Scleraxis-lineage cell depletion improves tendon healing and disrupts adult tendon homeostasis
Figures
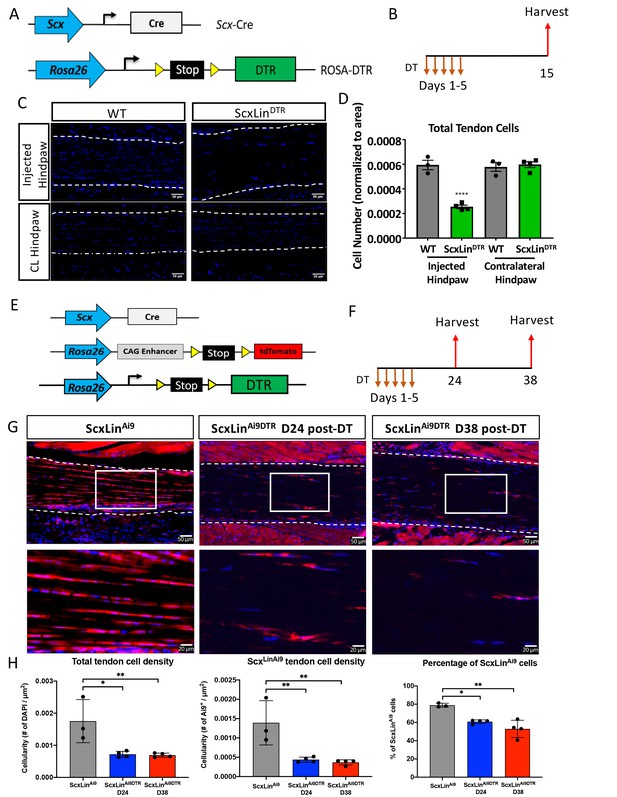
Efficiency of tendon cell and ScxLin cell depletion.
(A) To deplete ScxLin cells, Scx-Cre mice were crossed to the diphtheria toxin receptor mouse (ScxLinDTR). (B) Mice received five hind paw injections of DT and were harvested 10 days after the final injection. (C) Sections from injected and contralateral (CL) hind paws from WT and ScxLinDTR mice were stained with DAPI, and total DAPI+ cells within the tendon (white outline) were quantified (D). (E) To determine the depletion efficiency specifically of ScxLin cells, Scx-Cre; Rosa-DTRLSL; Rosa-Ai9 and Scx-Cre; Rosa-Ai9 reporter mice were given local, daily DT injections for 5 consecutive days and hind paws were harvested 24 and 38 days after the last injection (F) These are the contralateral control tendons from the mice in Figure 4 that underwent tendon injury and repair. (G) Hind paws from ScxLinAi9 and ScxLinAi9DTR were probed for Red Fluorescent Protein (RFP; Ai9) expression and counterstained with the nuclear dye DAPI. (H) Total tendon cell density (DAPI+), total ScxLinAi9+ cell density and the percentage of ScxLinAi9 cells (ScxLinAi9+ cells/ DAPI+ cells) were quantified in ScxLinAi9 and ScxLinAi9DTR tendons and demonstrate a significant reduction of ScxLinAi9 cells in ScxLinAi9DTR relative to ScxLinAi9 WT controls. N = 3–4 per genotype. Two-way ANOVA with Sidak’s multiple comparisons test used to assess statistical significance of tendon cell ablation between hind paw (injected with DT or contralateral) and genotype (ScxLinAi9 and ScxLinAi9DTR at 24 and 38 days). * indicates p<0.05 for the indicated comparison, ** indicates p<0.01 for indicated comparison, **** indicates p<0.0001 relative to all other groups.
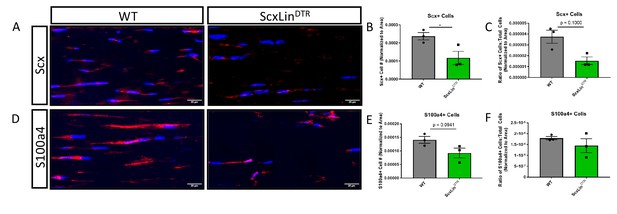
Scx+ and S100a4+ tendon cells following ScxLin depletion.
WT and ScxLinDTR hind paws were harvested uninjured 10 days following final DT injection and stained for either Scx (A–C) or S100a4 (D–F). Quantification of Scx+ cells normalized to either total area (B) or both area and total cell number (C). Quantification of S100a4+ cells normalized to either total area (E) or both area and total cell number (F). N = 3 per genotype. Statistical significance between genotypes determined using Student’s t-test when data was normal (B, E, F) or Mann-Whitney test when data not normal (C). * indicative of p<0.05.
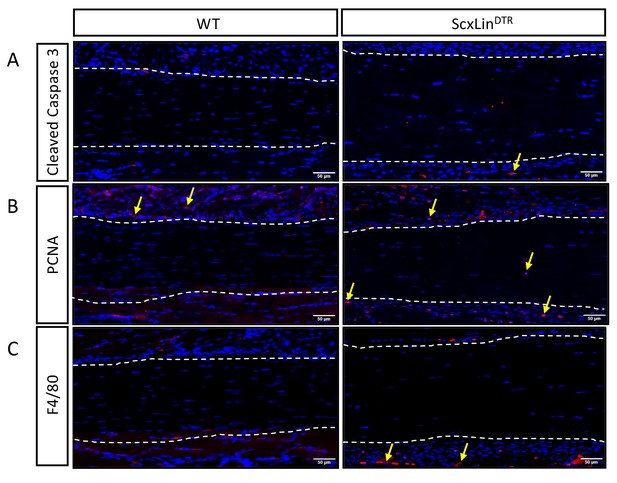
ScxLinDTR does not cause substantial effects on surrounding tissue.
Assessment of apoptosis (Cleaved Caspase 3, A), proliferation (PCNA, B), and Inflammation (F4/80, macrophages, C) for WT and ScxLinDTR uninjured tendons. Tendon outlined by white dotted line. Nuclei stained with DAPI. Examples of positive stain indicated by yellow arrows. N = 3 per genotype.
