Decoding the physical principles of two-component biomolecular phase separation
Figures
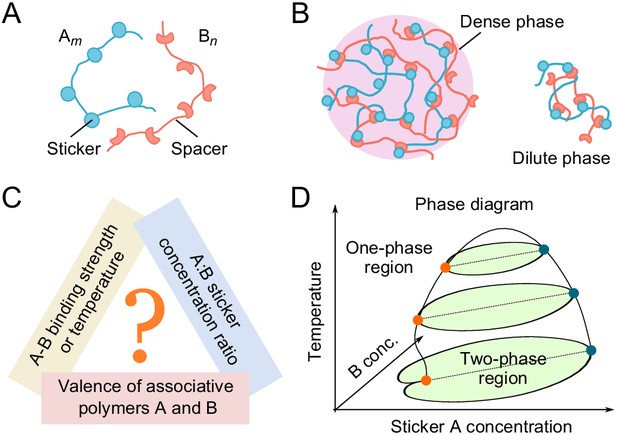
Phase behavior of sticker and spacer associative polymers.
(A) Schematic of multivalent associative polymers. Each polymer consists of complementary domains (stickers) connected by flexible linkers (spacers). A and B denote the polymer type and m and n denote their valences (number of stickers). (B) Association of stickers drives phase separation, leading to the formation of a dense, network phase coexisting with a dilute phase of small oligomers (depicted by a dimer). (C) The phase diagram depends on variety of biologically tunable parameters. In this study, we focus on the effects of sticker-sticker binding strength, sticker:sticker concentration ratio (i.e. stoichiometry), and polymer valences. (D) Schematic of a representative 3D phase diagram of an system as a function of temperature (inverse of binding strength) and A and B sticker concentrations. The dilute-phase concentration displays anomalous dependence on the binding strength and sticker concentrations in the strong binding regime. This is the ‘magic-ratio’ effect which we explore here in detail.
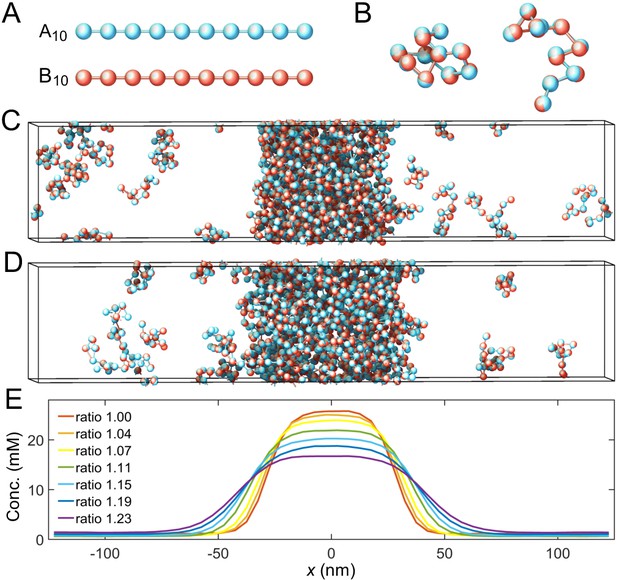
Coarse-grained molecular-dynamics simulations of two-component multivalent associative polymers.
(A) The system consists of two types of polymers A (blue) and B (red) of varying lengths and concentrations. Depicted are A and B polymers of length 10, denoted as A10 and B10. Each polymer is modeled as a linear chain of spherical particles connected by harmonic bonds. Stickers of different types interact pairwisely through an attractive potential, while repulsion between stickers of the same type prevents them from overlapping and thus ensures one-to-one binding of stickers of different types (see Appendix 1 for details). (B) Snapshots of dimers formed by A10 and B10 with one-to-one bonds. (C) Snapshot of a simulation with 125 A10 and 125 B10 polymers. The system phase separates into a dense phase (middle region) and a dilute phase (two sides) in a 250 nm×50 nm×50 nm simulation box with periodic boundary conditions. (D) Same as C but with 138 A10 and 112 B10 polymers, yielding an overall sticker concentration ratio 1.23. (E) Sticker concentration profiles of A10:B10 systems at various overall sticker stoichiometries (total global sticker concentration fixed at 6.64 mM), each with the center of the dense phase aligned at and averaged over time and over ten simulation repeats (see Appendix 1). All simulations performed in LAMMPS (Plimpton, 1995).
