Identification of host proteins differentially associated with HIV-1 RNA splice variants
Figures
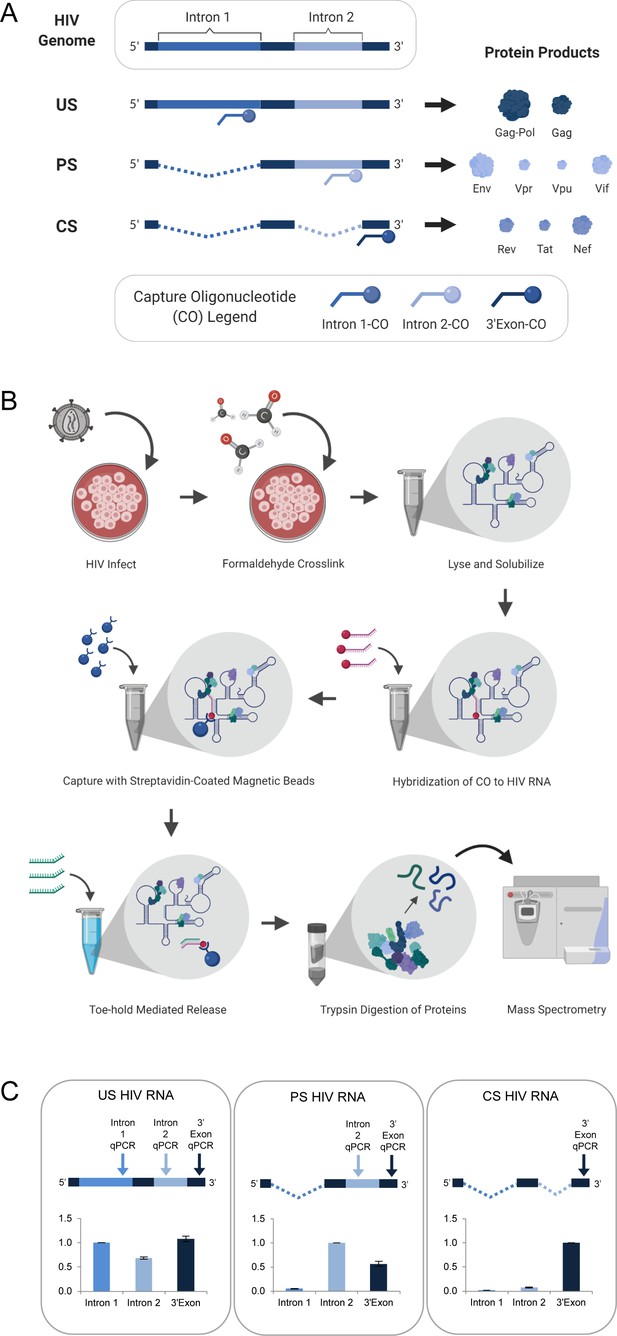
HyPR-MS for purification of HIV splice variant interactomes.
(A) Capture oligonucleotides (COs) were designed to complement specific regions of the HIV genome to make possible the isolation of the three HIV splice variant classes from a single- cell lysate. (B) Overview of Hybridization Purification of RNA-Protein Complexes Followed by Mass Spectrometry (HyPR-MSSV) procedure. (C) Purification of the HIV splice variant classes was verified using RT-qPCR assays specific to regions in intron 1, intron 2, and 3’-exon. The intensity data is normalized to the intron 1 assay for unspliced (US) capture, the intron 2 assay for partially spliced (PS) capture, and the 3’-exon assay for completely spliced (CS) capture. Error bars are the standard deviation for three biological replicates. Figures A and B created with BioRender.com.
-
Figure 1—source data 1
qPCR data for (A) capture specificity, (B) enrichment, and (C) capture efficiency calculations.
Related to Figure 1 and Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/62470/elife-62470-fig1-data1-v1.docx
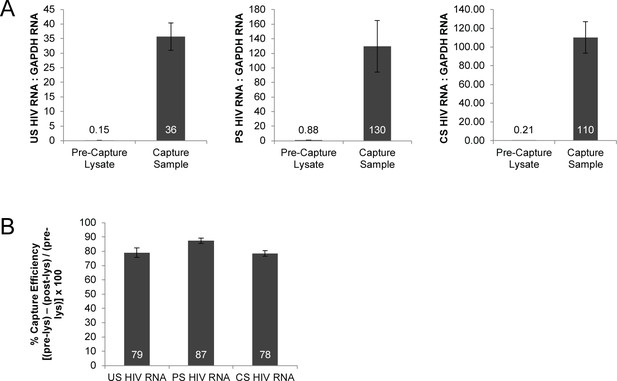
RT-qPCR confirmation of enrichment for HIV splice variants and efficiency of capture.
(A) Fold enrichment of each HIV splice variant class after capture was calculated using RT-qPCR assays to measure the ratio of HIV RNA: GAPDH RNA in the pre-capture, lysate sample, and the capture sample. The ratios are quantified in the charts and fold enrichment is (capture sample ratio/pre-capture ratio). Data in Supplementary file 1 and Figure 1—source data 1. (B) Capture efficiency calculated by measuring the amount of each HIV splice variant class in the lysate before capture (pre-lys) and in the lysate after the capture is completed (post-lys). (Pre-lys)-(Post-lys)/(Pre-lys) × 100 = % capture efficiency. Related to Figure 1C and data in Supplementary file 1 and Figure 1—source data 1.