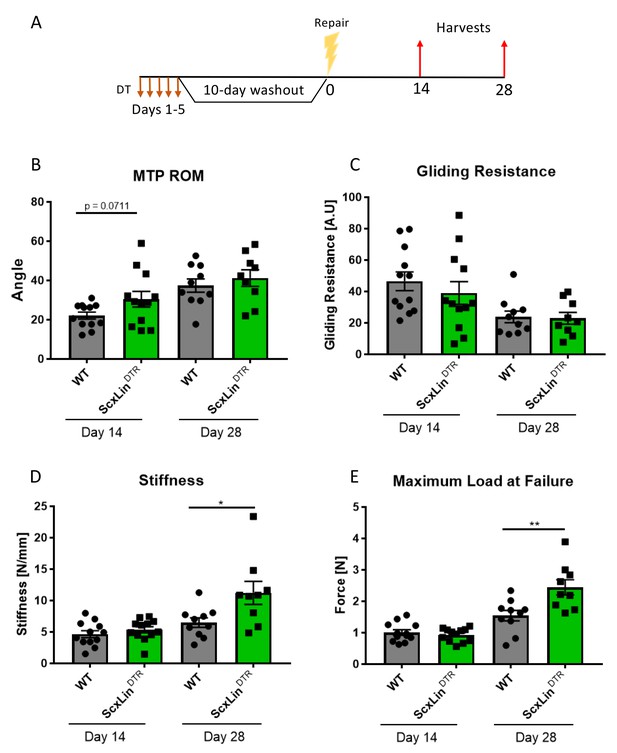
ScxLinDTR tendons heal with significantly increased biomechanical properties.
Mice received five hind paw injections of DT on consecutive days, underwent flexor tendon repair surgery 10 days after the final DT injection, and were harvested at 14- and 28 days post-repair (A). Measurement of metatarsophalangeal (MTP) joint flexion angle (B), gliding resistance (C), stiffness (D), and maximum load at failure (E) of WT and ScxLinDTR repaired tendons. N = 9–12 per genotype per timepoint. Students t-test used to assess statistical significance between genotypes at a given timepoint. * indicative of p<0.05, ** indicative of p<0.01.
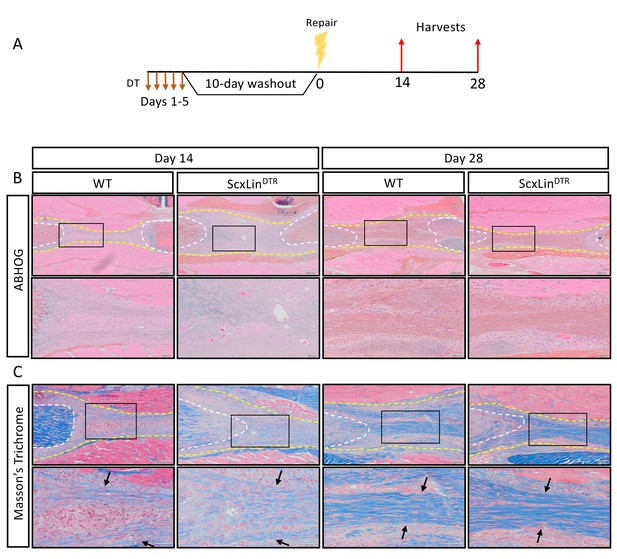
ScxLin cell depletion does not disrupt formation of a bridging collagen matrix.
Mice received five hindpaw injections of DT on consecutive days, underwent flexor tendon repair surgery 10 days after the final DT injection, and were harvested at 14 and 28 days post-repair (A). Alcian blue/hematoxylin and Orange G stain utilized to assess overall morphology (B). Masson’s trichrome stain used to visualize collagen content and organization (C). Tendon is outlined by white dotted line and scar tissue by yellow dotted line. Black boxes indicate location of higher magnification images. Boundaries of bridging collagen indicated by black arrows. N = 4 genotype per timepoint. Suture indicated by *.
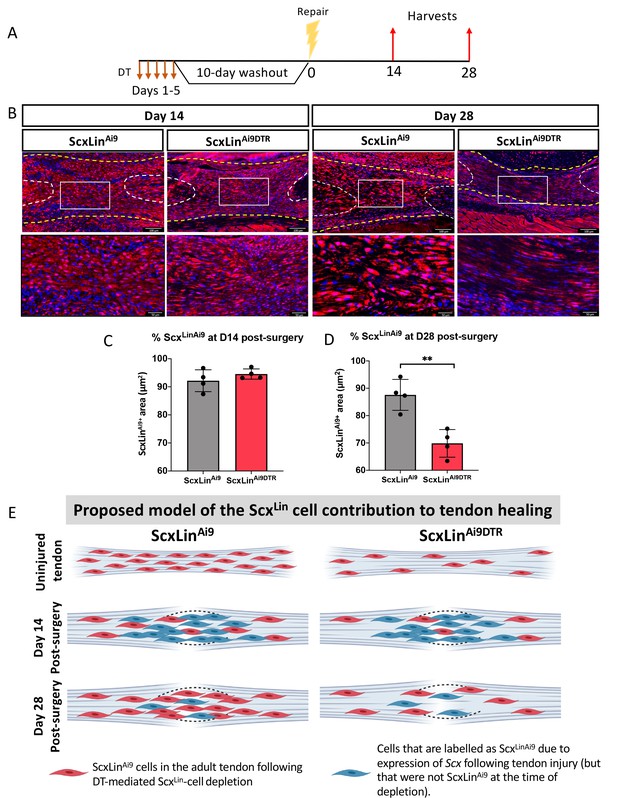
ScxLin cell depletion results in time-dependent changes in ScxLinAi9 cell presence during tendon healing.
(A) Mice received hind paw injections of DT on 5 consecutive days, underwent flexor tendon repair surgery 10 days after the final DT injection, and were harvested at 14 and 28 days post-repair. (B) Immunofluorescence for RFP (Ai9) in WT ScxLinAi9 and ScxLinAi9DTR tendon repairs at 14 and 28 days post-repair to define changes in ScxLinAi9 contribution following ScxLin cell depletion. Quantification of ScxLinAi9+ area in ScxLinAi9 WT repairs and ScxLinAi9DTR repairs at (C) D14 and (D) D28 post-surgery. Nuclei were stained with DAPI. N = 4 per genotype. Student’s t-test used to assess statistical significance between genotypes at a given timepoint. **indicates p<0.01. (E) Proposed model of the time-dependent contributions of ScxLinAi9 cells to the tendon healing process. During adult tendon homeostasis ScxLinAi9 cells are the predominant tenocyte population and ScxLinAi9DTR results in depletion of ~60% of these cells. Red cells indicate ScxLinAi9 cells that were present in the tendon when depletion was initiated. We hypothesize that no differences in the proportion of ScxLinAi9 cells is observed at D14 (concomitant with a lack of functional phenotypic differences) due to the predominance and functions of other cell populations, including those that express Scx in response to injury and are therefore labeled as ScxLin (blue cells). In contrast, we hypothesize that by D28 the contribution of ‘new’ ScxLin cells (blue cells) has waned, and that the ScxLinAi9 cells that were present in the tendon during adult tendon homeostasis (red cells) are now the predominant tenocyte population and exert their functions at this time as suggested by functional differences between WT and ScxLinDTR at this time. This schematic was made using http://www.biorender.com.