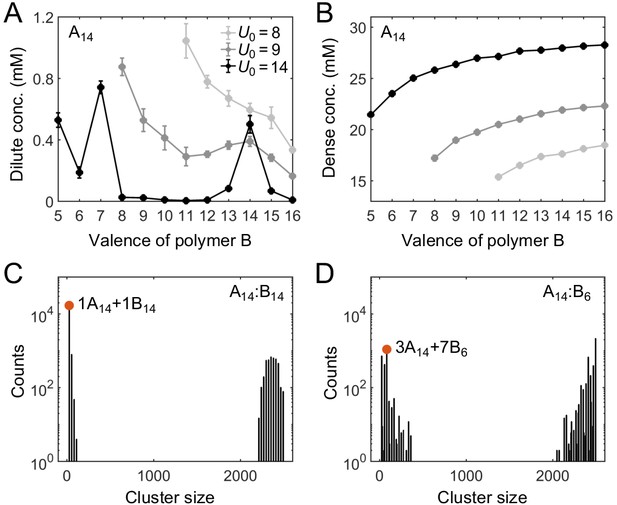
Simulations of associative polymers reveal a magic-number effect with respect to relative valence.
Total sticker concentrations (type A plus type B) in (A) dilute and (B) dense phases for simulated polymer systems at different binding strengths. U0 denotes the depth of the potential well, in units of (see Appendix 1 for details). The valence of polymer A is 14, and the valence of polymer B ranges from 5 to 16. Global sticker stoichiometry is one and total global sticker concentration is 6.64 mM. Histograms of cluster size in (C) A14:B14 and (D) A14:B6 systems, for . ‘Counts’ refer to number of clusters. Cluster size is measured in stickers. Red dots indicate the dominant oligomer in the dilute phase.
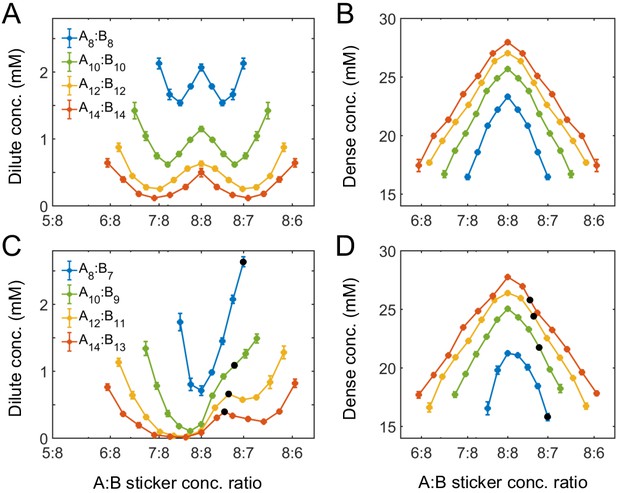
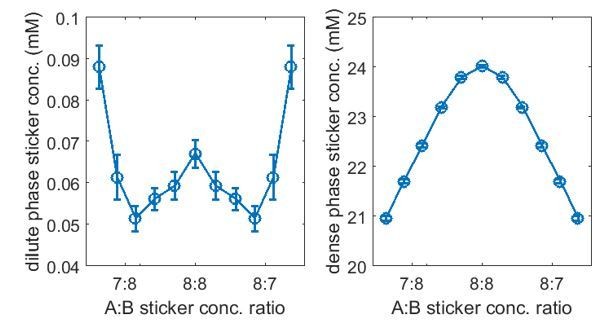
Simulations of associative polymers reveal a magic-ratio effect with respect to polymer stoichiometry.