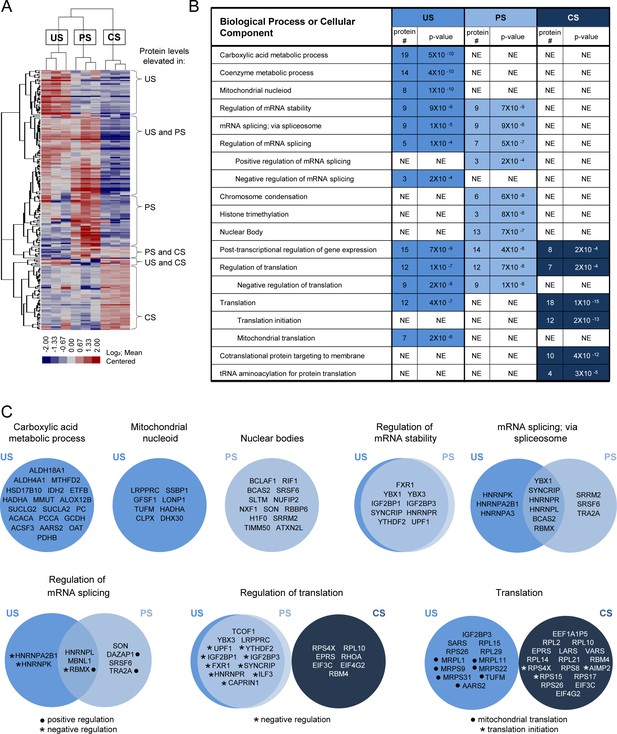
Determination and analysis of HIV splice variant protein interactomes.
(A) Heatmap depicts relative intensities for each of the 212 proteins (rows) in each of the three biological replicates of the unspliced (US), partially spliced (PS), and completely spliced (CS) (columns) differential interactomes. (B) Condensed list of gene ontology (GO) biological process or cellular component terms enriched in each of the HIV splice variant interactomes. The ‘protein #” column indicates the number of proteins in the interactome that are annotated with the biological process indicated. The ‘p-value’ column indicates the likelihood that the proteins of the biological process are present in each interactome by random chance and were provided by GO term enrichment software (Mi et al., 2017). A lower p-value suggests non-random over-representation of a biological process. ‘NE’ = not enriched. (C) Venn diagrams of proteins annotated for biological processes or cellular components enriched in the splice variant differential interactomes.
-
Figure 2—source data 1
Pairwise comparisons of mass spectrometric data for US, PS, and CS RNA interactomes.
- https://cdn.elifesciences.org/articles/62470/elife-62470-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Gene ontology term enrichment analysis.
- https://cdn.elifesciences.org/articles/62470/elife-62470-fig2-data2-v1.xlsx
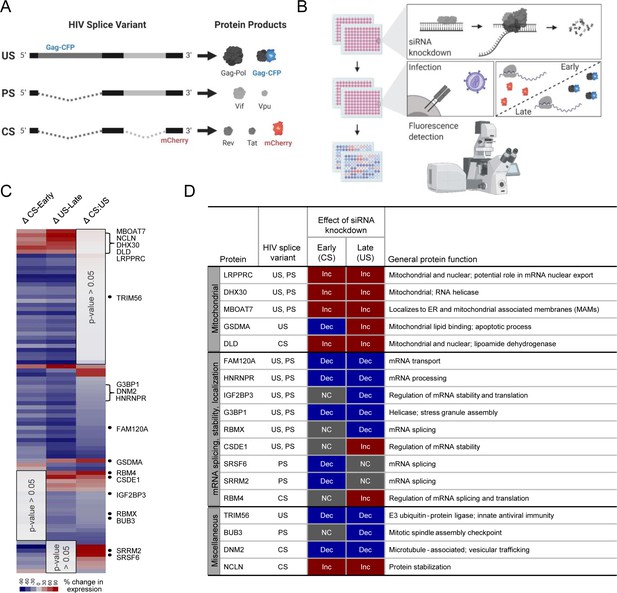
Screen for host protein effects on early and late HIV gene expression.
(A) The HIV-1 reporter virus expresses mCherry from the nef locus as a completely spliced (CS) RNA ‘early’ stage reporter and Gag fused to cyan fluorescent protein (CFP) as a unspliced (US) RNA ‘late’ stage reporter. (B) In 96-well plates, 293 T-cells engineered to stably express an F-actin-YFP fusion protein as a host gene control (293T-ACT-YFP cells) were transfected with gene-specific siRNAs, incubated for 48 hr to initiate knockdown (KD), and then transfected with siRNAs for a second time prior to infection with the HIV reporter virus at a multiplicity of infection (MOI) of ~1. Cells were fixed at 48 hr post-incubation. Fluorescence microscopy was used to quantify CFP and mCherry levels. (C) Heatmap of HIV gene expression changes after siRNA KD of host proteins, with 84 of 121 proteins showing statistically significant changes in early and/or late HIV gene expression (p-values<0.05). (D) Table summarizes Hybridization Purification of RNA-Protein Complexes Followed by Mass Spectrometry (HyPR-MS) and siRNA KD results for 18 genes of interest wherein KD was confirmed to be significant based on either quantitative immunoblot or immunofluorescence. Figures A and B created with BioRender.com.