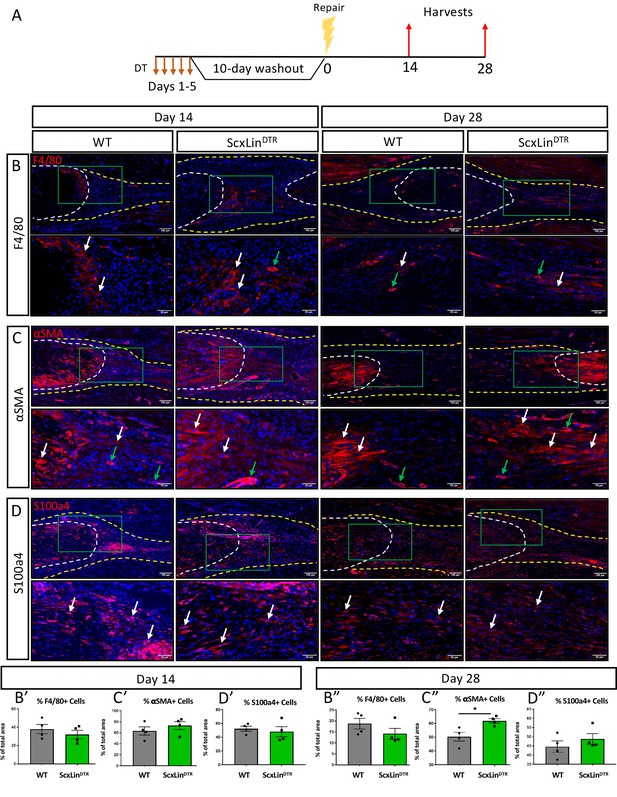
ScxLinDTR repaired tendons heal with increased presence of αSMA+ myofibroblasts.
Mice received five hindpaw injections of DT on consecutive days, underwent flexor tendon repair surgery 10 days after the final DT injection, and were harvested at 14 and 28 days post-repair (A). Immunofluorescence of WT and ScxLinDTR repair tendons 14 and 28 days post-repair to assess F4/80+ macrophages (B), αSMA+ myofibroblasts (C), and S100a4+ cells (D). Tendon is outlined by white dotted line and scar tissue by yellow dotted line. Green boxes indicate location of higher magnification images. Examples of positive stain indicated by white arrows, while examples of auto-fluorescent blood cells and α-SMA+ blood vessels indicated by green arrows. Quantification of F4/80 (A’ and A’’), αSMA (B’ and B’’), and S100a4 (C’ and C’’) fluorescence. N = 4 per genotype per timepoint. Student’s t-test used to assess statistical significance between genotypes at a given timepoint, except for D28 F4/80 and S100a4 which required a Mann-Whitney test. * indicates p<0.05.
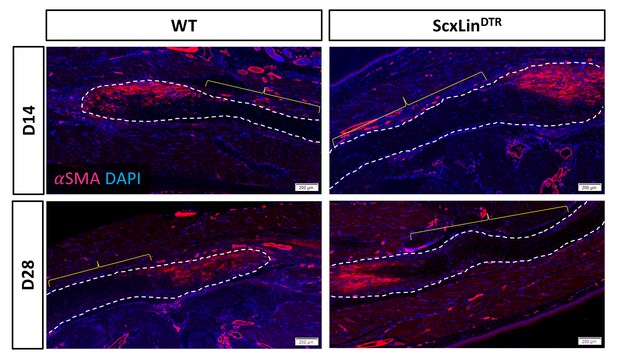
Specific localization of αSMA staining at the tendon repair site.
To demonstrate that the myotendinous release, which is incorporated as part of the surgical procedure to decrease the risk of repair rupture, does not lead to tendon degeneration, αSMA staining adjacent to the repair site was examined. An absence of αSMA was observed proximal/ distal to the repair site (yellow bracket). Tendon stubs are outlined in white. N = 4 per genotype per timepoint.
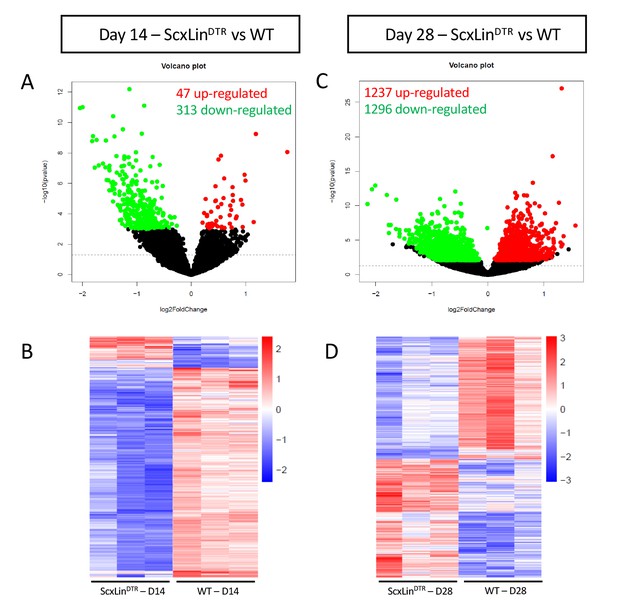
Bulk RNA sequencing reveals differences between ScxLinDTR and wild-type healing flexor tendons at 14 and 28 days post-repair.
Representation of differentially expressed genes (DEGs) at 14 (A, B) and 28 (C, D) days post-repair. Volcano plots (A, C) depict significantly upregulated DEGs as red dots and significantly downregulated DEGs as green dots. DEGs are consider significant when the multiple test corrected (adjusted) p-value is < 0.05. The dotted line represents the unadjusted p-value of 0.05. Heat maps (B, D) depict all significant DEGs, with the data representing the regularized log transformation of the normalized count data.
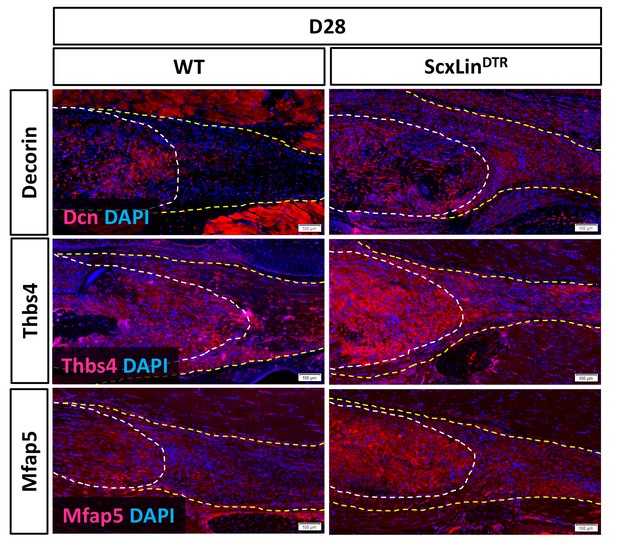
Enhanced expression specific matrix components is observed in ScxLinDTR tendon repairs at D28.
Based on the RNAseq data we examined the spatial localization of specific ECM components Decorin, Thbs4 and Mfap5 at D28 post-surgery. Sections were stained with the nuclear dye DAPI. N = 4 per genotype.