Sticker concentrations in (A) dilute and (B) dense phases for equal polymer length systems (i.e. ) at different global sticker stoichiometries. Sticker concentrations in (C) dilute and (D) dense phases for systems where polymer B is one sticker shorter than polymer A (i.e. ) at different global sticker stoichiometries; black dots indicate cases where the number of polymers of each type is the same. Interaction strength and total global sticker concentration 6.64 mM.
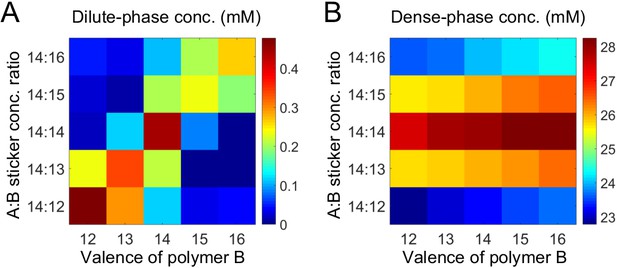
Simulations of associative polymers reveal a magic-ratio effect.
Sum of concentrations of stickers A and B in (A) dilute and (B) dense phases for systems A14:B12-16 at global sticker stoichiometries 14:12-16. Parameters: interaction strength and total global sticker concentration 6.64 mM.
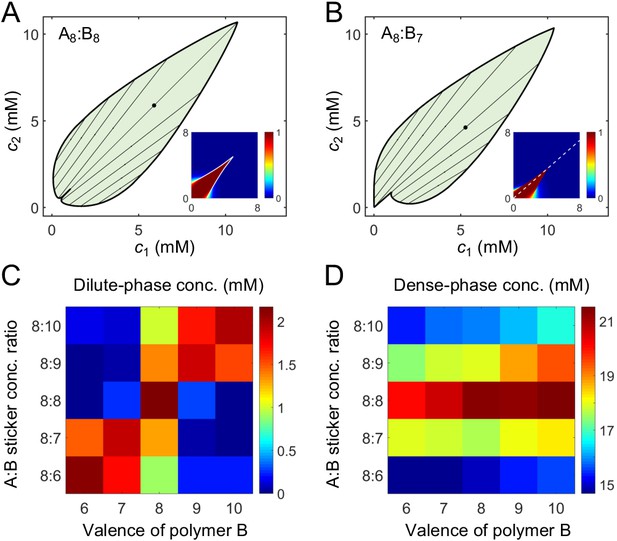
A dimer-gel theory predicts the magic-ratio effect.
Phase diagrams of (A) A8:B8 and (B) A8:B7 systems: one-phase region white, two-phase region green. The dilute- and dense-phase concentrations are connected by representative tie lines. The tie line along the direction of equal polymer stoichiometry is denoted with a black dot. Insets: fraction of stickers in dimers for (A) A8:B8 and (B) A8:B7 systems. White curve in A inset is the transition boundary between dimer- and independent bonds-dominated regions predicted by cs. Dashed white line in (B) inset denotes equal polymer stoichiometry. Sticker concentrations in (C) dilute and (D) dense phases for systems A8:B6-10 at global sticker stoichiometries 8:6-10. The total global sticker concentration is the same as in simulations, 6.64 mM. For details see Appendix 2. Parameters: , , , and values in Appendix 1—table 3.
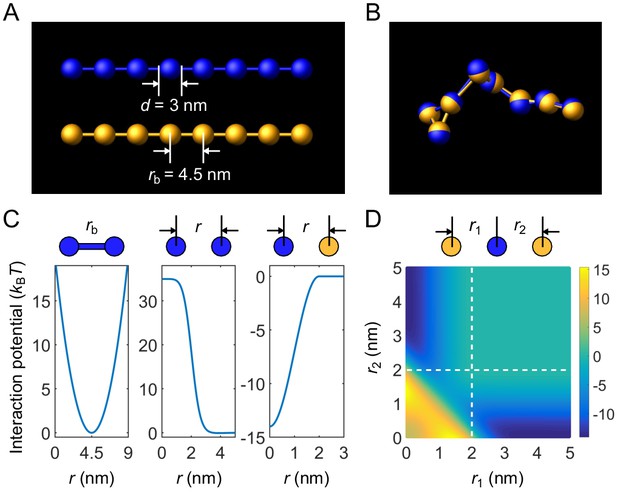
Coarse-grained molecular-dynamics simulations of two-component multivalent associative polymers.
