Alkylative damage of mRNA leads to ribosome stalling and rescue by trans translation in bacteria
Figures
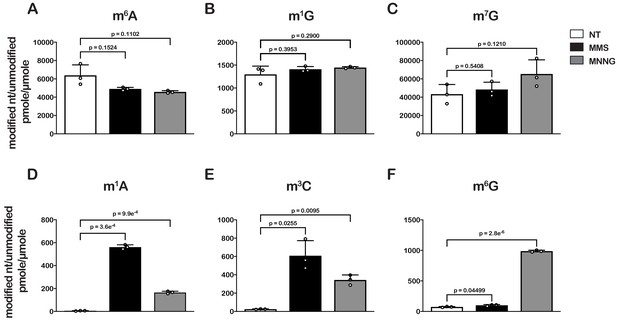
Treatment of E. coli with MMS and MNNG results in significant accumulation of alkylative-damage adducts in RNA.
(A–E) Bar graphs showing the amount of the indicated modified nucleotides relative to their unmodified parent in untreated (white bars), MMS-treated (black bars), and MNNG-treated (gray bars) cells. The values plotted are the averages of three biological repeats and the error bars represent standard deviations around the mean. Significant differences in mean, as denoted by p<0.05, were determined by Welch’s t-test.
-
Figure 1—source data 1
Quantification of alkylative-damage adducts.
- https://cdn.elifesciences.org/articles/61984/elife-61984-fig1-data1-v2.xlsx
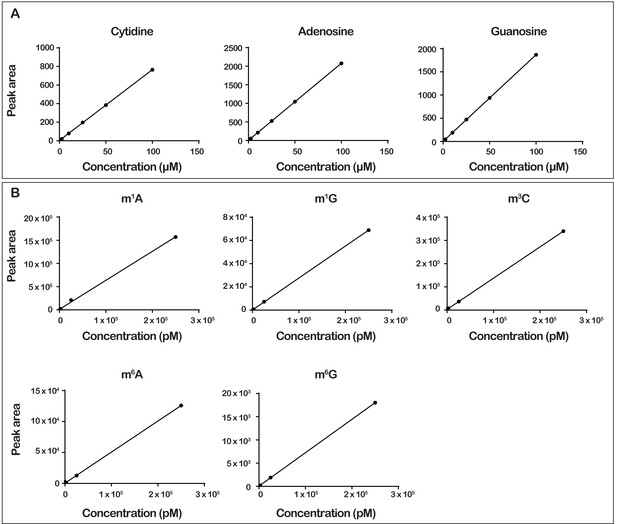
LC-MS calibration curves for modified and unmodified nucleosides.
(A) The integrated peak area for absorbance at 260 nm for each indicated unmodified nucleotide plotted against its concentration. (B) The integrated peak area for cps intensity plotted against concentrations of modified nucleoside standards. For all plots in A and B, the data were fit to linear-regression lines forced to have zero x- and y-intercepts. Retention times and mass transitions shown in Supplementary file 1, which were determined empirically, were used to identify the peak for each nucleotide.
-
Figure 1—figure supplement 1—source data 1
LC-MS calibration curve source data.
- https://cdn.elifesciences.org/articles/61984/elife-61984-fig1-figsupp1-data1-v2.xlsx
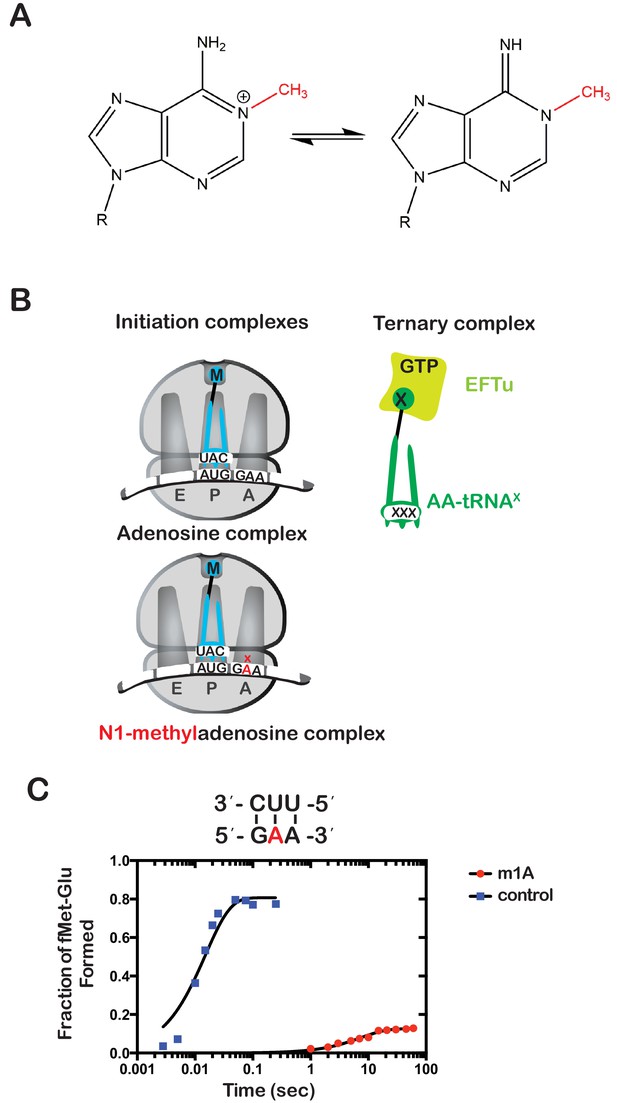
N(1)-methyladenosine (m1A) in mRNA significantly decreases the rate and endpoint of peptide bond formation in vitro.
