RII+ cells (n = 5). (D) The frequencies of Treg-related marker- or Th17-related marker-expressing cells on sorted IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- memory CD4+ T cells at day 7 post-stimulation (n = 4). Bar graphs show the mean ± SEM. * = p<0.05, ** = p<0.01, and *** = p<0.001 by two-tailed paired t-test.
-
Figure 6—source data 1
Figure 6A Frequency of IL-17 & IFN-γ producing sorted IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Figure 6B Concentraion of IL-17 & IFN-γ in the culture supernatant of sorted IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Figure 6C Frequency of Foxp3 & IFN-γ producing cells of IL-17 producing IL-1RI+IL-1RII- and IL-1RI+IL-1RII+ cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig6-data3-v2.xlsx
-
Figure 6—source data 4
Figure 6D Expression of Treg related markers on sorted L-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- cells.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig6-data4-v2.xlsx
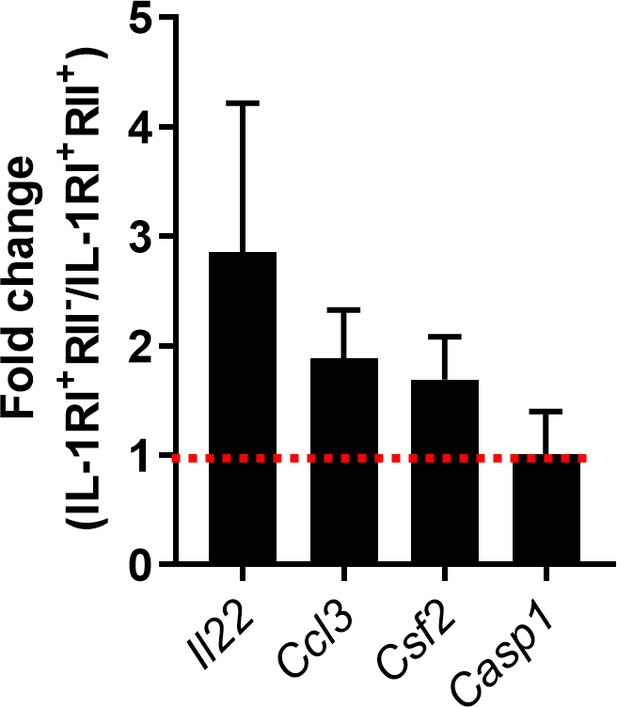
IL-1β-mediated changes in pathogenic Th17-cell-associated gene signature of TCR-activated memory CD4+ T cells in humans.
Treg-depleted memory CD4+ T cells were stimulated for 48 hr to induce the expression of IL-1RI and IL-1RII. IL-1RI+IL-1RII- and IL-1RI+IL-1RII+ cells were purified by cell sorting and stimulated through their TCR for 5 days in the presence of IL-1β. Expression of several pathogenic Th17 cell-associated genes in IL-1RI+IL-1RII- cells was presented relative to their expression in IL-1RI+IL-1RII+ cells. Bar graphs show the mean ± SEM (n = 3).
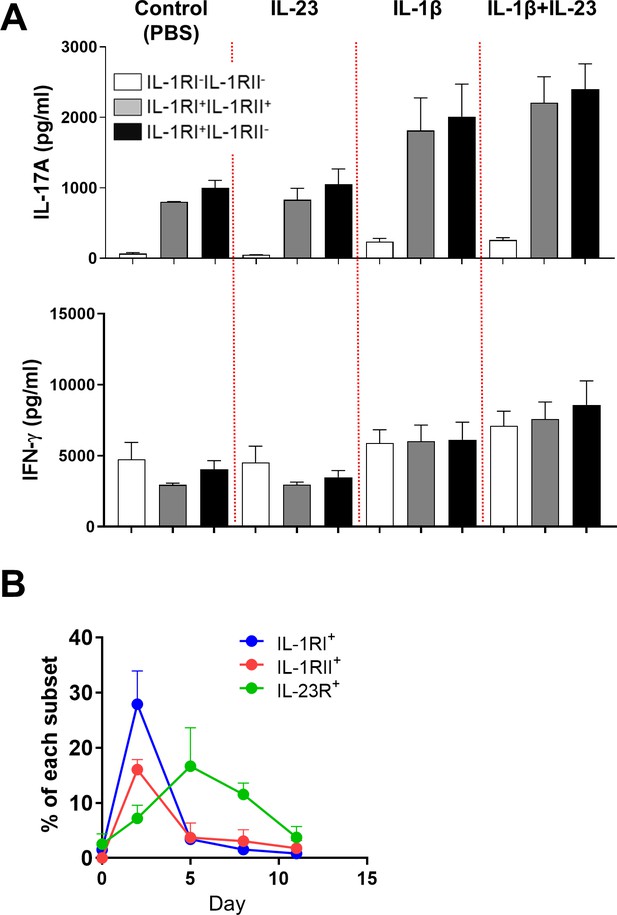
Effect of IL-23 on Th17 production in IL-1RI+IL-1RII- cells and IL-1RI+IL-1RII+ memory CD4 T cells.