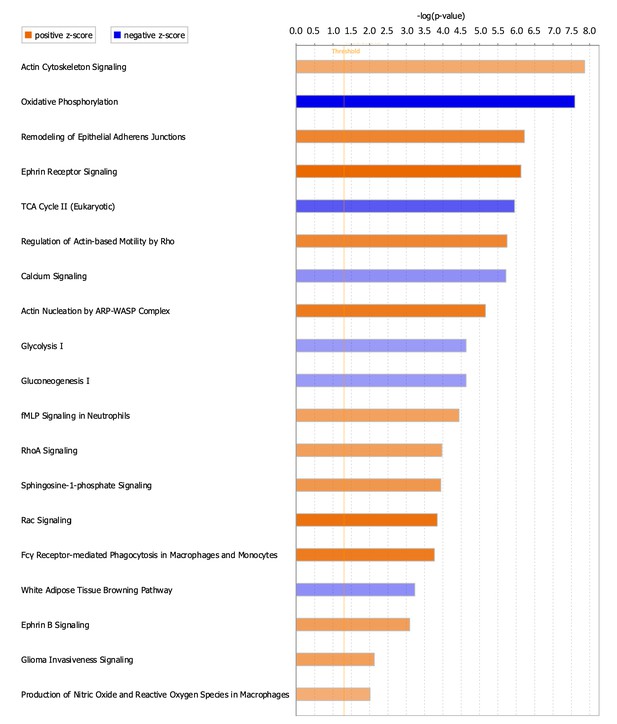
Canonical pathways positively and negatively enriched in ScxLinDTR healing tendons at day 28 post-repair.
Ingenuity pathway analysis was utilized to determine positively and negatively significantly enriched pathways in ScxLinDTR healing tendons at day 28 post-repair. Canonical pathways were considered significant if p<0.05 and ABS(Z-score)>2. The orange color indicates pathways that are significantly, positively enriched (‘activated’), while the blue color indicates significantly, negatively enriched (‘inhibited’) pathways. The orange dotted line represents -log (1.3)=0.05, indicating the p-value cut-off.
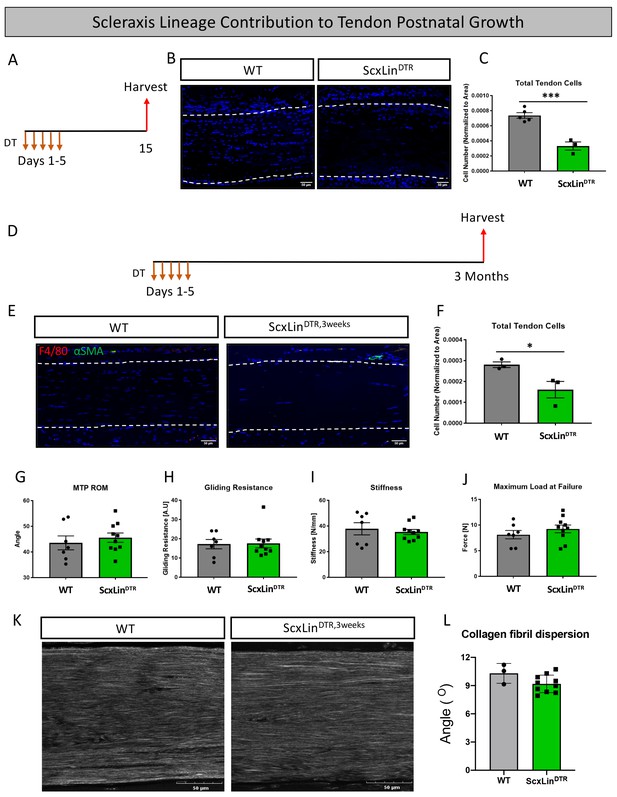
Tendon cell ablation does not negatively affect post-natal tendon growth 3 months post-ablation.
Pre-pubescent mice (3–4 weeks old) received five hindpaw injections of DT and were harvested 10 days after the final injection to assess tendon cell depletion (ScxLinDTR) (A). Hindpaw sections from both WT and ScxLinDTR hindpaws (B). Quantification of WT and ScxLinDTR,3weeks tendon cell number in pre-pubescent mice (C). To assess effects of tendon cell depletion on post-natal tendon growth, mice received five hindpaw injections of DT on consecutive days at 3–4 weeks of age and were harvested uninjured 3 months later for biomechanical, gliding, and histological evaluation (ScxLinDTR,3weeks) (D). Co-immunofluorescence of F4/80 (macrophages) and αSMA (myofibroblasts) in uninjured WT and ScxLinDTR,3weeks tendons (E). Quantification of WT and ScxLinDTR,3weeks tendon cell number (F). Measurement of metatarsophalangeal (MTP) joint flexion angle (G), gliding resistance (H), stiffness (I), and maximum load at failure (J) of WT and ScxLinDTR,3weeks uninjured tendons. N = 7–10 per genotype. Second harmonic generation (K) and quantification (L) of collagen fibril dispersion of WT and ScxLinDTR,3weeks. N = 3 per genotype. Nuclei stained with DAPI. Tendon is outlined by white dotted lines. Student’s t-test used to assess statistical significance between genotypes. * indicates p<0.05, *** indicates p<0.001.
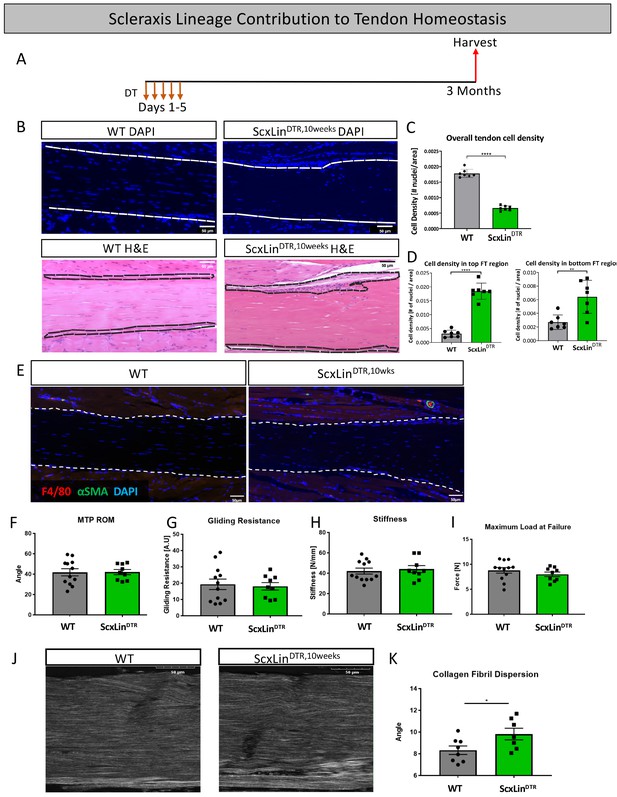
Tendon cell ablation negatively affected tendon homeostasis 3 months post-ablation.