-
Figure 3—source data 1
siRNA KD and fluorescence expression data analysis.
- https://cdn.elifesciences.org/articles/62470/elife-62470-fig3-data1-v1.xlsx
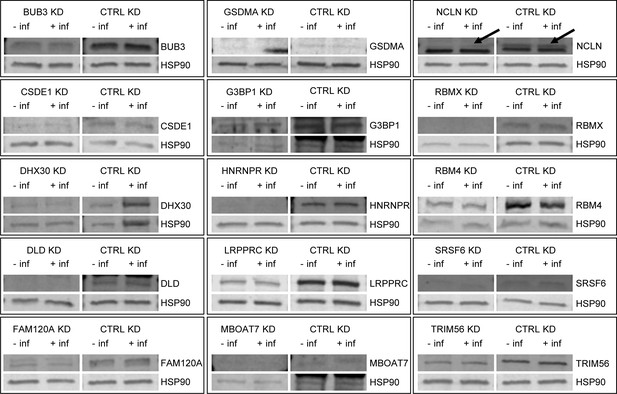
Western blots confirm efficacy of siRNA knockdown strategy.
Commercially available antibodies against the 20 host proteins were obtained to detect each protein in cells with and without siRNA knockdown of that protein. Cells were transfected with the appropriate siRNAs, infected with the HIV reporter virus previously described, then lysed at 48 hr post-incubation with the virus. The protein bands at the expected molecular weights were quantified for siRNA treated cells and untreated cells and the efficiency of protein knockdown calculated (Supplementary file 6). Of the 20 host proteins targeted, two (SRRM2, MW 300 kDa and DYNC1H1, MW 530 kDa) did not produce quantifiable results, likely due to molecular weight limitations of the western blot parameters, and three (DNM2, IGF2BP3, RPL15) did not produce data to indicate successful knockdown of the proteins. Related to Supplementary files 5 and 6.
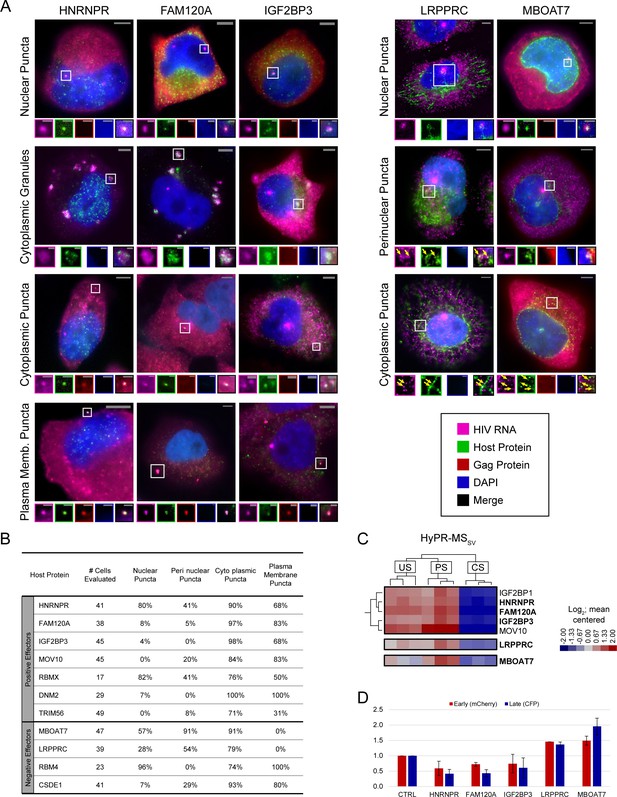
Unspliced (US) HIV RNA co-localizes with positive and negative effectors at multiple sites within the cell.
(A) Representative images of co-localization phenotypes observed using fluorescence in situ hybridization/immunofluorescence (FISH/IF). For each, a merged image of a cell highlighting a site of co-localization (white square) is shown. Enlarged regions of interest (ROIs) of each fluorescence channel are displayed in the associated small panels to separate overlapping US HIV RNA, host protein, HIV Gag polyprotein, and DAPI signals. Some images were obtained from experimental replicates that did not include Gag IF and therefore do not include images from the corresponding channel. Note: Brightness and contrast settings were adjusted individually for each color channel of the images to effectively show co-localization. These settings may be different for the ROIs. (B) Table showing the frequency of observing a particular co-localization phenotype of US HIV RNA with each of 11 host proteins. Frequencies are displayed as the percentage of cells observed with each co-localization phenotype. (C) ROIs from the Hybridization Purification of RNA-Protein Complexes Followed by Mass Spectrometry (HyPR-MS), hierarchically clustered heatmap (Figure 2A) showing the close relation of HNRNPR, FAM120A, and IGF2BP3 interaction profiles, each preferentially interacted with US and partially spliced (PS) HIV RNA. (D) Data for proteins of interest from the siRNA knockdown (KD) screen (Figure 3—source data 1). HIV gene expression decreases for HNRNPR, FAM120A, and IGF2BP3 upon KD with a greater decrease in late gene expression than in early. For mitochondria-related proteins LRPPRC and MBOAT7, HIV gene expression increases upon KD of the host protein.
