C. elegans orthologs MUT-7/CeWRN-1 of Werner syndrome protein regulate neuronal plasticity
Figures
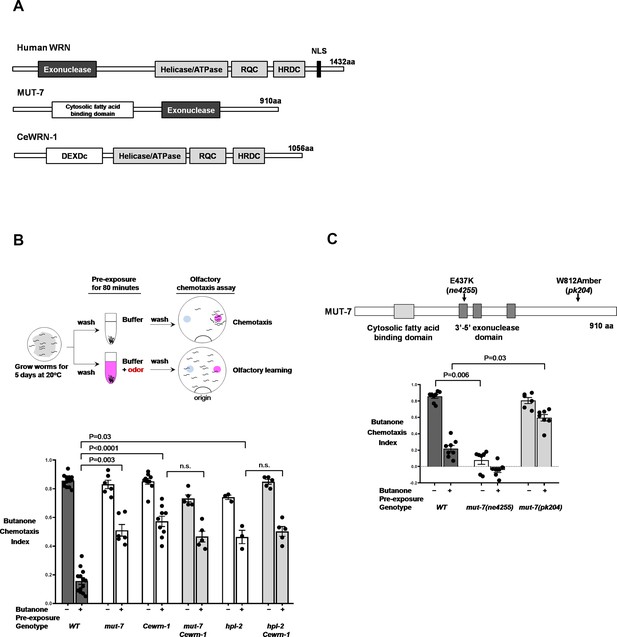
The two C. elegans orthologs of human WRN required for olfactory learning behavior.
(A) Alignment of C. elegans MUT-7 and CeWRN-1 with human WRN. The 3′−5′ exonuclease domain of nematode MUT-7 has been predicted to show 29% amino acid sequence identity to that of human WRN, as indicated by the dark gray regions. The three helicase domains of human WRN (helicase/ATPase, RecQ C-terminal domain [RQC], and helicase-and-RNaseD C-terminal [HRDC] domains) are conserved in nematode CeWRN-1, as indicated by light gray regions. The helicase/ATPase domain shares 43% identity, and the RQC and HRDC domains share 25% identity (Lee et al., 2004). (B) MUT-7 and CeWRN-1 function in AWC neurons to promote butanone-related learning at the time of odor exposure. (Upper) Scheme of olfactory learning. Five-day cultured adult animals were washed to remove bacteria, after which half of the population was pre-exposed to buffer alone (top), and the other half was pre-exposed to buffer with a diluted odor, such as butanone (bottom). After 80 min, the animals were placed at the ‘origin’ of a 9 cm assay plate containing a butanone spot (pink circle) and a control ethanol spot (blue circle). The animals allowed roaming around the dish for 2 hr at 20°C, and their olfactory behavior was quantified with the chemotaxis index (CI). (Bottom) The mean CIs are from the number of animals pre-exposed to buffer (−) or diluted odor (+). More than fifty animals were analyzed per assay. We used GraphPad Prism eight software to perform multiple comparisons and p-values from the two-way ANOVA results are presented for the indicated strains. Error bars represent SEM. (C) The enzymatic activity of the MUT-7 3′−5′ exonuclease affects olfactory behavior. (Upper) Schematic diagram of mut-7 alleles. Two alleles (ne4255 and pk204) are indicated by arrows. (Bottom) The ne4255 allele results in the loss of exonuclease activity and defective chemotaxis, whereas the pk204 allele results in low exonuclease activity and the loss of butanone-related learning, while chemotaxis remains normal. Bars represent mean CIs, error bars represent SEM, and p-values represent two-way ANOVA results obtained by using GraphPad software.