Mice received five hindpaw injections of DT on consecutive days at 10–12 weeks of age and were harvested uninjured 3 months later for biomechanical, gliding, and histological evaluation (ScxLinDTR,10weeks) (A). Cellularity was assessed using DAPI (B) and quantified (C) 3 months after ScxLin cell depletion. H and E staining was used to better define the hypercellular regions near the tendon epitenon. Cell density was quantified at the top and bottom boundaries of the tendon (D). N = 7 per genotype. Co-immunofluorescence of F4/80 (macrophages) and αSMA (myofibroblasts) in uninjured WT and ScxLinDTR tendons (E). N = 3 per genotype. Measurement of metatarsophalangeal (MTP) joint flexion angle (F), gliding resistance (G), stiffness (H), and maximum load at failure (I) of WT and ScxLinDTR,10weeks uninjured tendons. N = 9–12 per genotype. Second harmonic generation (SHG) (J) and quantification (K) of collagen fibril dispersion of WT and ScxLinDTR,10weeks. N = 7–8 per genotype. Nuclei stained with DAPI. Tendon is outlined by white dotted lines. Student’s t-test used to assess statistical significance between genotypes. * indicates p<0.05.
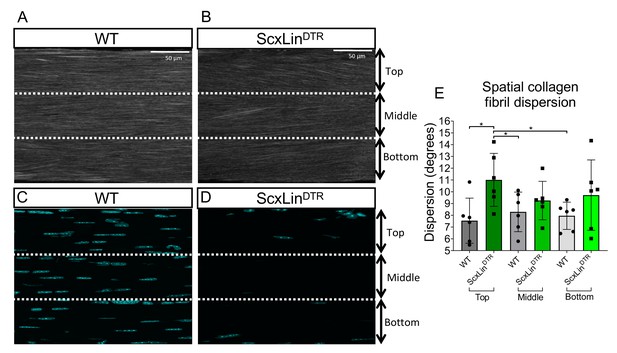
Collagen fibrils in the top third FT region have an altered organization at 3 months post-depletion.
Representative SHG images of collagen fibrils (A, B) and cell nuclei (C, D). Quantification of spatial collagen fibril dispersion on the top, middle, and bottom regions of WT and ScxLinDTR FTs at 3 months post-depletion (E). N = 6 per genotype. Two-way ANOVA used to assess statistical significance between genotypes and tendon regions. *indicates p<0.05.
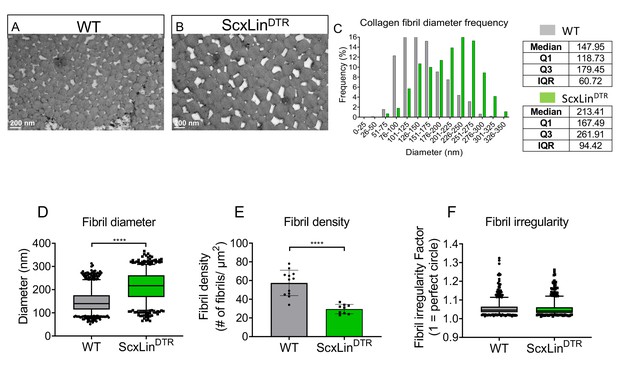
Collagen fibrils exhibit an altered diameter and density at 3 months post-depletion.
Representative TEM images of the WT and ScxLinDTR FTs at 3 months post-depletion (A, B). Collagen fibril diameter histogram demonstrates an increase in the median fibril diameter in DTR D90 FTs compared to WT (C). Collagen fibril diameter of the WT and DTR D90 FTs with boxplot whiskers spanning data between the 5th and 95th percentiles; data outside this range are plotted as individual points (D). Collagen fibril density of the WT and DTR D90 FTs (E). Collagen fibril irregularity of the WT and DTR D90 FTs (F). N = 4 for WT and N = 3 for DTR D90. Student’s t-test used to assess statistical significance between genotypes. ****indicates p<0.0001.
Tables
Ingenuity pathway analysis downstream effects - Disease and Functions.
Table of all disease and function annotations marked as significant (p<0.05 and ABS(Z-score)>2) using IPA core analysis for ScxLinDTR vs WT at day 28 post-repair.
| Disease or function annotation | p-value | Activation state | Z-Score | |
|---|---|---|---|---|
| Down-Regulated | Contractility of skeletal muscle | 4.96E-16 | Decreased | −3.595 |
| Abnormal bone density | 0.000000572 | Decreased | −3.299 | |
| Contractility of muscle | 8.91E-16 | Decreased | −2.636 | |
| Intestinal cancer | 6.76E-46 | Decreased | −2.561 | |
| Bleeding | 0.00000075 | Decreased | −2.424 | |
| Malignant neoplasm of large intestine | 8.42E-46 | Decreased | −2.343 | |
| Colorectal cancer | 6.67E-23 | Decreased | −2.343 | |
| Large intestine neoplasm | 2.97E-46 | Decreased | −2.256 | |
| Colorectal tumor | 1.28E-23 | Decreased | −2.256 | |
| Function of muscle | 6.93E-14 | Decreased | −2.245 | |
| Consumption of oxygen | 0.000000942 | Decreased | −2.237 | |
| Function of skeletal muscle | 1.59E-08 | Decreased | −2.186 | |
| Intestinal tumor | 9.72E-47 | Decreased | −2.144 | |
| Development of lung carcinoma | 0.000000663 | Decreased | −2.012 | |
| Up-regulated | Cell movement | 4.98E-22 | Increased | 4.735 |
| Migration of cells | 6.44E-17 | Increased | 4.733 | |
| Cell movement of tumor cell lines | 8.66E-10 | Increased | 4.343 | |
| Reorganization of cytoskeleton | 0.000000218 | Increased | 4.296 | |
| Migration of tumor cell lines | 0.00000032 | Increased | 4.162 | |
| Engulfment of cells | 1.05E-09 | Increased | 4.057 | |
| Endocytosis | 1.2E-11 | Increased | 3.821 | |
| Leukocyte migration | 4.37E-09 | Increased | 3.821 | |
| Cell movement of blood cells | 4.04E-09 | Increased | 3.818 | |
| Homing of cells | 0.00000055 | Increased | 3.792 | |
| Formation of cellular protrusions | 1.53E-14 | Increased | 3.669 | |
| Glucose metabolism disorder | 5.46E-10 | Increased | 3.516 | |
| Invasion of cells | 9.21E-08 | Increased | 3.396 | |
| Organization of cytoplasm | 3.25E-26 | Increased | 3.384 | |