(A) Chemical structure of m1A. The N1-methyl group is highlighted in red, and the resonance structure of the molecule is represented. (B) Schematic representation of adenosine and m1A initiation complexes encoding for the dipeptide Met-Glu. Both complexes contain the initiator fMet-tRNAfMet in the P site; the A complex displays a GAA codon, while the m1A complex displays a Gm1AA codon in the A site. (C) Representative time-courses of peptide bond-formation reactions between initiation complexes programmed with unmodified mRNA (blue) or an m1A-modified one (red) m1A, and Glu-tRNAGlu ternary complex.
-
Figure 2—source data 1
m1A kinetics source data.
- https://cdn.elifesciences.org/articles/61984/elife-61984-fig2-data1-v2.xlsx
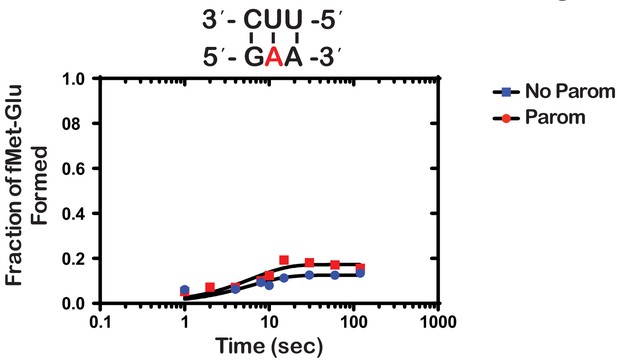
Paromomycin does not rescue the effect of m1A on peptide bond formation.
Representative time-courses of peptide bond-formation reactions between initiation complexes programmed with m1A mRNA and Glu-tRNAGlu ternary complexes in the absence (blue) and presence (red) of paromomycin.
-
Figure 3—source data 1
Paromomycin kinetics source data.
- https://cdn.elifesciences.org/articles/61984/elife-61984-fig3-data1-v2.xlsx
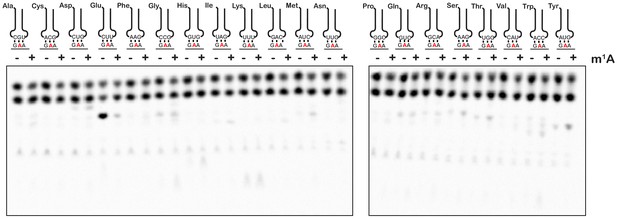
m1A does not alter the reactivity of ribosomes with near-cognate and non-cognate aa-tRNA.
Phosphorimager scan of electrophoretic TLCs used to follow dipeptide-formation reactions between unmodified and m1A-modified complexes with all canonical aa-tRNA ternary complexes.