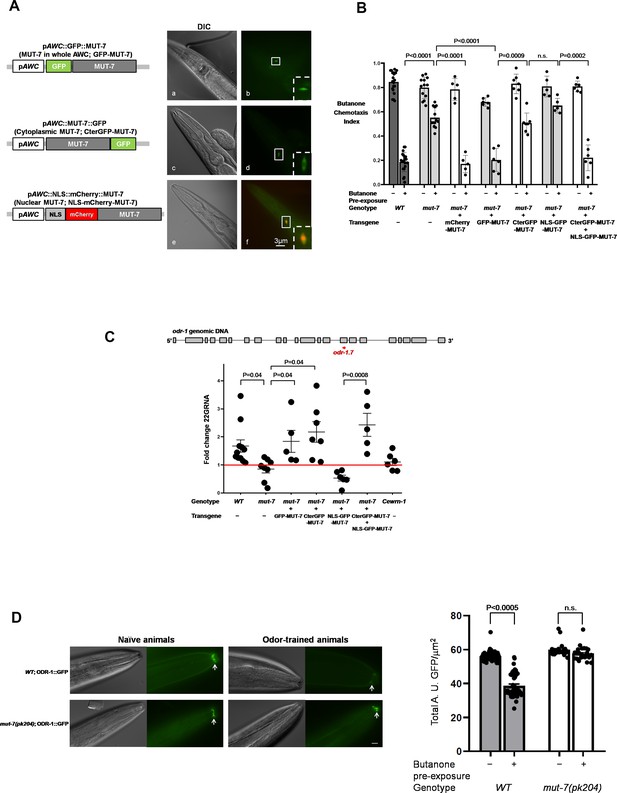
Different intracellular roles of MUT-7 mediate olfactory learning.
(A) Different localizations of MUT-7 in AWCs associated with different positions of the GFP tag. MUT-7 with an upstream (a and b) or downstream (c and d) GFP tag showed localization throughout the soma (b) or in the cytoplasm (d), respectively. A 4XNLS fragment was added upstream of N-terminal mCherry-tagged MUT-7 to cause nuclear accumulation in AWCs (e and f; the background green cytoplasmic signal comes from the coinjection marker pAWC::GFP). All constructs were expressed under an AWC-specific promoter (pAWC). (B) MUT-7 localization in AWCs affects olfactory behaviors. The individual constructs from (A) were introduced into mut-7(pk204) mutants, and the olfactory behavior of the transgenic animals was tested after 80 min of pre-exposure to either buffer alone (−) or diluted butanone (+). All strains analyzed in (B) and (C) were integrated lines obtained by using UV/TMP methods. The p-values come from two-way ANOVA results obtained by comparing the indicated odor-trained populations. n.s. indicates no significant difference. (C) Cytoplasmic MUT-7 is required for the synthesis of odr-1 22G RNAs after prolonged odor stimulation. (Upper) Total RNA was extracted from whole animals and odr-1 22G RNA was quantified by RT-qPCR with an odr-1.7 TaqMan probe. (Bottom) The expression of odr-1 siRNA was normalized to that of odor-insensitive sn2343 RNA, and the fold change between odor-trained and naïve animals of the indicated genotypes was then calculated. The red line indicates no change between odor-trained and naïve populations. The p-values displayed come from the comparison of the fold change between the indicated strains by using one-way ANOVA. (D) Prolonged odor exposure decreases endogenous ODR-1 expression. (Left) A gene encoding ODR-1::GFP under the control of an endogenous promoter was integrated into the worm genome by using a CRISPR-based method. GFP was observed in the flattened ciliated end of the AWC neuron indicated by the white arrows. All images were captured using an upright microscope (Leica DM6B) at 63X magnification. In odor-trained wild-type animals, ODR-1 expression decreased the fluorescence intensity by 30% compared to that in naïve wild-type animals. The fluorescence intensity in mut-7(pk204) mutant animals was not significantly different between the naïve and odor-trained populations. The fluorescence intensity in the naïve and odor-trained animals was quantified as shown in the right panel. The p-value comes from the comparison of fluorescence intensity between naïve and odor-trained worms by using two-way ANOVA. Error bars represent SEM, and n.s. indicates no significant difference.
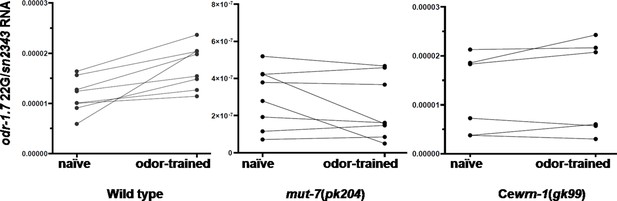
Pairwise comparison of odr-1 22G RNA levels.