-
Figure 4—source data 1
Quantitation of host protein and HIV RNA co-localization phenotypes.
- https://cdn.elifesciences.org/articles/62470/elife-62470-fig4-data1-v1.xlsx
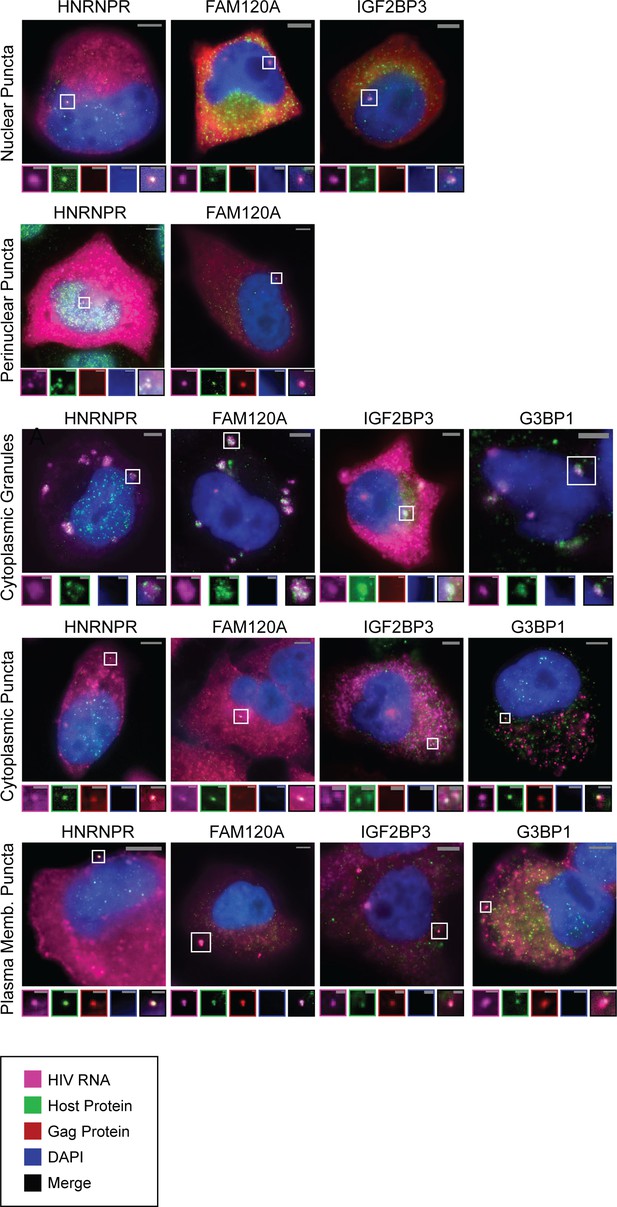
Representative images of host proteins co-localizing with unspliced (US) HIV RNA.
Analysis of host proteins (HNRNPR, FAM120A, IGF2BP3) that co-localize with US HIV RNA at cytoplasmic granules. HNRNPR primarily localizes to the nucleus while FAM120A and IGF2BP3 primarily localize to the cytoplasm. All three were observed to co-localize to transcription sites; however, only HNRNPR did so frequently (80% vs 8% and 4%, respectively; Figure 4B). All three co-localize with HIV RNA to cytoplasmic granules, cytoplasmic puncta, and plasma membrane puncta. We assayed, G3BP1, a known component of stress granules, and found it also co-localized with US RNA in these three cytoplasmic locations. An additional image of a large cytoplasmic granule is included for G3BP1 showing a spiral-like phenotype of small puncta around the granule at which US HIV RNA and G3BP1 also co-localize.
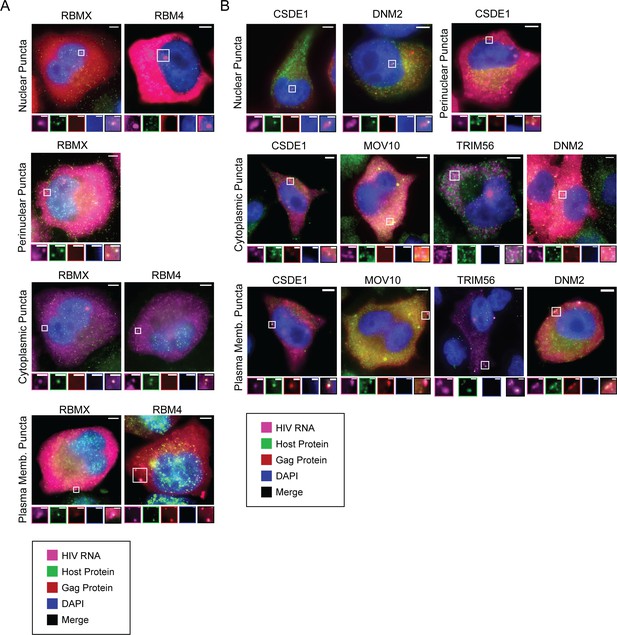
Representative images of host proteins co-localizing with unspliced (US) HIV RNA.