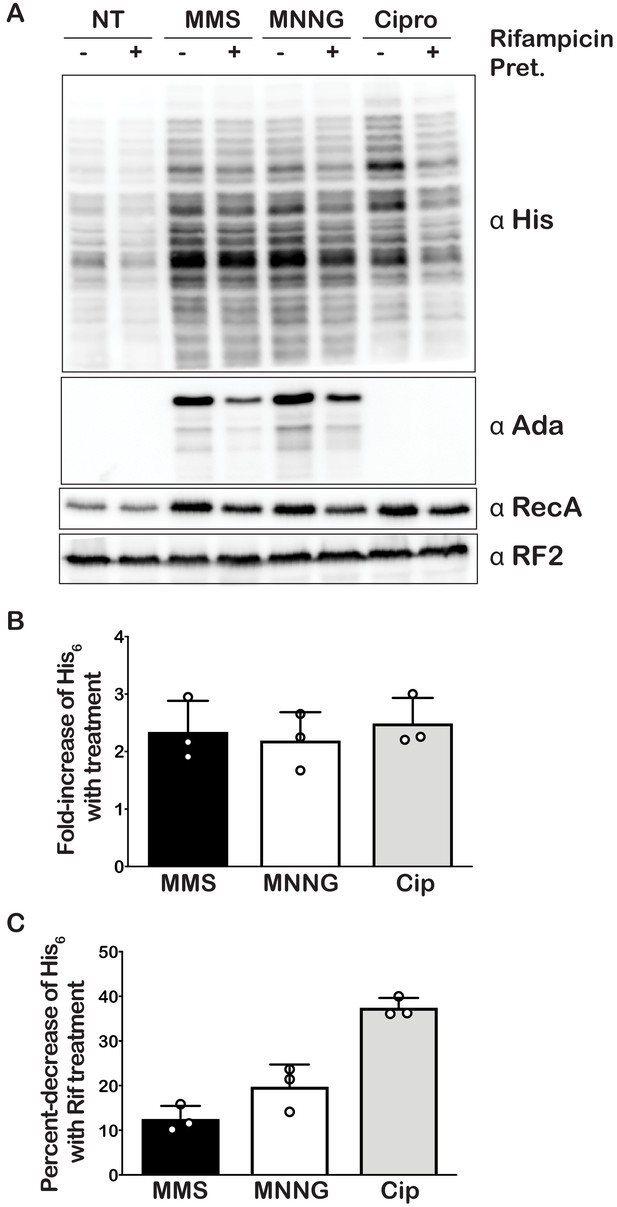
Alkylative stress activates trans translation in E. coli in a transcription-independent manner.
(A) Western blot analyses of total protein isolated from an E. coli strain expressing tmRNA-His6. Cells were either untreated or treated with MMS, MNNG, or ciprofloxacin. Additionally, for each condition, cells were either mock pretreated or received a rifampicin pretreatment. Blots were probed with the indicated antibodies. (B) Bar graph showing the relative change in His signal (tmRNA activity) as a result of the addition of each of the indicated compounds. (C) Bar graph used to depict the fold-decrease of His6 levels upon pre-treatment with rifampicin for each of the indicated treatments. In all cases, the initial His signal was normalized to that corresponding to RF2 levels before it was used to calculate the relative change. Three independent experiments were used to obtain the bar graphs, with the mean values plotted and the error bars representing the standard deviation around the mean.
-
Figure 5—source data 1
Quantification of tmRNA tagging.
- https://cdn.elifesciences.org/articles/61984/elife-61984-fig5-data1-v2.xlsx
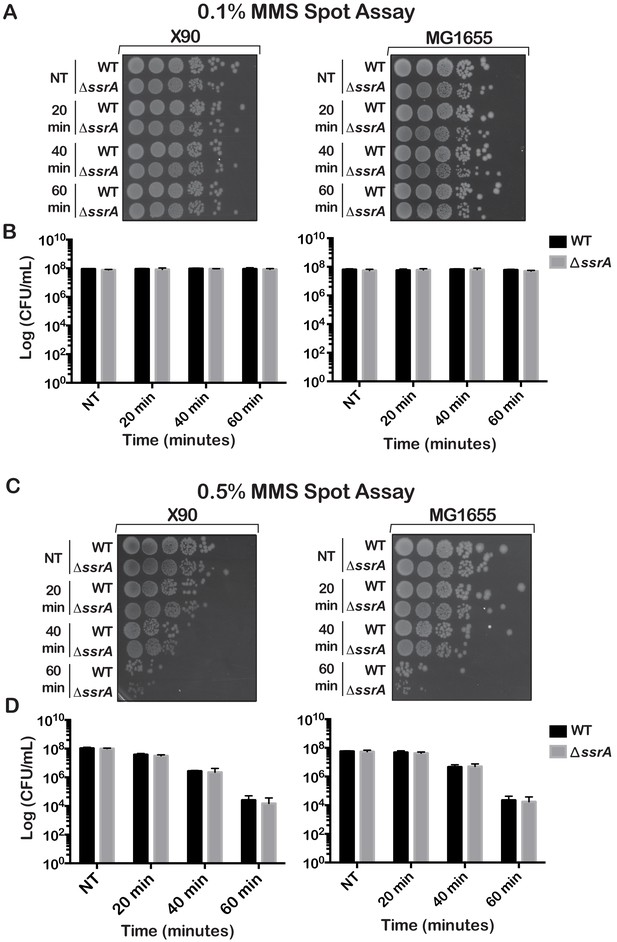
WT and ∆ssrA E. coli exhibit similar survival phenotypes after treatment with MMS.