(A) Polymers are modeled as linear chains of spherical particles connected by harmonic bonds. Depicted are A8 (blue) and B8 (yellow). (B) Snapshot of a dimer of A8 and B8 formed in the strong-binding regime. (C) Neighboring stickers in a polymer are connected through a harmonic potential (left). Stickers of the same type interact pairwisely through a repulsive potential (middle). Stickers of different types interact pairwisely through an attractive potential (right). (D) Interaction energy between three particles (one A and two B stickers) as a function of their separation distances. Simultaneous binding of two B stickers to one A sticker is energetically highly disfavored (lower left region) compared to one-to-one binding (dark blue regions).
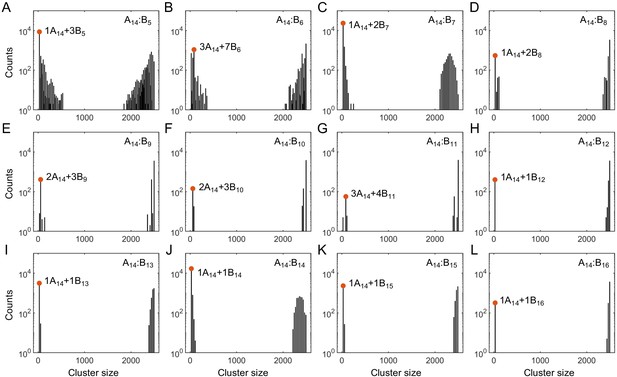
Histograms of cluster size in stickers in (A–L) A14:B5-16 systems at equal global sticker stoichiometry.
Parameters: specific binding strength and total global sticker concentration 6.64 mM. Red dots indicate the dominant oligomer in the dilute phase.
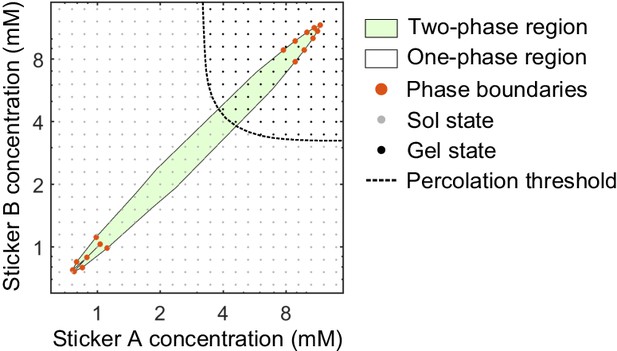
Phase diagram and percolation threshold of A8:B8 system.
Phase boundaries (red dots) are measured from MD simulations at . The complete two-phase region (green) and one-phase region (white) are extrapolated based on the phase boundaries from simulations. Sol- (gray dots) and gel- (black dots) states from simulations at where the system remains homogeneous are identified based on the gelation criterions. Percolation threshold (dashed line) is interpolated from the labeled sol- and gel-states.
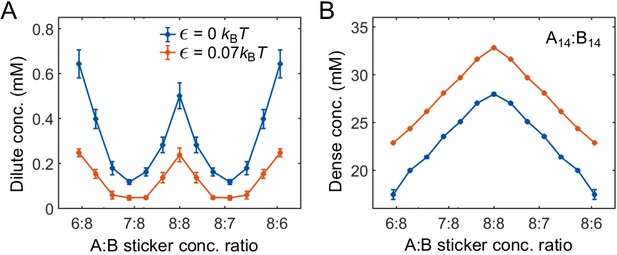
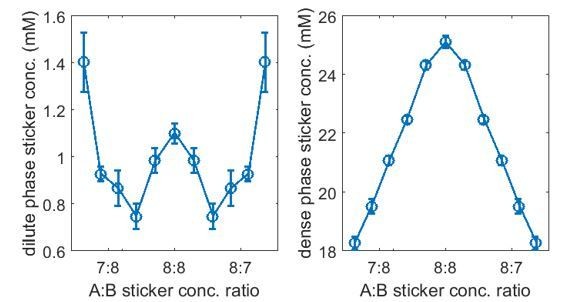
Strength of nonspecific attractive interactions strongly influences simulated dilute-phase boundary.