(A) Sorted IL-1RI+IL-1RII-, IL-1RI+IL-1RII+, and IL-1RI-IL-1RII- cells were stimulated with anti-CD3/28-coated microbeads and IL-2 (50 IU/ml) for 5 days under the indicated cytokine conditions (5 ng/ml of IL-1β and 25 ng/ml of IL-23). The amounts of IL-17 and IFN-γ in the culture supernatant were quantified by ELSIA (n = 3). (B) Time kinetics of IL-1RI, IL-1RII, and IL-23R expression on TCR-stimulated memory CD4+ T cells (n = 3). Bar graphs and line graphs show the mean ± SEM.
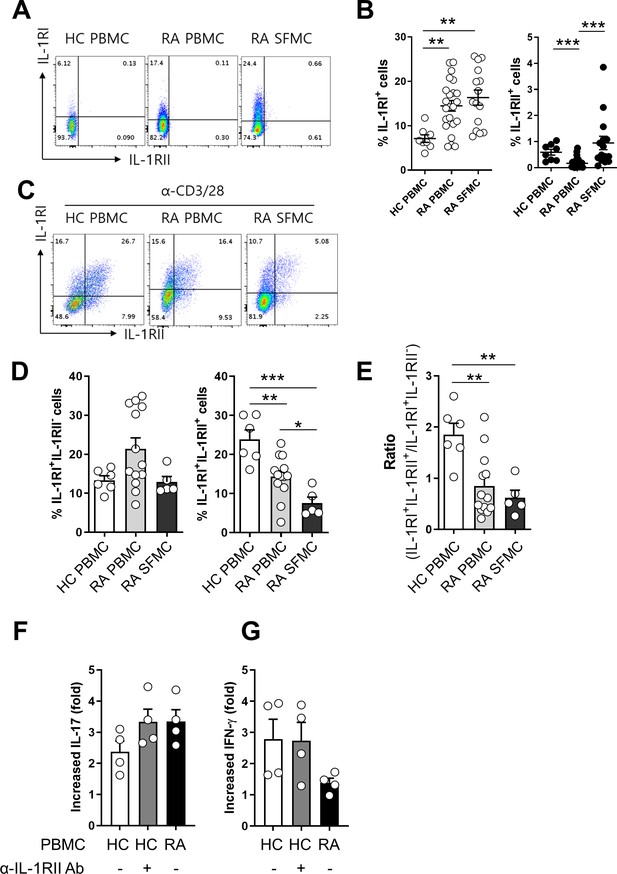
Aberrant expression of IL-1RI and IL-RII in synovial CD4 T cells in patients with rheumatoid arthritis (RA).