(A) Spot assay of WT and ∆ssrA cells either after a mock treatment or treatment with 0.1% MMS for the indicated amount of time. (B) Quantification of colony forming units in (A) performed in triplicates. (C) Spot assay of WT and ∆ssrA cells either after no treatment or treatment with 0.5% MMS for the indicated amount of time. (D) Quantification of colony forming units in (C) performed in at least duplicates. In all cases, the data plotted is the average of three experiments and the error bars represent standard deviations from the mean.
-
Figure 5—figure supplement 1—source data 1
Spot assay quantification.
- https://cdn.elifesciences.org/articles/61984/elife-61984-fig5-figsupp1-data1-v2.xlsx
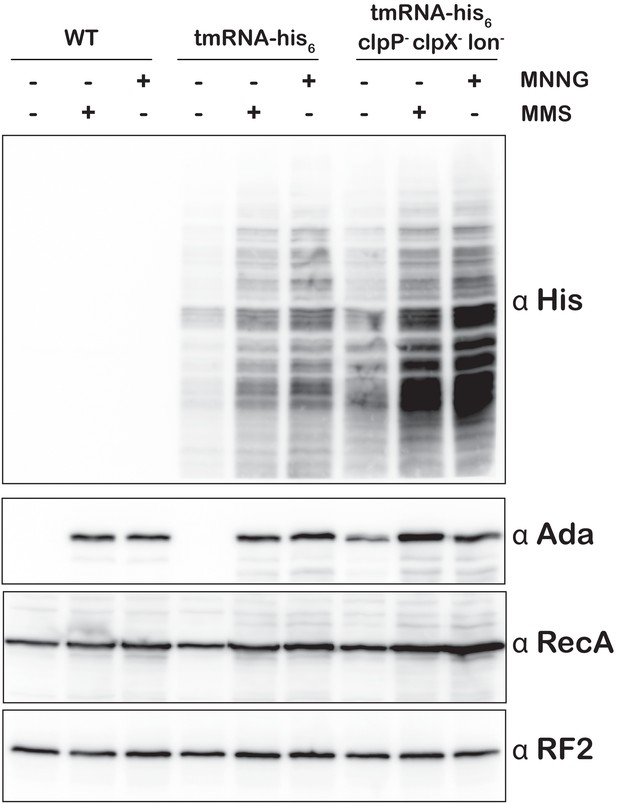
Deletion of ClpXP and Lon proteases results in further accumulation of tmRNA-induced His6 tagging of peptides upon alkylative stress.
Western blot analysis collected from the indicated E. coli strains grown in the absence or presence of the denoted compounds. Blots were probed with the depicted antibodies.
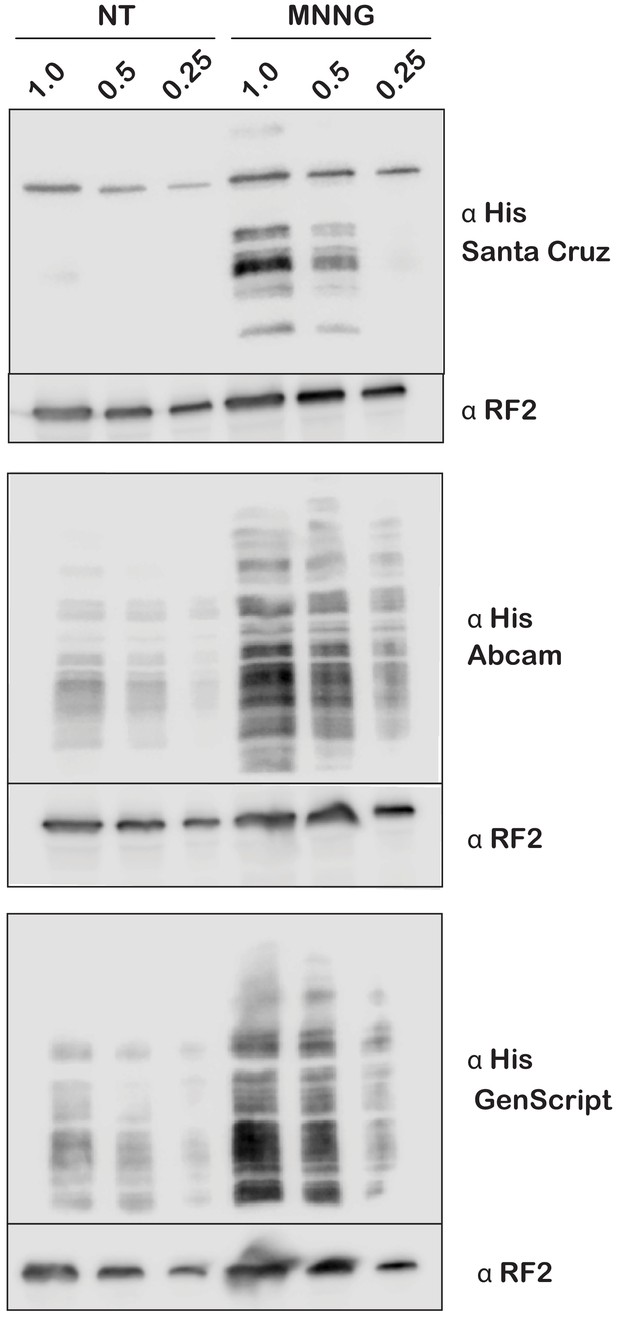
Different His antibodies display unique banding patterns on western blots.