The expression of odr-1 22G RNA in naïve and odor-trained populations was quantified by RT-qPCR by using the odr-1.7 probe. Each RT-qPCR result was normalized to the endogenous sn2343 gene, whose levels did not change with odor treatment. A line links the 22G values of each paired population (naïve vs. odor-trained) that was grown at the same time on the same plates. According to the scale on the y-axis, odr-1 22G RNA in wild-type and Cewrn-1(gk99) mutant animals showed higher expression levels (between 5 × 10−4 and 2 × 10−5) because total RNA was extracted from whole worms and some germ cells expressed odr-1 siRNAs (Gu et al., 2009). By contrast, odr-1 22G RNA in mut-7(pk204) showed very low expression (between 5 × 10−6 and 6 × 10−7).
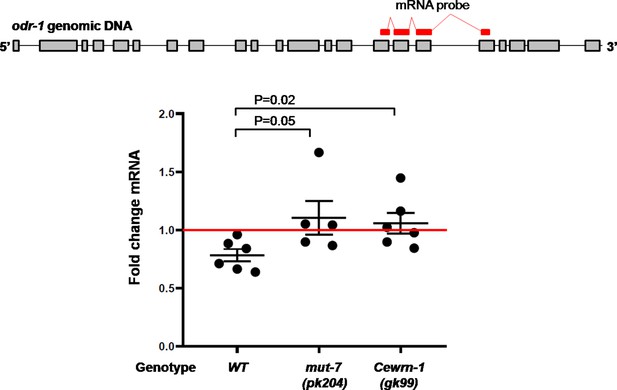
Prolonged odor exposure decreases odr-1 mRNA expression.
The expression of odr-1 mRNA in naïve and odor-trained worms was quantified by SYBR Green based quantitative PCR. The fold change is the ratio calculated between the mRNA level in odor-trained worms and the mRNA level in naïve worms. p-values are from t-test results obtained by comparing the indicated strains.
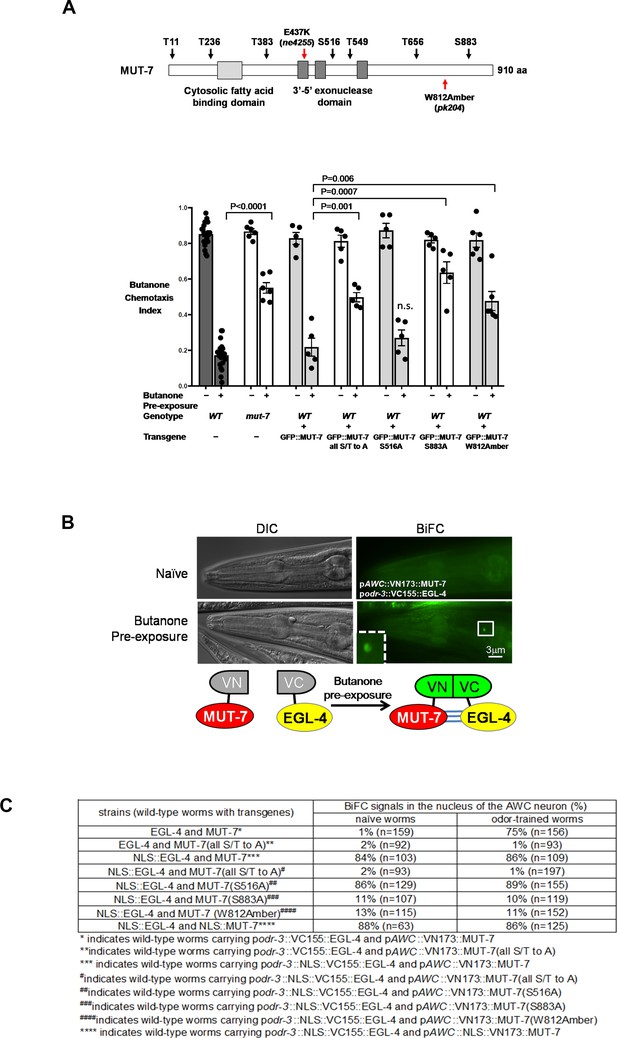
MUT-7 is phosphorylated by EGL-4 in the nucleus after prolonged odor exposure.