(A and B) Representative flow cytometric plot (A) and the frequencies (B) of IL-1RI and IL-1RII expression on ex vivo memory CD4+ T cells from peripheral blood (n = 23) and synovial fluid (n = 15) of RA patients and peripheral blood of HCs (n = 8). SFMC: synovial fluid mononuclear cells. (C and D) Representative flow cytometric plot (C) and the frequencies (D) of IL-1RI and IL-1RII expression on TCR-stimulated memory CD4+ T cells from peripheral blood (n = 13) and synovial fluid (n = 5) of RA patients and peripheral blood of HCs (n = 6) at day 2 post-stimulation. (E) The ratio of IL-1RI+IL-1RII+ to IL-1RI+IL-1RII- cells in (D). (F and G) The effect of anti-IL-1RII neutralizing Ab treatment on IL-1β-mediated IL-17 production by TCR-stimulated memory CD4+ T cells of HCs (n = 4). Fold change indicates the ratio of IL-1β-mediated IL-17 production between the anti-IL-1RI Ab-treated group and control isotype-treated group. Scatter plot and bar graphs show the mean ± SEM. * = p<0.05, ** = p<0.01, and *** = p<0.001 by two-tailed uppaired or paired t-test.
-
Figure 7—source data 1
Figure 7B Ex vivo expression of IL-1RI and IL-1RII on CD4+ T cells between HC PBMC, RA PBMC, and RA SFMC.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig7-data1-v2.xlsx
-
Figure 7—source data 2
Figure 7D Expression of IL-1RI and IL-1RII on stimulated memory CD4+ T cells between HC PBMC, RA PBMC, and RA SFMC.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig7-data2-v2.xlsx
-
Figure 7—source data 3
Figure 7E Expression of IL-1RI and IL-1RII on stimulated memory CD4+ T cells between HC PBMC, RA PBMC, and RA SFMC (ratio).
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig7-data3-v2.xlsx
-
Figure 7—source data 4
Figure 7F and G IL-1β-mediated IL-17 & IFN-γ production in response to TCR stimulation compared with HC and RA.
- https://cdn.elifesciences.org/articles/61841/elife-61841-fig7-data4-v2.xlsx
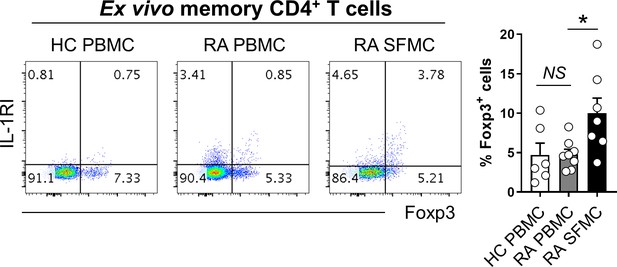
Foxp3 Tregs cells in peripheral blood and synovial fluid of patients with rheumatoid arthritis (RA) and peripheral blood of HCs.
Representative flow cytometric plot and the frequencies of IL-1RI and Foxp3 expression on ex vivo memory CD4+ T cells from peripheral blood and synovial fluid of RA patients and peripheral blood of HCs. SFMC: synovial fluid mononuclear cells. Bar graphs show the mean ± SEM. *** = p<0.005 by two-tailed paired t-test. NS indicates not significant.
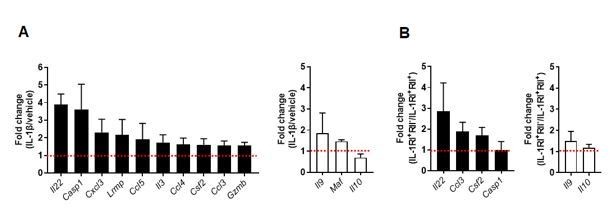
IL-1β-mediated changes in pathogenic and nonpathogenic gene signature of TCR-activated memory CD4+ T cells in humans.
(A) Purified memory CD4+ T cells were stimulated with anti-CD3/28 coated microbeads in the presence of PBS (as vehicle) or IL-1β (5 ng/ml) for 7 days. Expression of pathogenic or nonpathogenic genes in IL-1β-treated CD4 T cells was presented relative to their expression in PBS-treated CD4 T cells. (B) Treg-depleted memory CD4+ T cells were stimulated for 48 h to induce the expression of IL-1RI and IL-1RII. IL-1RI+IL-1RII- and IL-1RI+IL-1RII+ cells were purified by cell sorting and stimulated through their TCR for 5 days in the presence of IL-1β. Expression of pathogenic or nonpathogenic genes in IL-1RI+IL-1RII- cells was presented relative to their expression in IL-1RI+IL-1RII+ cells. Bar graphs show the mean ± SEM.