| Organization of cytoskeleton | 2.94E-20 | Increased | 3.384 | |
| Cell movement of leukocytes | 7.74E-08 | Increased | 3.38 | |
| Endocytosis by eukaryotic cells | 1.16E-08 | Increased | 3.371 | |
| Engulfment of tumor cell lines | 0.00000031 | Increased | 3.348 | |
| Proliferation of neuronal cells | 3.67E-09 | Increased | 3.313 | |
| Metabolism of carbohydrate | 1.94E-13 | Increased | 3.285 | |
| Formation of lamellipodia | 0.000000932 | Increased | 3.121 | |
| Cell movement of breast cancer cell lines | 0.000000603 | Increased | 3.103 | |
| Cell movement of fibroblast cell lines | 3.42E-08 | Increased | 3.083 | |
| Growth of neurites | 6.25E-09 | Increased | 2.981 | |
| Microtubule dynamics | 5.21E-18 | Increased | 2.974 | |
| Cell spreading | 3.31E-11 | Increased | 2.875 | |
| Formation of filopodia | 0.000000177 | Increased | 2.873 | |
| Cell movement of connective tissue cells | 0.000000415 | Increased | 2.792 | |
| Concentration of lipid | 3.14E-08 | Increased | 2.779 | |
| Progressive neurological disorder | 6.74E-10 | Increased | 2.671 | |
| Outgrowth of neurites | 7.31E-08 | Increased | 2.662 | |
| Production of reactive oxygen species | 6.22E-12 | Increased | 2.625 | |
| Quantity of macropinosomes | 0.000000865 | Increased | 2.621 | |
| Neuromuscular disease | 1.47E-15 | Increased | 2.619 | |
| Progressive myopathy | 1.28E-11 | Increased | 2.611 | |
| Synthesis of carbohydrate | 0.000000499 | Increased | 2.553 | |
| Outgrowth of cells | 0.000000025 | Increased | 2.52 | |
| Advanced malignant tumor | 0.000000208 | Increased | 2.517 | |
| Differentiation of connective tissue cells | 1.48E-09 | Increased | 2.512 | |
| Secondary tumor | 0.000000643 | Increased | 2.461 | |
| Arrhythmia | 2.14E-08 | Increased | 2.4 | |
| Fibrosis | 0.000000337 | Increased | 2.397 | |
| Extension of cellular protrusions | 0.00000098 | Increased | 2.371 | |
| Invasive tumor | 2.47E-08 | Increased | 2.345 | |
| Synthesis of reactive oxygen species | 3.6E-14 | Increased | 2.312 | |
| Organization of actin cytoskeleton | 4.37E-08 | Increased | 2.298 | |
| Disassembly of filaments | 0.000000544 | Increased | 2.27 | |
| Metabolism of reactive oxygen species | 2.61E-15 | Increased | 2.269 | |
| Cancer of cells | 9.31E-14 | Increased | 2.254 | |
| Response of tumor cell lines | 2.31E-08 | Increased | 2.231 | |
| Morphogenesis of neurons | 1.44E-12 | Increased | 2.224 | |
| Neuritogenesis | 1.86E-12 | Increased | 2.224 | |
| Neoplasia of cells | 1.76E-16 | Increased | 2.221 | |
| Quantity of metal | 0.0000002 | Increased | 2.198 | |
| Ruffling | 0.000000297 | Increased | 2.157 | |
| Tubulation of cells | 0.00000126 | Increased | 2.132 | |
| Angiogenesis | 4.02E-15 | Increased | 2.109 | |
| Hereditary myopathy | 1.58E-23 | Increased | 2.104 | |
| Dystrophy of muscle | 1.85E-11 | Increased | 2.104 | |
| Development of vasculature | 2.67E-16 | Increased | 2.06 | |
| Growth of axons | 0.000000724 | Increased | 2.017 | |
| Migration of fibroblast cell lines | 0.000000461 | Increased | 2.002 | |
Regulation of matrix components in ScxLinDTR healing tendons at day 28.
Expression level, fold change, and adjusted p-value of key matrix-related genes in ScxLinDTR tendons vs WT at day 28 post-repair generated from bulk RNA-seq. Orange color indicative of increased expression and blue color indicative of decreased expression.
| Gene | BaseMean | Fold change (Log2) | p-adj |
|---|---|---|---|
| Collagens | |||
| Col1a1 | 742982.883 | 0.275 | 0.117206 |
| Col1a2 | 720257.233 | 0.359 | 0.033355 |
| Col2a1 | 461.965 | 0.176 | 0.700754 |
| Col3a1 | 748380.28 | 0.645 | 0.005567 |
| Col4a1 | 35181.773 | 0.17 | 0.559295 |
| Col4a2 | 31627.065 | 0.265 | 0.206116 |
| Col4a3 | 49.144 | −0.009 | 0.988635 |
| Col4a4 | 143.52 | 0.232 | 0.589161 |
| Col4a5 | 284.059 | 0.372 | 0.440755 |
| Col4a6 | 52.693 | −0.093 | 0.904823 |
| Col5a1 | 118821.857 | 0.495 | 0.031814 |
| Col5a2 | 105087.042 | 0.46 | 0.092164 |
| Col5a3 | 35303.666 | 0.418 | 0.06355 |
| Col6a1 | 122410.423 | 0.406 | 0.00366 |
| Col6a2 | 122396.322 | 0.455 | 0.003183 |
| Col6a3 | 55660.629 | 0.545 | 0.021485 |
| Col6a4 | 33.599 | −0.879 | 0.0385 |
| Col6a5 | 72.046 | 0.599 | 0.286724 |
| Col6a6 | 51.234 | −0.07 | 0.901424 |
| Col7a1 | 2215.503 | −0.532 | 0.06829 |
| Col8a1 | 6553.562 | 0.983 | 0.025451 |
| Col8a2 | 4787.689 | −0.128 | 0.727708 |
| Col9a1 | 390.971 | −1.328 | 0.000857 |
| Col9a2 | 50.18 | −0.071 | 0.911757 |
| Col9a3 | 52.911 | 0.177 | 0.713852 |
| Col10a1 | 4.585 | 0.254 | N/A* |
| Col11a1 | 13080.117 | −0.239 | 0.496276 |