Western blots of total protein isolated from E. coli expressing tmRNA-His6 in the absence or presence of MNNG. For each of the two treatments, lysates were serially diluted by two- and fourfold before being resolved by SDS-PAGE next to their undiluted sample. Following the transfer, the blots were probed with one of three different anti-His antibodies (Santa Cruz Biotechnology, Abcam, or GenScript). α-RF2 was used as a loading control.
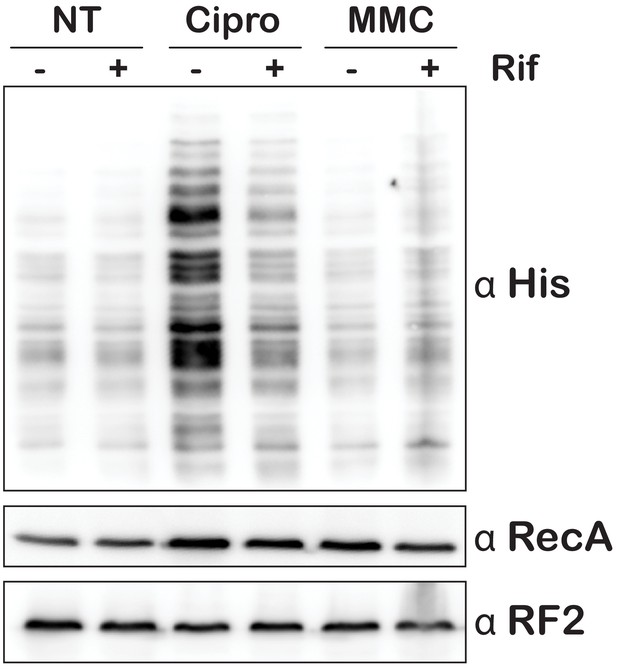
Ciprofloxacin, but not mitomycin C, increases His6 tagging by tmRNA.
Western blot analysis of total protein collected from E. coli expressing tmRNA-His6. Cells were either untreated or treated with ciprofloxacin (Cipro) or mitomycin C (MMC). Additionally, cells either received (+) or did not receive (-) a pre-treatment with rifampicin before treatment with the indicated damaging agent. The blot was probed with the indicated antibodies.
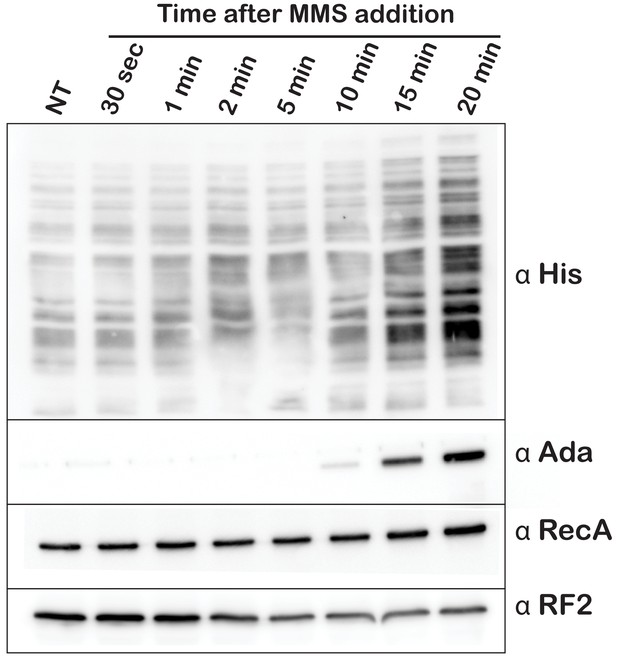
Optimal His6 tagging and activation of Ada and RecA levels are achieved after 20 min of MMS treatment.