(A) Schematic representation of the seven predicted phosphorylation sites of PKG in MUT-7 in the top panel. Different point mutations were introduced by site-directed mutagenesis, and the constructs were transferred to wild-type animals to generate dominant-negative strains for behavioral assays. The p-value comes from the results of two-way ANOVA for the comparison of the indicated strains. n.s. indicates no significant difference between the wild type and the indicated mutant animals. (B) The BiFC assay revealed an in vivo interaction between EGL-4 and MUT-7 in the AWC nucleus. (Upper) BiFC florescence signals were observed in odor-trained worms (bottom) but not in naïve worms (top). (Bottom) Schematic representation of the BiFC constructs. (C) The BiFC screen showed the critical residues within MUT-7 for the specific interaction with nuclear EGL-4. The BiFC screen revealed that the specific interaction between nuclear MUT-7 and EGL-4 was consistent with the adaptation results shown in (B). Since nuclear EGL-4 induced adaptation of the odor-seeking behavioral response in naive worms, the percentage of BiFC signals in naïve worms expressing NLS::EGL-4 with different versions of MUT-7 is similar to that in the odor-trained worms.
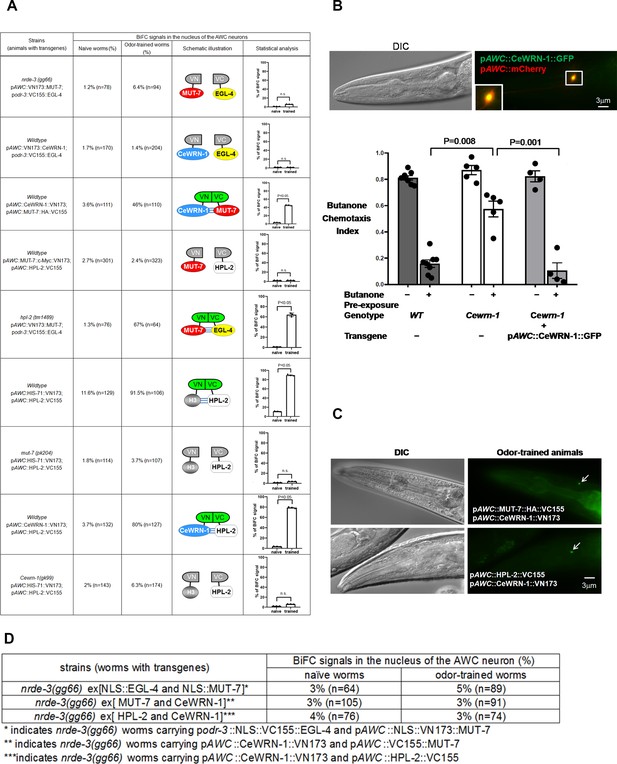
CeWRN-1 function in the AWC nucleus is required for olfactory learning.
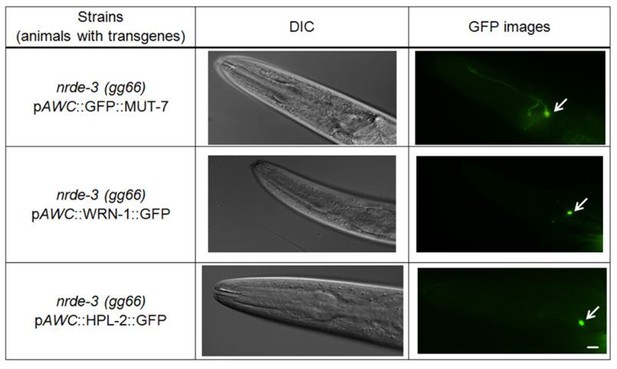
(A) The BiFC screen showed in vivo specific protein interactions in the AWC nucleus after prolonged odor exposure. The BiFC fluorescent signals were scored in naïve and odor-trained worms. Each strain was examined in three separate experiments, and all the data were statistically analyzed by two-way ANOVA. (B) (Upper) CeWRN-1 is expressed in the AWC nucleus. Fluorescent images of wild-type animals expressing Cewrn-1 cDNA with a GFP tag at the C-terminus showed protein accumulation in the nucleus. (Bottom) The expression of CeWRN-1 in AWC rescues the learning defects of Cewrn-1(gk99) mutant animals. GFP-tagged CeWRN-1 was introduced into the Cewrn-1(gk99) null mutant and restored olfactory learning ability. The p-values come from two-way ANOVAs between the specified groups. (C) CeWRN-1 interacts with MUT-7 and HPL-2. An influenza hemagglutinin (HA) peptide was inserted between MUT-7 and GFP, and fluorescent images showed that MUT-7 was distributed throughout the AWC cell body, as shown in Figure 4—figure supplement 3. Thus, the same strategy was applied to generate the pAWC::MUT-7::HA::VC155 construct (top) in the BiFC assay. (D) NRDE-3 is required for the MUT-7 and EGL-4 interaction, the CeWRN-1 and HPL-2 interaction, and the MUT-7 and CeWRN-1 interaction in the AWC nucleus. We expressed the indicated BiFC constructs in nrde-3(gg66) mutants and scored the BiFC signals in naïve and odor-trained worms.