(A) Two host proteins (RBMX and RBM4), in addition to HNRNPR discussed above, primarily localize to the nucleus and are observed to co-localize to nuclear puncta frequently (82% and 96%, respectively). The image showing RBM4 (negative effector) co-localization with US HIV RNA at nuclear puncta is enlarged to show co-localization at small puncta near the putative HIV transcription site. (B) Proteins (CSDE1, MOV10, TRIM56, DNM2) primarily localize to the cytoplasm and frequently co-localize with US HIV RNA at cytoplasmic puncta (>71%) and plasma membrane puncta (>31%).
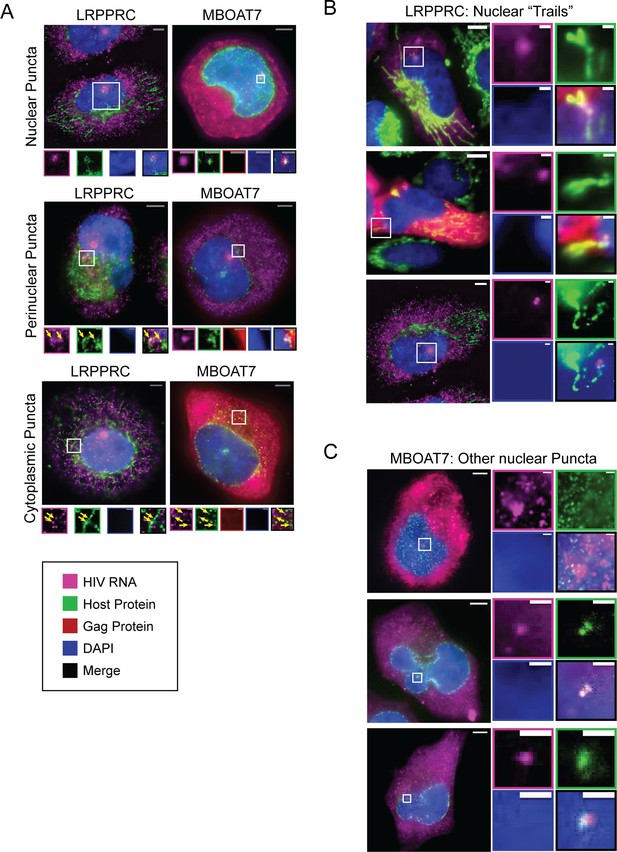
Representative images of host proteins co-localizing with unspliced (US) HIV RNA.
(A) Analysis of host proteins (LRPPRC and MBOAT7) with general localization to the nuclear membrane or the cytoplasm and co-localization at nuclear puncta, puncta proximal to the nuclear membrane, and cytoplasmic puncta. Both are negative effectors of HIV gene expression. LRPPRC trailed from the nuclear perimeter to the putative transcription sites in 28% of cells evaluated. MBOAT7 was at putative transcription sites but also, less frequently, associated with smaller US HIV RNA puncta in the nucleus, shown in the right-hand frame. (B) Additional examples of the LRPPRC nuclear co-localization phenotype as described in D. Images in E were prepared from merged 3–4 z-stacks for each cell. (C) Top: image shows multiple peri-nuclear co-localizations of MBOAT7 and US HIV RNA on a z-plane that runs approximately parallel to the nucleus perimeter. Middle and bottom: images zoom in on co-localization of MBOAT7 and US HIV RNA at small puncta in the nucleus. The US HIV RNA puncta here are likely not transcription sites because the intensity of fluorescence in situ hybridization (FISH) is very low. Note: Brightness and contrast settings were adjusted individually for each color channel of the images to effectively show co-localization. These settings may be different for the regions of interest (ROIs). Related to Figure 3 and Figure 4—source data 1.
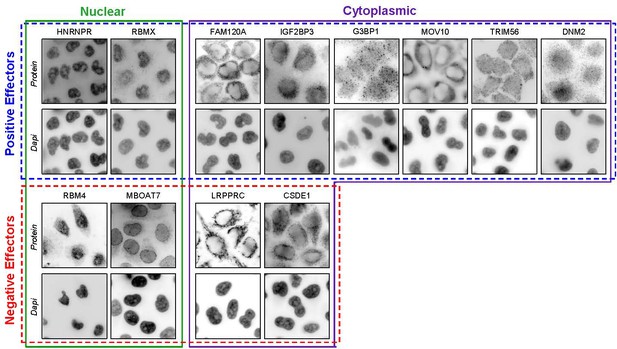
Representative images of host protein general cellular localization.
Uninfected HeLa cells were treated with host protein specific antibodies and DAPI for immunofluorescence detection. Host protein images are organized by general primary cellular localization and the siRNA knockdown determined effect that host protein has on HIV gene expression. MBOAT7 has a prominent peri-nuclear localization that has not been definitively shown to be inside or outside of the nucleus. This protein appears to localize in both the nucleus and cytoplasm but is grouped as a nuclear protein here.