Total sticker concentrations in (A) dilute and (B) dense phases for A14:B14 system at different global sticker stoichiometries with the additional attractive interaction strength (blue) and (red) ( and in Equation (32)). Parameters: specific interaction strength , total global sticker concentration 6.64 mM.
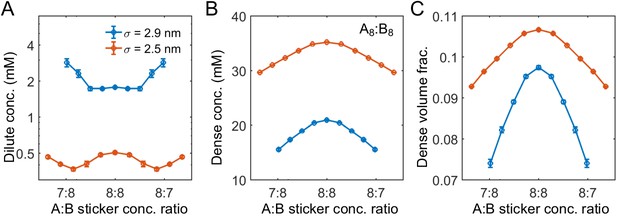
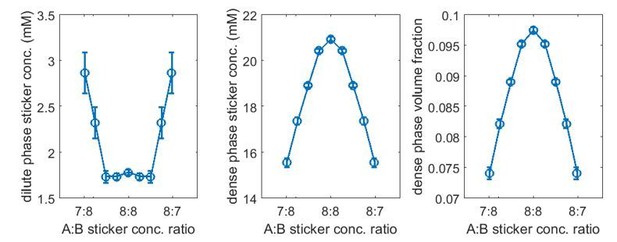
Range of nonspecific repulsive interactions strongly influences phase boundaries in simulations.
Sticker concentrations in (A) dilute and (B) dense phases and (C) volume fraction of polymers in dense phase for A8:B8 system at different global sticker stoichiometries with standard repulsive Lennard-Jones potential (Equation (33)) σ = 2.9 nm (blue) and σ = 2.5 nm (red). Parameters: repulsive interaction strength , specific interaction strength with cut-off distance r0 = 1.5 nm, and total global sticker concentration is 6.64 mM. Note that the overlapping volume between two bound stickers is counted once in the volume fraction calculation, that is, two perfectly overlapping stickers only occupy a volume of one sticker.
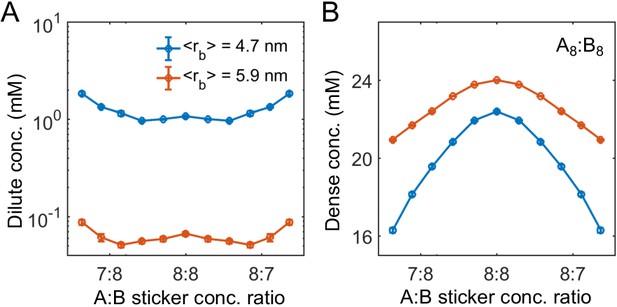
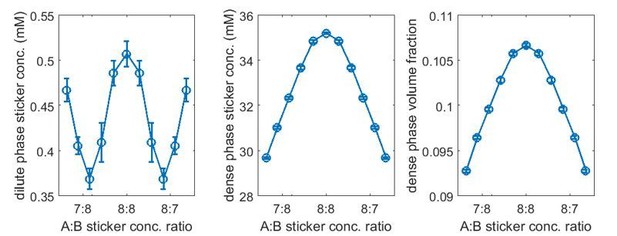
Linker length strongly influences phase boundaries in simulations.