| Col11a2 | 1974.589 | −0.671 | 0.028981 |
| Col12a1 | 34791.768 | −0.204 | 0.545342 |
| Col13a1 | 308.833 | −0.085 | 0.853468 |
| Col14a1 | 7279.653 | 0.693 | 0.00124 |
| Col15a1 | 8889.517 | 0.385 | 0.296359 |
| Col16a1 | 24279.347 | 0.237 | 0.252556 |
| Col17a1 | 269.42 | 0.002 | N/A* |
| Col18a1 | 14754.886 | 0.391 | 0.169961 |
| Col19a1 | 4.738 | 0.133 | N/A* |
| Col20a1 | 438.526 | −0.7 | 0.060808 |
| Col22a1 | 2064 | −0.797 | 0.022217 |
| Col23a1 | 4457.185 | −0.636 | 0.025351 |
| Col24a1 | 966.961 | −0.35 | 0.047102 |
| Col25a1 | 118.033 | 0.022 | 0.975523 |
| Col26a1 | 78.9 | 0.371 | 0.475453 |
| Col27a1 | 4062.784 | 0.406 | 0.253217 |
| Col28a1 | 579.441 | 0.291 | 0.653743 |
| ECM proteoglycans | |||
| Hspg2 | 47213.867 | 0.248 | 0.108795 |
| Aspn | 19143.191 | 0.839 | 0.007556 |
| Bgn | 151131.153 | 0.251 | 0.256881 |
| Dcn | 95817.718 | 0.654 | 1.26E-05 |
| Fmod | 132295.748 | 0.172 | 0.683103 |
| Kera | 5207.231 | 0.522 | 0.154489 |
| Lum | 43114.099 | 0.353 | 0.130133 |
| Omd | 65.324 | 0.56 | 0.151367 |
| Prelp | 18853.124 | 0.381 | 0.057946 |
| Epyc | 133.935 | 1.187 | 0.003793 |
| Ogn | 5228.656 | 0.636 | 0.004079 |
| Optc | 30.193 | 0.042 | 0.957221 |
| Chad | 2558.426 | −0.133 | 0.813626 |
| Chadl | 146.381 | 0.377 | 0.295306 |
| Nyx | 21.048 | 0.443 | 0.360179 |
| Podn | 1763.369 | 0.432 | 0.131578 |
| Podnl1 | 216.612 | −0.087 | 0.872241 |
| Acan | 8738.435 | −0.407 | 0.194754 |
| Bcan | 125.867 | −0.764 | 0.014902 |
| Ncan | 1.472 | −0.047 | N/A* |
| Vcan | 5214.463 | 0.431 | 0.165678 |
| Hapln1 | 109.201 | 0.036 | 0.955062 |
| Hapln2 | 8.84 | −0.024 | N/A* |
| Hapln3 | 31.92 | 0.039 | 0.949531 |
| Hapln4 | 128.699 | 0.053 | 0.926101 |
| Prg2 | 2.717 | −0.083 | N/A* |
| Spock1 | 12.387 | 0.674 | N/A* |
| Spock2 | 621.489 | 0.027 | 0.956371 |
| Spock3 | 34.16 | −0.034 | 0.963822 |
| Prg4 | 26463.024 | −0.191 | 0.759532 |
| Srgn | 1500.611 | −0.08 | 0.843956 |
| Impg2 | 49.211 | −0.196 | 0.676547 |
| Esm1 | 120.466 | −0.02 | 0.971642 |
| Basement membrane components | |||
| Lama1 | 3.29 | −0.445 | N/A* |
| Lama2 | 3430.887 | 0.361 | 0.201711 |
| Lama3 | 104.957 | −0.303 | 0.484039 |
| Lama4 | 7706.819 | 0.34 | 0.081572 |
| Lama5 | 3390.382 | −0.237 | 0.430648 |
| Lamb1 | 11272.507 | 0.236 | 0.389409 |
| Lamb2 | 13730.5 | 0.343 | 0.092701 |
| Lamb3 | 77.394 | −0.06 | 0.924993 |
| Lamc1 | 15292.13 | 0.313 | 0.119095 |
| Lamc2 | 424.266 | 0.393 | 0.064032 |
| Lamc3 | 21.597 | 0.093 | 0.886633 |
| Nid1 | 12717.306 | 0.539 | 0.02301 |
| Nid2 | 2799.817 | 0.179 | 0.586124 |
| Colq | 358.747 | −0.379 | 0.341156 |
| Major ECM glycoproteins | |||
| Eln | 17607.464 | 0.518 | 0.270402 |
| Emilin1 | 6960.539 | 0.124 | 0.684821 |
| Emilin2 | 4405.463 | 0.512 | 0.061758 |
| Emilin3 | 595.356 | 0.857 | 0.005562 |
| Emid1 | 550.685 | 0.252 | 0.631487 |
| Fbln1 | 2214.023 | 0.23 | 0.320082 |
| Fbln2 | 48333.486 | 0.127 | 0.615495 |
| Fbln5 | 2725.675 | 0.326 | 0.023116 |
| Fbln7 | 4562.409 | 0.784 | 0.00233 |
| Efemp1 | 1308.939 | 0.543 | 0.077648 |
| Efemp2 | 4858.991 | 0.222 | 0.11327 |
| Fbn1 | 36959.196 | 0.668 | 0.005087 |
| Fbn2 | 2856.248 | −0.008 | 0.983466 |
| Fn1 | 510510.053 | 0.307 | 0.223595 |
| Fras1 | 430.729 | −0.837 | 0.013797 |
| Gldn | 464.82 | 0.424 | 0.390168 |
| Hmcn1 | 1034.806 | 0.589 | 0.036404 |
| Hmcn2 | 6034.922 | 0.252 | 0.456036 |
| Ibsp | 2.365 | −0.125 | N/A* |
| Matn1 | 0.671 | 0.074 | N/A* |
| Matn2 | 6213.381 | 0.503 | 0.015668 |
| Matn3 | 201.369 | −0.581 | 0.155595 |
| Matn4 | 2796.498 | −0.245 | 0.6134 |
| Mfap1a | 536.887 | 0.101 | 0.639732 |
| Mfap1b | 398.958 | 0.03 | 0.902813 |
| Mfap2 | 3608.727 | 0.465 | 0.013072 |
| Mfap3 | 1213.924 | 0.119 | 0.519005 |
| Mfap4 | 4217.93 | 0.4 | 0.210242 |
| Mfap5 | 14548.008 | 0.753 | 5.34E-05 |
| Mmrn1 | 439.183 | 0.492 | 0.262418 |
| Mmrn2 | 1557.776 | −0.201 | 0.519886 |
| Npnt | 1084.388 | −0.453 | 0.209896 |
| Papln | 16.561 | −0.122 | 0.859483 |
| Postn | 102294.871 | 0.591 | 0.048873 |
| Sparc | 331616.177 | 0.479 | 0.000296 |
| Sparcl1 | 12872.967 | −0.027 | 0.959288 |
| Spp1 | 29368.724 | 0.234 | 0.637831 |
| Srpx2 | 5778.794 | 0.362 | 0.077229 |
| Tnc | 28609.297 | 0.378 | 0.326135 |
| Tnn | 5098.501 | −0.623 | 0.126873 |
| Tnr | 19.271 | 0.482 | 0.385953 |
| Tnxa | 6.323 | 0.944 | N/A* |
| Tnxb | 18888.425 | 0.781 | 0.015263 |
| Thbs1 | 14477.798 | 0.49 | 0.099587 |
| Thbs2 | 36613.787 | 0.39 | 0.149809 |
| Thbs3 | 17595.855 | 0.267 | 0.193149 |
| Thbs4 | 203095.542 | 0.841 | 5.69E-05 |
| Comp | 35501.759 | 0.293 | 0.10314 |
Ingenuity pathway analysis canonical pathways.