Western blot analysis of total protein collected from E. coli expressing tmRNA-His6. Cells were either untreated or treated with MMS for the indicated lengths of time. The blot was probed with anti His, Ada, RecA, and RF2 antibodies.
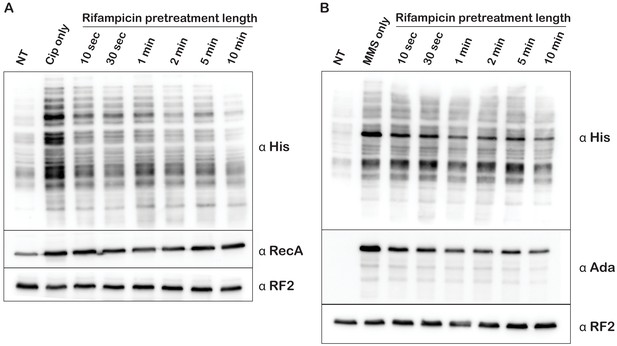
Significant transcriptional runoff is achieved after 10 s of rifampicin treatment.
Western blot analysis used to follow the effect of rifampicin pretreatment on ciprofloxacin (Cipro)- (A) and MMS-induced (B) activation of His tagging by tmRNA. The time on top of the blots indicate the amount of time cells were incubated in the presence of rifampicin before ciprofloxacin and MMS were added. The blots were probed with the depicted antibodies.
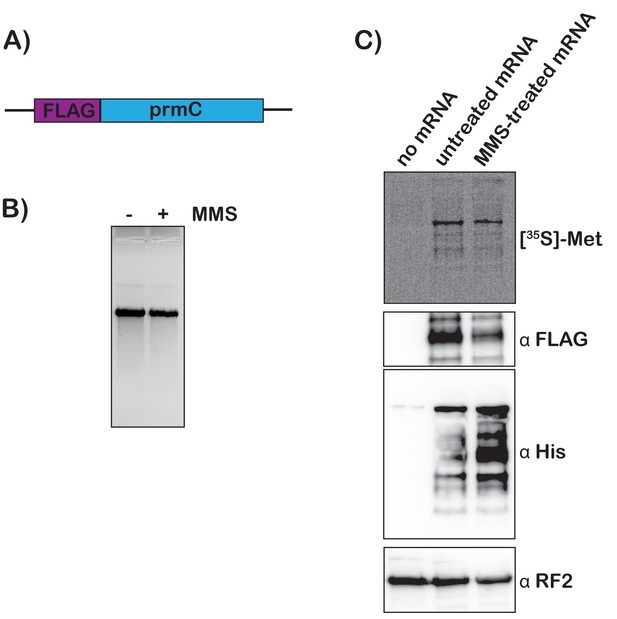
Translation of MMS-treated mRNA results in tmRNA tagging in an S30 extract.
(A) Schematic of the mRNA used in the in vitro translation assays. (B) Fluorescence image of EtBr-stained gel was used to visualize the mock-treated and MMS-treated RNAs. (C) Top shows a phosphor-imager scan of a PVDF-transfer membrane of a bis-tricine gel that was used to separate products from the in-vitro-translation reactions containing the indicated mRNAs. Bottom is western-blotting analyses of the same membranes with the indicated antibodies. Shown is a representative of two assays.
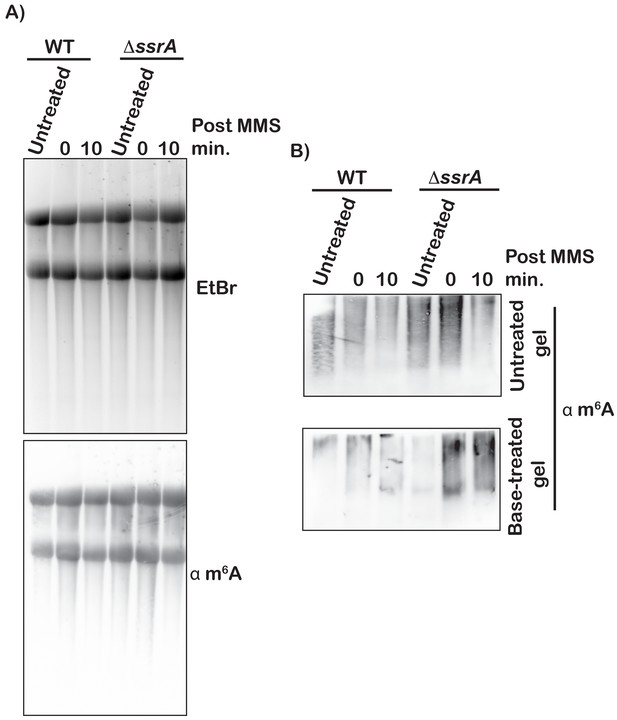
Deletion of ssrA gene results in stabilization of m1A-modified mRNA.