Sticker concentrations in (A) dilute and (B) dense phases for A8:B8 system at different global sticker stoichiometries with linkers modeled by a FENE potential (Equation (34)) with and R0 = 7 nm (mean linker length 4.7 nm, blue) and and R0 = 14 nm (mean linker length 5.9 nm, red). Parameters: repulsive interaction strength and length scale σ = 3 nm, specific interaction strength with cut-off distance r0 = 1.5 nm, and total global sticker concentration 6.64 mM.
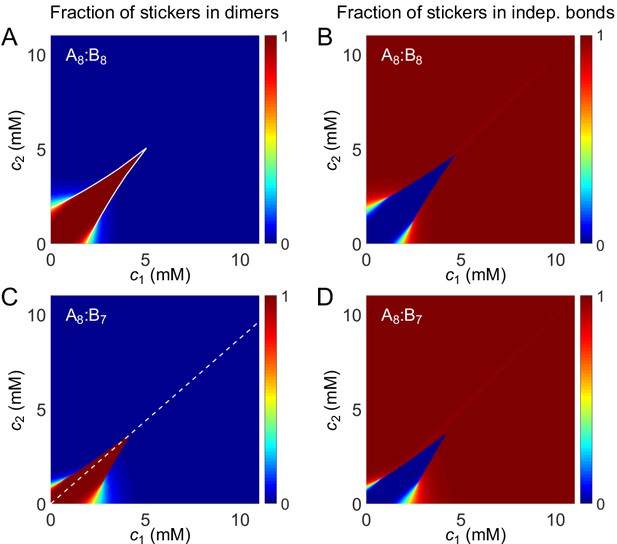
Fraction of stickers in dimers (A) and in independent bonds (B) for A8:B8 system.
White curve in (A) is the transition boundary between dimer- and independent bonds-dominated regions predicted by and . Fraction of stickers in dimers (C) and in independent bonds (D) for A8:B7 system. Dashed white line denotes equal polymer stoichiometry.
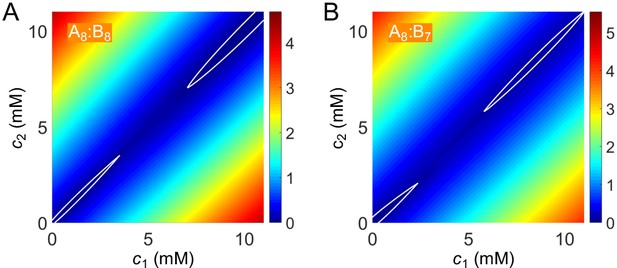
Free-energy density as a function of global sticker concentrations for (A) A8:B8 and (B) A8:B7 systems.
White curves highlight the basins in the dilute dimer-dominated and dense gel-dominated regions.
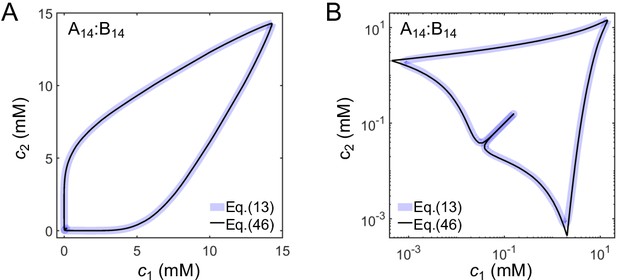
Comparison of phase diagrams derived from the full expression for F (Equation (13)) and the approximate expression (Equation (46)) shown on a (A) linear and (B) log scale for A14:B14 system.
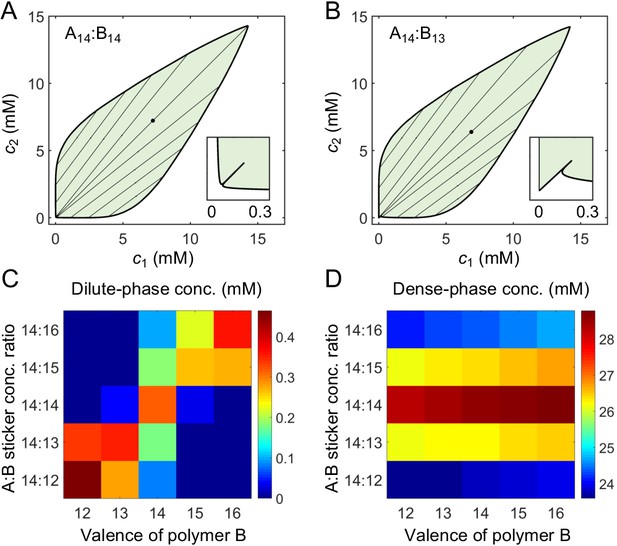
A dimer-gel theory predicts the magic-ratio effect.