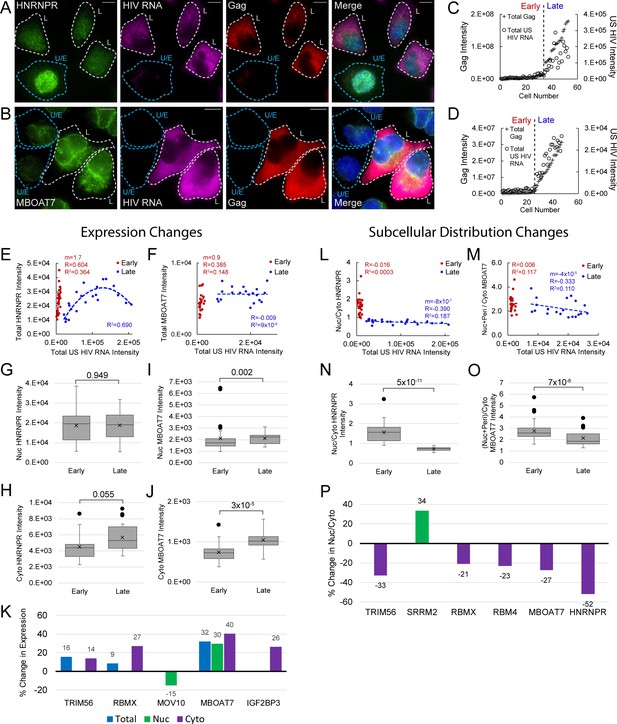
Host protein expression and cellular distribution.
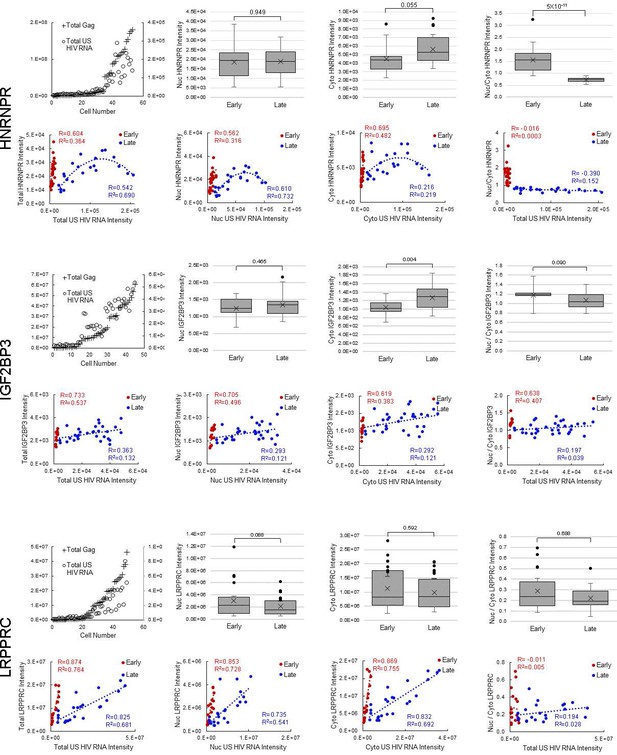
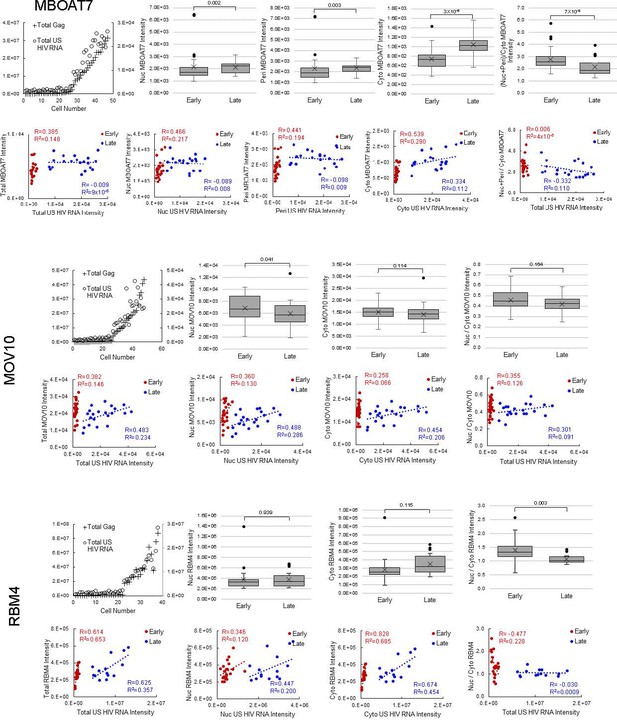
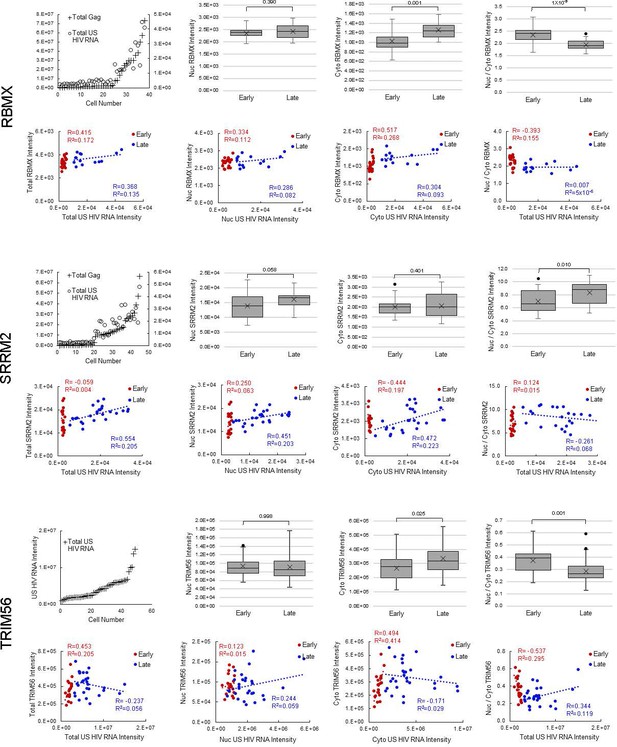
(A) HNRNPR cellular distribution appears to be different in uninfected/early stage infected cells (U/E; blue outlines) than in late stage infected cells (L; white outlines). (B) MBOAT7 expression appears greater in late stage infected cells than in uninfected/early stage infected cells. (C–D) Plots of cellular Gag-immunofluorescence (IF) and unspliced (US) HIV RNA-fluorescence in situ hybridization (FISH) intensities for cells analyzed for HNRNPR (C) and MBOAT7 (D). Cells prior to the inflection points in each plot are termed ‘early cells’ as they are either uninfected or at stages of HIV replication prior to late gene expression (i.e., high amounts of Gag in the cytoplasm). Cells after the inflection point are termed ‘late cells’ as they express high, IF-detectable levels of Gag in the cytoplasm. (E–F) For each cell in the early and late cell sub-groups, the intensity of US HIV RNA vs the intensity of HNRNPR (E) and MBOAT7 (F) is plotted. Linear or polynomial regressions (R2) are fit to each early and late sub-group and the Pearson’s correlation coefficient (R) calculated for linear regressions. This demonstrates the extent of correlation between US HIV RNA expression and the expression of each host protein. (G–J) A Student’s t-test is applied to determine if the host protein intensities in early cells are significantly different from those in late cells in the nucleus and the cytoplasm. (K) The percent change in the median expression for all host proteins with early vs late p-values<0.05. Calculations were made for total cell, nuclear, and cytoplasmic differences. (L–M) Total cellular US HIV RNA intensities vs host protein nuclear to cytoplasmic (nuc/cyto) or nuclear plus perinuclear (nuc+peri)/cyto ratios. This demonstrates the extent of correlation of host protein cellular distribution with US HIV RNA expression. (N–O) A Student’s t-test measures