(A) Top: fluorescence image of a denaturing agarose gel used to separate total RNA isolated from the indicated cells under the denoted conditions. Bottom: immunoblot analysis of the same samples using m6A antibody. (B) m6A-immunoblot analysis of the indicated RNA samples. Top blot shows analysis of samples that were transferred directly following electrophoresis. Bottom blot shows analysis of the same samples except that the agarose gel was soaked in an alkaline buffer (50 mM NaOH, 1.5 M NaCl) before transfer.
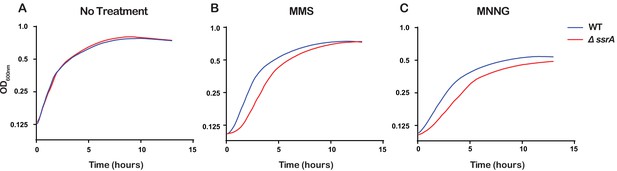
Ribosome rescue by tmRNA is important for cellular recovery after treatment with alkylating agents.
(A–C) Growth curves, as measured by change in OD600nm as a function of time, for the indicated cells following a treatment with the denoted compound. Average of three replicate growth assays is plotted.
-
Figure 7—source data 1
Growth curve source data.
- https://cdn.elifesciences.org/articles/61984/elife-61984-fig7-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Escherichia coli) | ssrA | UniProt | P0A832 | |
| Gene (Escherichia coli) | smpB | UniProt | P0A832 | |
| Gene (Escherichia coli) | ClpX | UniProt | P0A6H1 | |
| Gene (Escherichia coli) | ClpP | UniProt | P0A6G7 | |
| Gene (Escherichia coli) | Lon | UniProt | P0A9M0 | |
| Strain, strain background (Escherichia coli) | MG1655 | PMID:6271456 | Wild type | |
| Strain, strain background (Escherichia coli) | X90 | PMID:2651442 | ara∆(lac-pro) nalA argE(Am) rif thi-1/F’ lacIq lac+ pro+ | |
| Strain, strain background (Escherichia coli) | SM20 | PMID:16194232 | Gift from Sean Moore; X90, ∆ssrA, cam | |
| Strain, strain background (Escherichia coli) | SM694 | PMID:16194232 | Gift from Sean Moore; X90, ssrA::his6 - kan | |
| Strain, strain background (Escherichia coli) | SM876 | PMID:16194232 | Gift from Sean Moore; X90, ssrA::his6 - kan, clpPX-lon::cam | |
| Strain, strain background (Escherichia coli) | SKEC4 | Other | Gift from Kazuki Saito and Allen Buskirk; MG1655, ∆ssrA, ∆smpB, kan | |
| Strain, strain background (Escherichia coli) | ET1 | This Paper | MG1655, ssRA::his6 - kan | |
| Antibody | Goat polyclonal anti-mouse IgG-HRP | Thermo Scientific | Cat#: 31430; RRID:AB_228307 | WB: (1:10000) |
| Antibody | Goat polyclonal anti-mouse IgG-HRP | Thermo Scientific | Cat#: 31460; RRID:AB_228341 | WB: (1:10000) |
| Antibody | Rabbit polyclonal anti-RF2-His | PMID:22000017 | WB: (1:1000) | |
| Antibody | Mouse monoclonal anti-6× His tag antibody [HIS.H8] | Abcam | Cat#: ab18184; RRID:AB_444306 | WB: (1:2500) |
| Antibody | Mouse monoclonal anti-His-tag antibody (H-3) | Santa Cruz Biotechnology | Cat#: sc-8036; RRID:AB_627727 | WB: (1:2500), HRP-conjugated |
| Antibody | Mouse monoclonal THE His Tag Antibody | Genscript | Cat#: A00186; RRID:AB_914704 | WB: (1:2500) |
| Antibody | Mouse monoclonal anti-Ad (ADA-1) | Santa Cruz Biotechnology | Cat#: sc-53152; RRID:AB_772487 | WB: (1:500) |
| Antibody | Rabbit polyclonal anti-RecA | Abcam | Cat#: ab63797; RRID:AB_1142554 | WB: (1:10000) |
| Antibody | Rabbit polyclonal anti-m6A | Synaptic Systems | Cat#: 202 003; RRID:AB_2279214 | IB: (1:5000) |
| Commercial assay or kit | SuperSignal West Pico chemiluminescent substrate | Thermo Scientific | Cat#: 30480 | |