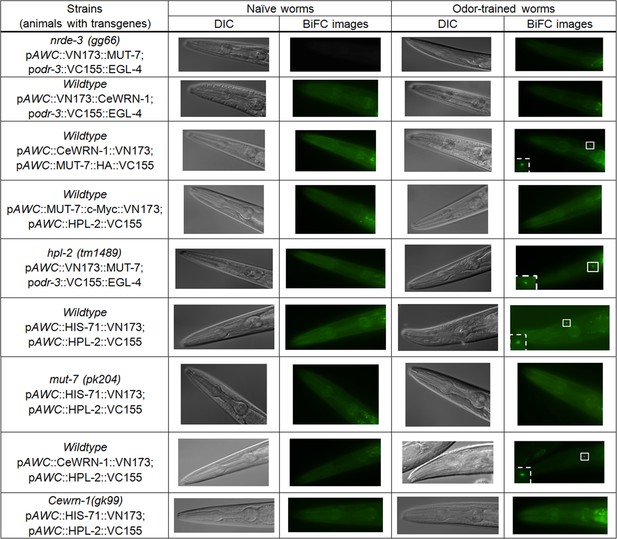
Images of BiFC screening results.
All representative fluorescent images related to Figure 4A from naïve and odor-trained worms are shown.
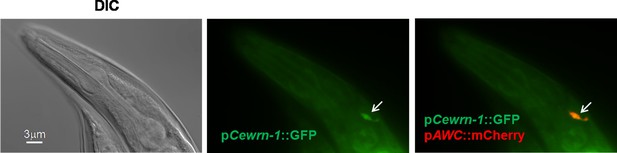
Localization of CeWRN-1 in AWC neurons.
Fluorescent images of wild-type animals expressing GFP under a putative 2.5 kb Cewrn-1 promoter showed the same localization obtained with the AWC promoter-driving mCherry.
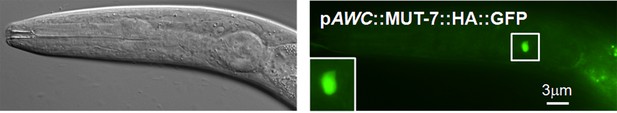
Expression of MUT-7::HA::GFP in the whole cell body of the AWC neuron.
An influenza hemagglutinin (HA) peptide was inserted between MUT-7 and GFP in the CterGFP-MUT-7 construct. This construct was introduced into wild-type animals, and GFP was observed in the whole AWC soma region.