significant differences in the cellular distribution between the early and late cells for HNRNPR and MBOAT7. (P) The percent change in the median nuc/cyto or nuc+peri/cyto ratio for all host proteins with p-values<0.05. Purple indicates the late cells have a higher proportion of the host protein in the cytoplasm than do the early cells. Green indicates the late cells have a higher proportion in the nucleus.
-
Figure 5—source data 1
Quantitation of expression and distribution changes of host proteins in early and late HIV infection.
- https://cdn.elifesciences.org/articles/62470/elife-62470-fig5-data1-v1.xlsx
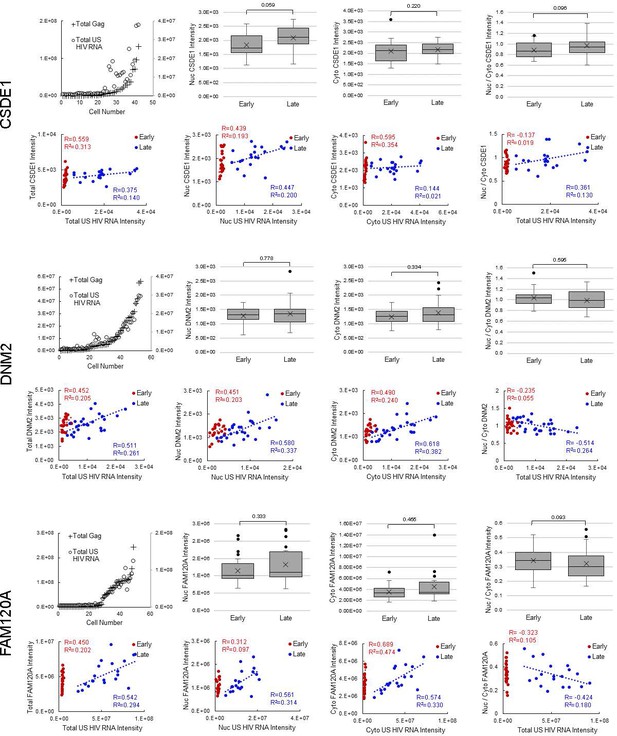
Host protein expression and cellular distribution.
Data displayed for all 12 host proteins analyzed by fluorescence in situ hybridization/immunofluorescence (FISH/IF). The host protein intensities in early and late cells for each host protein, in the nucleus and the cytoplasm are plotted. A Student’s t-test is applied to determine if the host protein intensities in early cells are significantly different from those in late cells. The ratios of nuclear to cytoplasmic (nuc/cyto) host protein intensity for early and late cells are plotted (nuclear plus perinuclear [nuc+peri] for MBOAT7). A Student’s t-test measures significant differences in the cellular distribution between the sub-groups for each host protein. For each cell in the early and late cell sub-groups, the intensity of unspliced (US) HIV RNA vs the intensity of the host protein is plotted. Values for the nucleus and the cytoplasm are in separate graphs. Linear regressions (R2) are fit to each early and late, nuclear and cytoplasmic sub-group and the Pearson’s correlation coefficient (R) calculated. This demonstrates the extent of correlation between US HIV RNA expression and the expression of each host protein. Total cellular US HIV RNA intensities vs host protein nuc/cyto or nuc+peri/cyto ratios. This demonstrates the extent of correlation of host protein cellular distribution with US HIV RNA expression. Related to Figure 5 and Figure 5—source data 1.