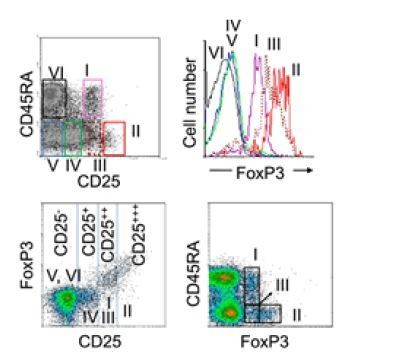
Efficiency of sorting-based Treg depletion from CD4+ T cells.
Six subsets of CD4+ T cells defined by the expression of CD45RA and CD25: (I), CD25++CD45RA+ cells → resting Treg cells; (II), CD25+++CD45RA− cells → activated Treg cells; (III), CD25++CD45RA− cells → non-suppressive cytokine-producing Foxp3low T cells; (IV), CD25+CD45RA− cells; (V), CD25−CD45RA− cells; (VI), CD25−CD45RA+ cells (Miyara et al., 2009).

Foxp3 Tregs cells in peripheral blood and synovial fluid of patients with rheumatoid arthritis (RA) and peripheral blood of HCs.
(A) Representative flow cytometric plot of IL-1RI, IL-1RII, and Foxp3 expression on TCR-stimulated memory CD4+ T cells from peripheral blood of RA patients and peripheral blood of HCs at day 2 post-stimulation. The frequency of Foxp3+, IL-1RI+IL-1RII+, IL-1RI+IL-1RII- cells and ratio of IL-1RI+IL-1RII+ to IL-1RI+IL-1RII- cells. (B) Representative flow cytometric plot and frequency of IL-1RI and IL-1RII expression on TCR-stimulated memory CD4+ T cells from HCs with plasma and synovial fluid of RA patients and plasma of HCs at day 2 post-stimulation. Bar graphs show the mean ± SEM. * = p < 0.05 by two-tailed paired or unpaired t-test. NS indicates not significant.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo-sapiens) | Jurkat (clone E6-1) | ATCC | Cat# TIB-152, RRID:CVCL_0367 | |
| Cell line (Homo-sapiens) | Foxp3+ Jurkat | This paper | Foxp3-expressing cell line | |
| Transfected construct (Homo-sapiens) | pLECE3 | Lee et al., 2020 | Lentiviral expression vector | |
| Biological sample (Homo-sapiens) | Rheumatoid Arthritis patient (Blood and Synovial fluid sample) | Chungnam National University Hospital | Department of Internal Medicine | |
| Antibody | Anti-human-NFATc2 (mouse monoclonal) | Santacruz biotech | Cat# Sc-7296, RRID:AB_628012 | ChIP (2 μg/test) |
| Antibody | Anti-human-GITR PE-Cy7 (mouse monoclonal) | BioLegend | Cat# 371223, RRID:AB_2687170 | FACS (1:50) |
| Antibody | Anti-human-CD73 APC-Cy7 (mouse monoclonal) | BioLegend | Cat# 344021, RRID:AB_2566755 | FACS (1:25) |
| Antibody | Anti-human-CD39 BV421 (mouse monoclonal) | BioLegend | Cat# 328213, RRID:AB_10933084 | FACS (1:50) |
| Antibody | Anti-human-CTLA-4 PerCP-Cy5.5 (mouse monoclonal) | BioLegend | Cat# 369607, RRID:AB_2629673 | FACS (1:25) |
| Antibody | Anti-human-CXCR3 PE-Cy7 (mouse monoclonal) | BioLegend | Cat# 353719, RRID:AB_11218804 | FACS (1:50) |
| Antibody | Anti-human-CD127 BV421 (mouse monoclonal) | BioLegend | Cat# 351309, RRID:AB_10898326 | FACS (1:50) |
| Antibody | Anti-human-Foxp3 Alexa Fluor 647 (mouse monoclonal) | BioLegend | Cat# 320014, RRID:AB_439750 | FACS (1:25) |
| Antibody | Anti-human-IL-10 PE (mouse monoclonal) | eBioscience | Cat# 12-7108-81, RRID:AB_466178 | FACS (1:25) |
| Antibody | Anti-human-IL-17 PE-Cy7 (mouse monoclonal) | eBioscience | Cat# 25-7179-41, RRID:AB_11042972 | FACS (1:25) |