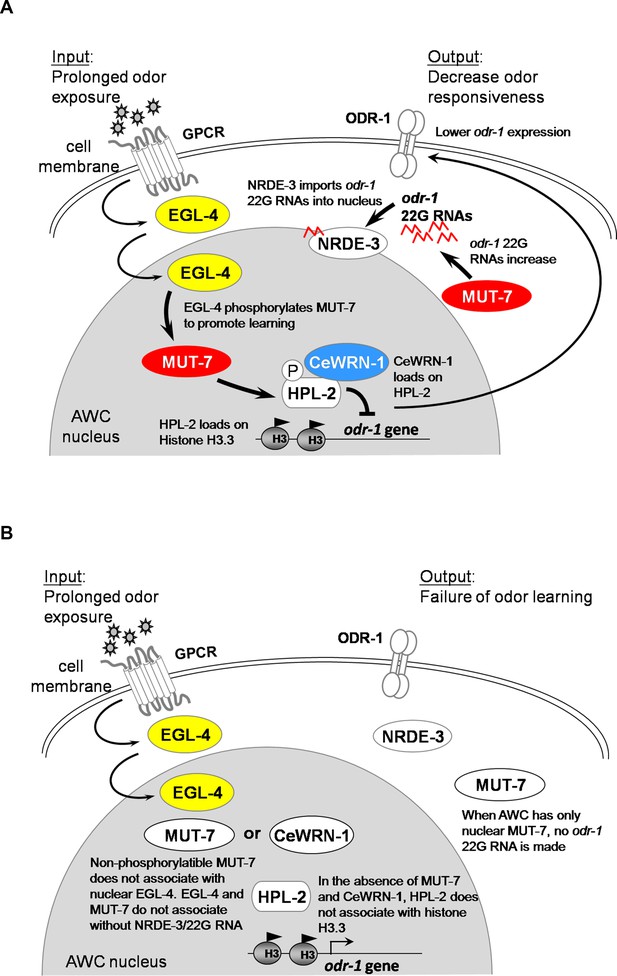
Models of the involvement of MUT-7 and CeWRN-1 in long-term olfactory learning in AWC.
(A) In wild-type AWC neurons, prolonged odor exposure causes EGL-4 to enter the nucleus. odr-1 22G RNAs are generated by cytoplasmic MUT-7 and shuttled into the nucleus by NRDE-3. Once the small RNA enters the nucleus, EGL-4 phosphorylates nuclear MUT-7, which directs the association between CeWRN-1 and HPL-2. HPL-2 then associates with methylated H3.3, thereby downregulating odr-1 transcription. Lower levels of the ODR-1 protein are predicted to promote olfactory learning. (B) Loss of MUT-7 from the cytoplasm inhibits odr-1 22G RNA production. Although EGL-4 accumulates in the nucleus, the lack of induced small RNA import leads to the disruption of the MUT-7 and EGL-4 interaction, the MUT-7 and CeWRN-1 interaction, and the CeWRN-1 and HPL-2 interaction. The lack of these associations affects the odr-1 mRNA-downregulating signal because of the failure of HPL-2 loading on histones. The continued expression of ODR-1 results in defects in olfactory learning. Supplemental Information titles and legends.
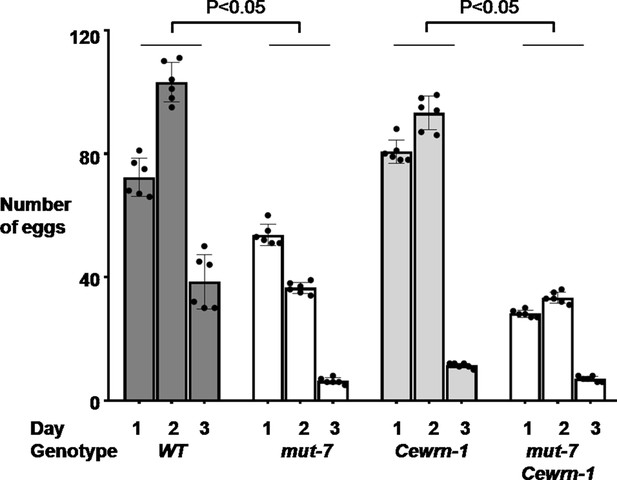
Loss of MUT-7 significantly decreases brood size.
Animals were synchronized in the larva one stage and seeded on NGM culture plates. The number of eggs was scored from the young adult stage onward every 24 hr for 3 days. A t-test was used to compare total eggs between the indicated strains. Error bars represent SEM.