Phase diagrams of (A) A14:B14 and (B) A14:B13 systems: one-phase region white, two-phase region green. The dilute- and dense-phase concentrations are connected by representative tie lines. The tie line along the direction of equal polymer stoichiometry is denoted with a black dot. Inset: enlarged dilute-phase boundaries. Sticker concentrations in (C) dilute and (D) dense phases for systems A14:B12-16 at global sticker stoichiometries 14:12-16 and total sticker concentration 6.64 mM. Parameters: , , , and in Appendix 1—table 3.
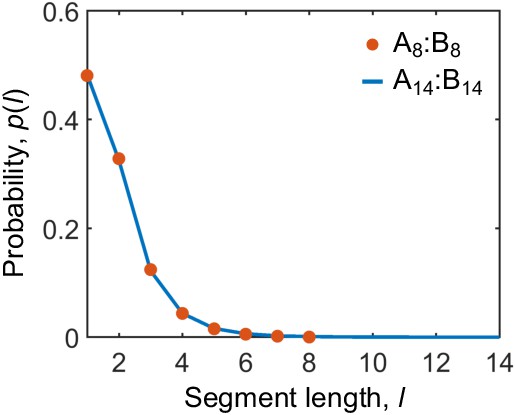
Binding between stickers of different types in the dense phase is correlated.
Probability distribution for finding a bound sticker to be in a consecutively bound segment of length l in the dense phase of A8:B8 and A14:B14 systems at equal stoichiometry.
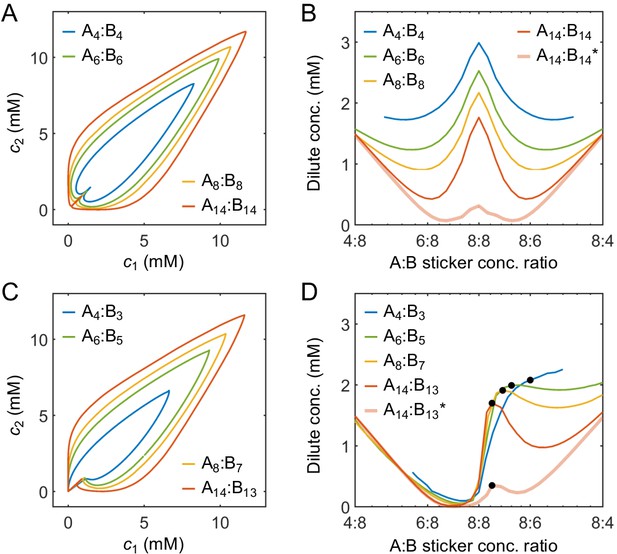
Effects of model parameters on the phase boundaries in the strong-binding regime.
(A) Phase diagrams and (B) dilute-phase concentrations at different global stoichiometries for A4:B4 to A14:B14 systems. (C) Phase diagrams and (D) dilute-phase concentrations at different global stoichiometries for A4:B3 to A14:B13 systems. Black dots indicate equal polymer stoichiometries. Parameters: , values of in Appendix 1—table 3 for binding strength , and for all systems except for A14:B14* and A14:B13* where and . Total global sticker concentration 6.64 mM.