| Antibody | Anti-human-IFN-γ Alexa Fluor 700 (mouse monoclonal) | eBioscience | Cat# 56-7319-42, RRID:AB_2574509 | FACS (1:200) |
| Antibody | Anti- Human IL-23R Biotinylated (Goat polyclonal) | R and D systems | Cat# BAF1400, RRID:AB_355982 | FACS (1:50) |
| Antibody | Anti-human IL-1 RI PE (Goat polyclonal) | R and D systems | Cat# FAB269P, RRID:AB_2124912 | FACS (1:10) |
| Antibody | Anti-Human IL-1 RII Fluorescein-conjugated (mouse monoclonal) | R and D systems | Cat# FAB663F, RRID:AB_1964612 | FACS (1:10) |
| Antibody | Anti- Human IL-1RII (mouse monoclonal) | R and D systems | Cat# MAB263, RRID:AB_2125174 | Neutralization (20 μg/ml) |
| Antibody | Mouse IgG2A Isotype control (mouse monoclonal) | R and D systems | Cat# MAB003, RRID:AB_357345 | Neutralization control (20 μg/ml) |
| Antibody | Anti-human-CD4 v500 (mouse monoclonal) | BD Bioscience | Cat# 560768, RRID:AB_1937323 | FACS (1:50) |
| Antibody | Anti-human-CD4 APC (mouse monoclonal) | BD Bioscience | Cat# 555349, RRID:AB_398593 | FACS (1:25) |
| Antibody | Anti-human-CD3 APC-Cy7 (mouse monoclonal) | BD Bioscience | Cat# 557832, RRID:AB_396890 | FACS (1:50) |
| Antibody | Anti-human-CD25 PE-Cy5 (mouse monoclonal) | BD Bioscience | Cat# 555433, RRID:AB_395827 | FACS (1:25) |
| Antibody | Anti-human-CD25 APC (mouse monoclonal) | BD Bioscience | Cat# 561399, RRID:AB_10643029 | FACS (1:25) |
| Antibody | Anti-human-CD45RA PE-Cy7 (mouse monoclonal) | BD Bioscience | Cat# 560675, RRID:AB_1727498 | FACS (1:50) |
| Antibody | Anti-human-CD161 BV421 (mouse monoclonal) | BD Bioscience | Cat# 562615, RRID:AB_2737678 | FACS (1:25) |
| Antibody | Anti-human-CCR6 APC (mouse monoclonal) | BD Bioscience | Cat# 560619, RRID:AB_1727439 | FACS (1:25) |
| Antibody | Streptavidin conjugated Pacific Blue | BD Bioscience | Cat# 560797, RRID:AB_2033992 | FACS (1:25) |
| Antibody | Streptavidin conjugated Alexa Fluor 488 | Life technologies | Cat# S11223 | FACS (1:500) |
| Antibody | anti-human CD3 functional grade purified(mouse monoclonal) | eBioscience | Cat# 16-0037-81, RRID:AB_468854 | Functional T cell assay (0.1 ~ 10 µg/ml) |
| Antibody | Anti-human CD28 Functional Grade (mouse monoclonal) | eBioscience | Cat# 16-0289-85, RRID:AB_468927 | Functional T cell assay (1.5 µg/ml) |
| Recombinant DNA reagent | pGL4/IL-1RII-a1414 (luciferase plasmid) | This paper | Containing the IL-1RII promoter region (−1314 to +100) | |
| Recombinant DNA reagent | pGL4/IL-1RII-a814 (luciferase plasmid) | This paper | Containing the IL-1RII promoter region (−714 to +100) | |
| Recombinant DNA reagent | pGL4/IL-1RII-a215 (luciferase plasmid) | This paper | Containing the IL-1RII promoter region (−115 to +100) | |
| Recombinant DNA reagent | pGL4.74 (Renilla control) | Promega | Cat# E6921 | |
| Recombinant DNA reagent | pGL4.10 (luciferase plasmid) | Promega | Cat# E6651 | |
| Recombinant DNA reagent | pLEF-puro-FoxP3 | This paper | Foxp3 overexpression plasmid | |
| Peptide, recombinant protein | Recombinant human IL-1β | R and D systems | Cat# 201-LB-005 | |