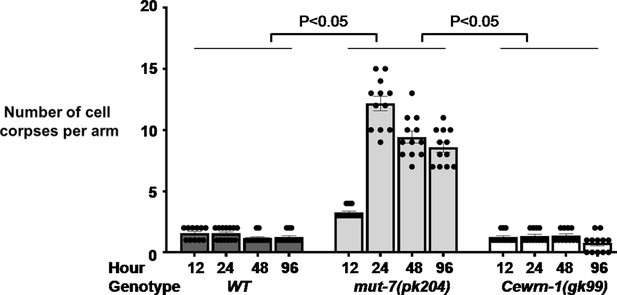
Loss of MUT-7 significantly affects germline cell death.
Oocyte development was visualized from the young adult stage onward at 12, 24, 48, and 96 hr. The t-test was used to compare the total numbers of cell corpses per gonad arm between the indicated strains. Error bars represent SEM.
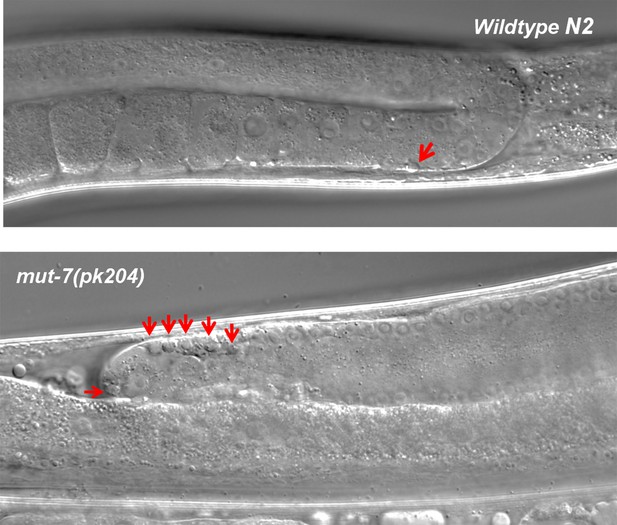
Images of cell corpses in wild-type and mut-7(pk204) animals.
Cell corpses in each strain are indicated by red arrows.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Caenorhabditis elegans) | N2 | Caenorhabditis Genetics Center | RRID:WB-STRAIN:WBStrain00000001 | Genotype: wild type |
| Strain, strain background (C. elegans) | Cewrn-1(gk99) | Caenorhabditis Genetics Center | RRID:WB-STRAIN:WBStrain 00035559 | Genotype: wrn-1(gk99) II |
| Strain, strain background (C. elegans) | hpl-2(tm1489) | Caenorhabditis Genetics Center | RRID:WB-STRAIN:WBStrain00030670 | Genotype: hpl-2(tm1489) III |
| Strain, strain background (C. elegans) | mut-7(ne4255) | Caenorhabditis Genetics Center | RRID:WB-STRAIN:WBStrain00040468 | Genotype: mut-7(ne4255) III |
| Strain, strain background (C. elegans) | mut-7(pk204) | Caenorhabditis Genetics Center | RRID:WB-STRAIN:WBStrain00028942 | Genotype: mut-7(pk204) III |
| Strain, strain background (C. elegans) | nrde-3(gg66) | Caenorhabditis Genetics Center | RRID:WB-STRAIN:WBStrain00040756 | Genotype: nrde-3(gg66) X |
| Recombinant DNA reagent | pceh-36prom3-pPD95.75 vector | Oliver Hobert Lab | AWC-specific promoter ceh-36prom3 | |
| Recombinant DNA reagent | pCewrn-1::GFP | This study | Bi-Tzen Juang Lab | 2.5 kb upstream of the Cewrn-1 start site of translation |
| Recombinant DNA reagent | pAWC::GFP::MUT-7 | Noelle D. L'Etoile Lab Juang et al., 2013 | ||
| Recombinant DNA reagent | pAWC::4XNLS::mCherry::MUT-7 | This study | Bi-Tzen Juang Lab | 4XNLS::mCherry obtained from pGC302 (Addgene) |
| Recombinant DNA reagent | pAWC::4XNLS::GFP::MUT-7 | This study | Bi-Tzen Juang Lab | 4XNLS::GFP obtained from pGC240 (Addgene) |
| Recombinant DNA reagent | pAWC::MUT-7::GFP | This study | Bi-Tzen Juang Lab | MUT-7 was amplified from yk443 plasmid |
| Recombinant DNA reagent | pAWC::CeWRN-1::GFP | This study | Bi-Tzen Juang Lab | Cewrn-1 cDNA from yk1276 plasmid |
| Recombinant DNA reagent | ODR-1::GFP | This study | Noelle D. L'Etoile Lab | AID::3xFLAG, 1 kb downstream of odr-1.b stop codon |
| Recombinant DNA reagent | pAWC::GFP::MUT-7(W812Amber) | This study | Bi-Tzen Juang Lab | Site-directed mutagenesis reaction Technologies |
| Recombinant DNA reagent | podr-3::NLS::VC155::EGL-4 | This study | Bi-Tzen Juang Lab | Add NLS sequence and BiFC analysis |