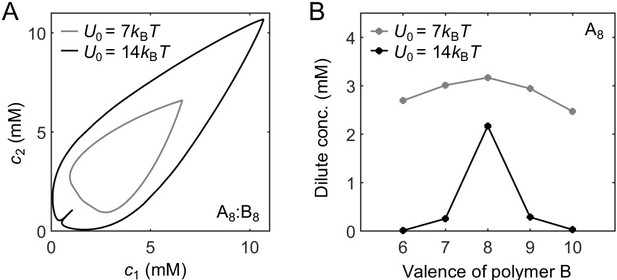
The magic-ratio effect disappears in the weak-binding regime.
(A) Phase diagrams for A8:B8 system with different binding strengths (gray) and (black). (B) Dilute-phase concentrations for A8:B6-10 systems at equal global sticker stoichiometry with different binding strengths (gray) and (black). Parameters: for , for , values of in Appendix 1—table 3, , and . Total global sticker concentration 6.64 mM.
Tables
List of all dilute-phase components and sticker percentages for A14:B14 system at global sticker stoichiometry 1.21.
| Comp | 1A14 | 1A14+1B14 | 2A14+1B14 | 2A14+2B14 | 3A14+2B14 | 4A14+3B14 | 5A14+3B14 |
|---|---|---|---|---|---|---|---|
| frac | 0.10% | 37.0% | 7.67% | 0.38% | 19.1% | 16.6% | 0.38% |
| comp | 5A14+4B14 | 6A14+4B14 | 6A14+5B14 | 7A14+5B14 | 8A14+6B14 | 10A14+7B14 | 17A14+13B14 |
| frac | 10.8% | 0.85% | 0.42% | 0.80% | 3.45% | 0.48% | 1.99% |
Comparison of dilute- and dense-phase concentrations for systems with different total numbers of particles.
| Valence (Stoichiometry) | A14:B14 (14:14) | A14:B13 (14:14) | A14:B13 (14:13) |
|---|---|---|---|
| cdil (mM) for | 0.50 ± 0.06 | 0.08 ± 0.02 | 0.39 ± 0.02 |
| cdil (mM) for | 0.45 ± 0.03 | 0.12 ± 0.02 | 0.34 ± 0.03 |
| cdil (mM) for | 28.0 ± 0.2 | 27.76 ± 0.09 | 25.80 ± 0.08 |
| cden (mM) for | 27.93±0.05 | 27.82±0.06 | 25.82±0.06 |
List of dissociation constants from simulations.
| A1:B1 (t) | A1:B1 (d) | A1:B1 (r) | A2:B2 (d) | A2:B2 (r) | |
|---|---|---|---|---|---|
| 49.6 | * | 48.9 | * | 12.4 | |
| 1.88 | * | 1.90 | * | 0.31 | |
| 5.7e-3 | 5.9e-3 | 5.8e-3 | 7.7e-6 | 7.1e-6 | |
| A4:B3 (r) | A4:B4 (r) | A6:B5 (r) | A6:B6 (r) | ||
| 4.32 | 3.29 | 1.81 | 1.52 | ||
| 4.5e-2 | 2.2e-2 | 4.3e-3 | 2.2e-3 | ||
| 3.77e-9 | 1.38e-11 | 6.95e-15 | 3.00e-17 | ||
| A8:B6 (r) | A8:B7 (r) | A8:B8 (r) | A8:B9 (r) | A8:B10 (r) | |
| 1.15 | 1.01 | 0.90 | 0.79 | 0.73 | |
| 9.0e-4 | 4.5e-4 | 2.5e-4 | 1.5e-4 | 1.0e-4 | |
| 4.22e-18 | 1.26e-20 | 7.07e-23 | 1.77e-23 | 7.59e-24 | |
| A14:B12 (r) | A14:B13 (r) | A14:B14 (r) | A14:B15 (r) | A14:B16 (r) | |
| 0.37 | 0.32 | 0.30 | 0.32 | 0.27 | |
| 1.4e-6 | 6.7e-7 | 3.8e-7 | 2.6e-7 | 1.5e-7 | |
| 2.74e-35 | 1.08e-37 | 8.85e-40 | 1.80e-40 | 5.16e-41 |
-
*footnotetext: theoretical value, direct simulation, and reweighting method.