| Chemical compound, drug | Methyl methanesulfonate | Millipore Sigma | Cat#: 129925 | |
| Chemical compound, drug | 1-methyl-3-nitro-1-nitrosoguanidine | Fisher | Cat#: M05275G | Mfr: TCI America |
| Chemical compound, drug | Ciprofloxacin | Millipore Sigma | Cat#: 17850 | |
| Chemical compound, drug | Mitomycin C | Millipore Sigma | Cat#: M4287 | |
| Chemical compound, drug | Rifampicin | Millipore Sigma | Cat#: R3501 | |
| Chemical compound, drug | Nuclease P1 fromPenicillium citrinum | Sigma | Cat#: N8630 | |
| Chemical compound, drug | Alkaline phosphatase, calf intestinal (CIP) | NEB | Cat#: M0290 | |
| Chemical compound, drug | Paromomycin sulfate | VWR | Cat#: AAJ61274-06 | |
| Chemical compound, drug | EasyTag L-[35S]-Methionine | PerkinElmer | Cat#: NEG709A005MC | |
| Chemical compound, drug | Transfer ribonucleic acid: glutamic acid specific | Chemical Block | ||
| Chemical compound, drug | Transfer ribonucleic acid: N-Formyl-methionine specific | Chemical Block | ||
| Chemical compound, drug | tRNA from E. coli MRE 600 | Millipore Sigma | Cat#: 10109541001 | Mfr: Roche |
| Chemical compound, drug | N6-methyladenosine | Berry and Associates | Cat#: PR3732 | |
| Chemical compound, drug | O6-methylguanosine | Berry and Associates | Cat#: PR3757 | |
| Chemical compound, drug | 1-methyladenosine | Cayman chemical | Cat#: 16937 | |
| Chemical compound, drug | N1-methylguanosine | Carbosynth | Cat#: NM08574 | |
| Chemical compound, drug | N3-methylcytidine | Carbosynth | Cat#: NM05757 | |
| Chemical compound, drug | N7-methylguanosine | Carbosynth | Cat#: NM08037 | |
| Chemical compound, drug | Adenosine | Fisher | Cat#: AC164040050 | Mfr: ACROS Organics |
| Chemical compound, drug | Guanosine | Fisher | Cat#: AC411130250 | Mfr: ACROS Organics |
| Chemical compound, drug | Cytidine | Fisher | Cat#: AC111810100 | Mfr: ACROS Organics |
| Chemical compound, drug | Uridine | Tokyo Chemical Industry | Cat#: U0020 | |
| Sequence-based reagent | m1A oligo | Midland Certified Reagent Company | C AGA GGA GGU AAA AAA AUG G(1-methyl-A)A UUG UAC AAA | |
| Sequence-based reagent | Control oligo | This Paper | C AGA GGA GGU AAA AAA AUG GAA UUG UAC AAA | |
| Software, algorithm | Graphpad Prism | GraphPad Prism (http://www.graphpad.com/) | RRID:SCR_002798 | |
| Software, algorithm | Agilent MassHunter WorkStation - Qualitative Analysis for GC/MS | Agilent (https://www.agilent.com/en/products/software-informatics/masshunter-suite/masshunter-qualitative-analysis-gcms) | RRID:SCR_016657 | Version B.08.00 |
| Software, algorithm | Image Quant TL | Cytivia (https://www.cytivalifesciences.com/en/is/shop/protein-analysis/molecular-imaging-for-proteins/imaging-software/imagequant-tl-8-2-image-analysis-software-p-09518) | RRID:SCR_018374 | Version 8.2 |
| Software, algorithm | Fiji | Fiji (http://fiji.sc) | RRID:SCR_002285 | |
| Software, algorithm | Quantity One 1-D Analysis Software | Bio-Rad (http://www.bio-rad.com/en-us/product/quantity-one-1-d-analysis-software) | RRID:SCR_014280 |
Additional files
-
Supplementary file 1
Mass transitions, retention times, and collision energies for nucleoside standards.
- https://cdn.elifesciences.org/articles/61984/elife-61984-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61984/elife-61984-transrepform-v2.pdf