| Peptide, recombinant protein | Recombinant human IL-6 | R and D systems | Cat# 206-IL-010 | |
| Peptide, recombinant protein | Recombinant human IL-23 | R and D systems | Cat# 1290-IL | |
| Peptide, recombinant protein | Recombinant human TGF-β1 | R and D systems | Cat# 240-B | |
| Peptide, recombinant protein | Recombinant human IL-2 | PeproTech | Cat# AF-200–02 | |
| Peptide, recombinant protein | Foxp3 393–403 | Lozano et al., 2015 (PMID:26324768) | NFAT/Foxp3 interaction inhibition peptide | |
| Peptide, recombinant protein | Foxp3 399A | Lozano et al., 2015 (PMID:26324768) | Control peptide | |
| Peptide, recombinant protein | dNP2-VIVIT | Lee et al., 2019 (PMID:31737742) | NFAT inhibition peptide (1 μM) | |
| Peptide, recombinant protein | dNP2-VEET | Lee et al., 2019 (PMID:31737742) | Control peptide (1 μM) | |
| Commercial assay or kit | QuikChange II XL Site-Directed Mutagenesis Kit, 10 Rxn | genomics agilent | Cat# 200523 | |
| Commercial assay or kit | EasySep Human memory CD4+ T cell enrichment kit | STEMCELL | Cat# 19157 | |
| Commercial assay or kit | EasySep Human Naïve CD4+ T Cell Isolation Kit | STEMCELL | Cat# 19555 | |
| Commercial assay or kit | CD4+CD25+CD127dim/- Regulatory T Cell Isolation Kit II, human | Miltenyi Biotec | Cat# 130-094-775 | |
| Commercial assay or kit | CD3/CD28 activation Dynabeads | Thermo Fisher | Cat# 11161D | |
| Commercial assay or kit | EZ-ChIP | Merck Millipore | Cat# 17–371 | |
| Commercial assay or kit | NucleoSpin gDNA clean-up kit | MACHEREY-NAGEL | Cat# 740230.50 | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | Cat# E1910 | |
| Commercial assay or kit | GoScript Reverse Transcriptase | Promega | Cat# A5001 | |
| Commercial assay or kit | Power SYBR Green Master Mix | Bio-Rad | Cat# 4367659 | |
| Commercial assay or kit | Human IL-17A ELISA Ready-set-go | eBioscience | Cat# 88-7176-88, RRID:AB_2575036 | |
| Commercial assay or kit | Human IL-10 ELISA Ready-SET-Go! | eBioscience | Cat# 88-7106-86, RRID:AB_2575004 | |
| Commercial assay or kit | Human IFN-γ ELISA MAX Deluxe | Biolegend | Cat# 430104 | |
| Commercial assay or kit | Foxp3 Fix/Perm buffer set | Biolegend | Cat# 421403 | |
| Chemical compound, drug | Cyclosprorin A | Sigma | Cat# 30024 | 1 μM |
| Chemical compound, drug | 1a,25-dihydroxyvitamin D3 | Sigma | Cat# D1530 | 10 μM |
| Chemical compound, drug | Phobol 12-myristate 13-acetate (PMA) | Sigma | Cat# p8139 | 50 ng/ml |
| Chemical compound, drug | Ionomycin calcium salt | Sigma | Cat# I0634 | 1 μg/ml |
| Sequence-based reagent | All sequences are listed in Materials and methods. | |||
| Software, algorithm | Prism | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | Promo 3.0 | ALGGEN | RRID:SCR_016926 | |
| Software, algorithm | VISTA | Joint genome institute | RRID:SCR_011808 | |
| Software, algorithm | Primer designing tool | NCBI | ||
| Other | Neon Transfection System | Thermo Fisher |