| Recombinant DNA reagent | podr-3::VC155::EGL-4. | This study | Bi-Tzen Juang Lab | BiFC analysis |
| Recombinant DNA reagent | pAWC::VN173::MUT-7 | This study | Bi-Tzen Juang Lab | BiFC analysis |
| Recombinant DNA reagent | pAWC::NLS::VN173::MUT-7 | This study | Bi-Tzen Juang Lab | Add NLS sequence and BiFC analysis |
| Recombinant DNA reagent | pAWC::VN173::MUT-7 | This study | Bi-Tzen Juang Lab | BiFC analysis |
| Recombinant DNA reagent | pAWC::MUT-7::HA::VC155 | This study | Bi-Tzen Juang Lab | Add HA taq |
| Recombinant DNA reagent | pAWC::MUT-7::c-Myc::VN173 | This study | Bi-Tzen Juang Lab | Add c-Myc taq |
| Recombinant DNA reagent | pAWC::HPL-2::VC155 | Noelle D. L'Etoile Lab | Juang et al., 2013 | |
| Recombinant DNA reagent | pAWC::HIS-71::VN173 | Noelle D. L'Etoile Lab | Juang et al., 2013 | |
| Recombinant DNA reagent | pAWC::CeWRN-1::VN173 | This study | Bi-Tzen Juang Lab | BiFC analysis |
| Commercial kit | QuikChange Lightning Site-Directed Mutagenesis Kit | Agilent Technologies | Agilent:210518 | |
| Commercial kit | TURBO DNA-free Kit | Thermo Fisher: Invitrogen | Catalog number::AM1907 | |
| Commercial kit | Multiscribe Reverse Transcriptase | Thermo Fisher: Applied Biosystems | Catalog number: 4311235 | |
| Commercial kit | TaqMan Universal PCR Master Mix | Thermo Fisher: Applied Biosystems | Catalog number: 4326708 | |
| Commercial kit | iScrpt cDNA Synthesis Kit | Bio-Rad | Catalog number: 1708890 | |
| Commercial kit | iTaq Universal SYBR Green Supermix | Bio-Rad | Catalog number: 1725120 | |
| Software, algorithm | GraphPad | Prism | GraphPad Prism 8.0.1 | |
| Software, algorithm | MetaMorph | Molecular Devices | Version 7.8.13.0 |
| Strains (worms with transgenes) | BiFC signals in the nucleus of the AWC neuron (%) | |
|---|---|---|
| naïve worms | odor trained worms | |
| Wildtype ex[CeWRN-1 and MUT-7(all S/T to A)]* | 3% (n=68) | 4% (n=81) |
Additional files
-
Supplementary file 1
Asymmetric expression of str-2 in AWC neurons.
The cell fate of the AWC neuron was examined by quantifying the asymmetric expression of pstr-2::DsRed. Animals were scored in three categories according to pstr-2::DsRed expression either in AWC (0AWCpstr-2 ON), in only one AWC (1AWCpstr-2 ON), or in both AWCs (2AWCpstr-2 ON).
- https://cdn.elifesciences.org/articles/62449/elife-62449-supp1-v2.docx
-
Supplementary file 2
Nuclear accumulation of EGL-4 after prolonged butanone exposure.
Integrated GFP-tagged EGL-4 (termed pyIs500) was expressed in wild-type, hpl-2(tm1489), and Cewrn-1(gk99) animals. The nuclear expression of EGL-4 was scored in naïve and odor-trained animals.
- https://cdn.elifesciences.org/articles/62449/elife-62449-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62449/elife-62449-transrepform-v2.docx