All enriched pathways marked as significant (-log(p-value)>1.3 and ABS(Z-score)>2) using IPA core analysis for ScxLinDTR vs WT at day 28 post-repair.
| Canonical pathway | -log(p) | Z-Score | |
|---|---|---|---|
| Negatively enriched | Oxidative phosphorylation | 7.6 | −5.303 |
| TCA Cycle II (Eukaryotic) | 5.96 | −3.464 | |
| White Adipose Tissue Browning Pathway | 3.23 | −2.353 | |
| Calcium Signaling | 5.72 | −2.335 | |
| Glycolysis I | 4.63 | −2.111 | |
| Gluconeogenesis I | 4.63 | −2.111 | |
| Positively enriched | Ephrin Receptor Signaling | 6.13 | 3.888 |
| Rac Signaling | 3.85 | 3.674 | |
| Actin Nucleation by ARP-WASP Complex | 5.16 | 3.441 | |
| Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes | 3.77 | 3.411 | |
| Remodeling of Epithelial Adherens Junctions | 6.23 | 3.162 | |
| Regulation of Actin-based Motility by Rho | 5.75 | 3.128 | |
| Sphingosine-1-phosphate Signaling | 3.94 | 2.711 | |
| Ephrin B Signaling | 3.09 | 2.53 | |
| RhoA Signaling | 3.97 | 2.502 | |
| Glioma Invasiveness Signaling | 2.13 | 2.496 | |
| fMLP Signaling in Neutrophils | 4.44 | 2.449 | |
| Actin Cytoskeleton Signaling | 7.86 | 2.214 | |
| Production of Nitric Oxide and Reactive Oxygen Species in Macrophages | 2.01 | 2.117 | |
Ingenuity pathway analysis upstream regulators.
All possible upstream regulators where expression log ratio > 0.5, ABS(Z-score)>2, p-value<0.05, and agreement between predicted activation state and directionality of regulator’s gene expression, compiled using IPA core analysis for ScxLinDTR vs WT at day 28 post-repair.
| Upstream regulator | Expression log ratio | Predicted activation state | Activation Z-score | p-value of overlap |
|---|---|---|---|---|
| Activated in ScxLinDTR | ||||
| S100A4 | 0.518 | Activated | 2.946 | 0.00456 |
| F2 | 0.533 | Activated | 2.606 | 0.000134 |
| BTNL2 | 0.805 | Activated | 2.324 | 0.012 |
| EBF2 | 0.657 | Activated | 2.223 | 0.000738 |
| F2R | 0.599 | Activated | 2.22 | 0.0243 |
| NTRK2 | 0.608 | Activated | 2.137 | 0.012 |
| SOX2 | 1.138 | Activated | 2.071 | 0.0000143 |
| FGF2 | 0.544 | Activated | 2.017 | 0.000393 |
| Inhibited in ScxLinDTR | ||||
| FOXO4 | −0.501 | Inhibited | −2.697 | 0.0236 |
| MEF2C | −0.905 | Inhibited | −2.577 | 3.06E-08 |
| SMYD1 | −0.836 | Inhibited | −2.219 | 2.63E-12 |
| LDHB | −0.609 | Inhibited | −2.219 | 0.000241 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Scx-Cre | Dr. Ronen Schweitzer | MGI:5317938 | Referred to as ScxLinin manuscript |
| Genetic reagent (Mus musculus) | C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J (Rosa-DTRLSL) | Jackson Laboratory | Stock #: 007900 RRID:IMSR_JAX:007900 | Referred to as ScxLinDTRin manuscript |
| Genetic reagent (Mus musculus) | B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (ROSA-Ai9) | Jackson Laboratory | Stock #: 007909 RRID:IMSR_JAX:007909 | Referred to as ScxLinAi9or ScxLinAi9DTR in manuscript |
| Antibody | Anti-SCXA (rabbit polyclonal) | Abcam | Catalog #: ab58655 RRID:AB_882467 | (1:500) |
| Antibody | Anti-S100a4 (rabbit monoclonal) | Abcam | Catalog #: ab197896 RRID:AB_2728774 | (1:2000) |
| Antibody | Anti-cleaved caspase 3 (rabbit polyclonal) | Cell Signalling Technology | Catalog #: 9661 RRID:AB_2341188 | (1:100) |
| Antibody | Anti-PCNA (mouse monoclonal) | Abcam | Catalog #: ab29 RRID:AB_303394 | (1:100) |
| Antibody | Anti-F4/80 (rabbit polyclonal) | Santa Cruz Biotechnology | Catalog #: sc-26643 RRID:AB_2098331 | (1:500) |
| Antibody | Anti-THBS4 (rabbit monoclonal) | Abcam | Catalog #: ab263898 | (1:250) |
| Antibody | Anti-MFAP5 (rabbit monoclonal) | Abcam | Catalog #: ab203828 | (1:2000) |
| Antibody | Anti-Decorin (Rabbit polyclonal) | Abcam | Catalog #: ab175404 | (1:250) |
| Antibody | Anti-alpha-SMA-Cy3 (mouse monoclonal) | Sigma-Aldrich | Catalog #: C6198 RRID:AB_476856 | (1:200) |
| Antibody | Anti-alpha-SMA-FITC (mouse monoclonal) | Sigma-Aldrich | Catalog #: F3777 RRID:AB_476977 | (1:500) |
| Antibody | Rhodamine Red-X (RRX) AffiniPure F(ab')₂ Fragment Donkey Anti-Rabbit IgG (H+L) (Donkey polyclonal) | Jackson ImmunoResearch | Catalog #: 711-296-152 | (1:200) |
| Antibody | Alexa Fluor 488 AffiniPure F(ab')₂ Fragment Donkey Anti-Goat IgG (H+L) (Donkey polyclonal) | Jackson ImmunoResearch | Catalog #: 705-546-147 RRID:AB_2340430 | (1:200) |
| Antibody | Rhodamine Red-X (RRX) AffiniPure F(ab')₂ Fragment Donkey Anti-Mouse IgG (H+L) (Donkey polyclonal) | Jackson ImmunoResearch | Catalog #: 715-296-150 RRID:AB_2340834 | (1:200) |
| Chemical Compound, Drug | Diphtheria Toxin (DT) | Millipore Sigma | D0564-1MG | 20 ng DT / injection |
| Software, algorithms | GraphPad Prism software | GraphPad Prism (https://graphpad.com) | Version 7.02 | |
| Software, algorithms | OlyVIA software | Olympus (https://www.olympus-lifescience.com/en/support/downloads/) | Version 2.9 | |
| Software, algorithms | ImageJ software | ImageJ (http://imagej.nih.gov/ij/) |