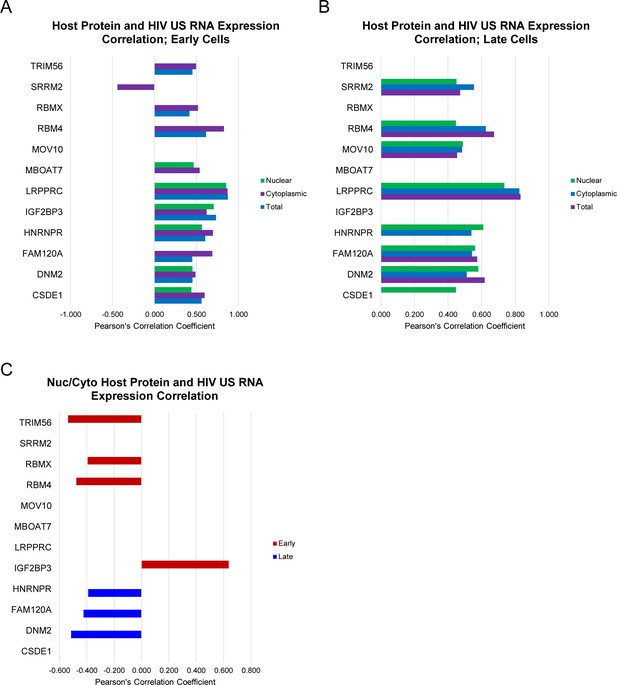
Summary of host protein expression and cellular distribution.
(A-B) Pearson’s correlation coefficient for host protein expression and unspliced (US) HIV RNA expression in early cells (A) and late cells (B). (C) Pearson’s correlation coefficients for the ratio of nuclear/cytoplasmic host protein intensity and US HIV RNA expression in early and late cells. For A, B, and C, only proteins with R>0.35 and R2>0.15 are displayed. Related to Figure 5 and Figure 5—source data 1.
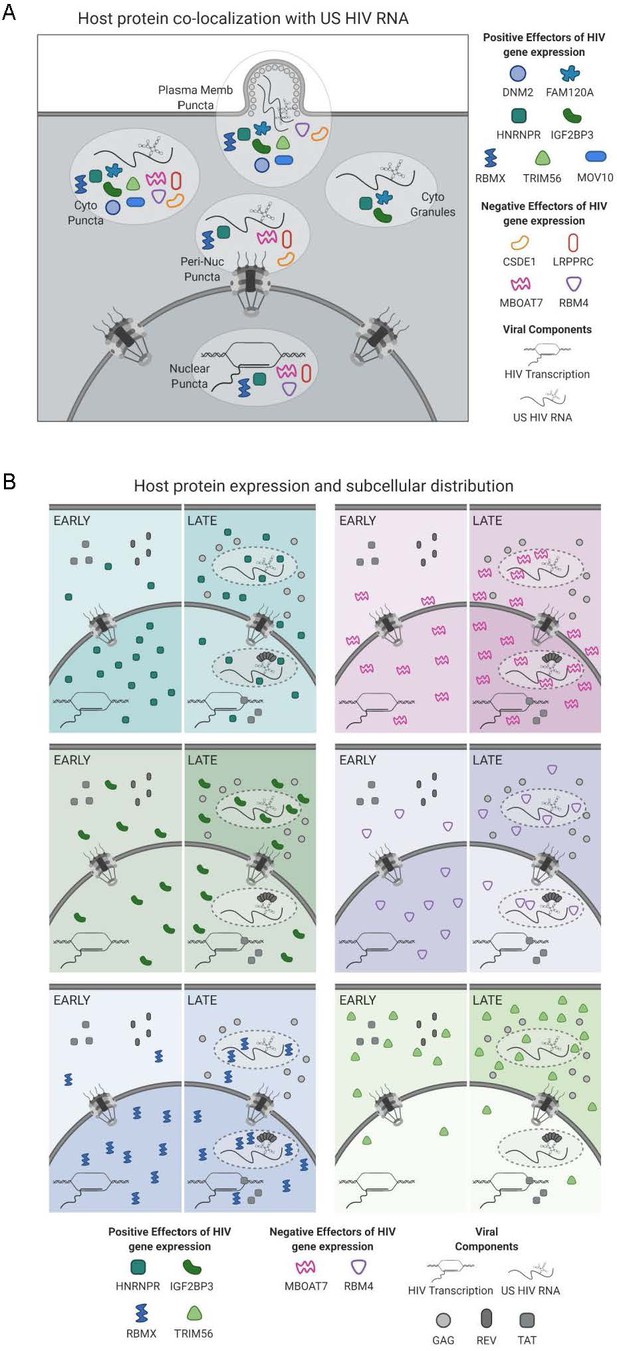
Models for subcellular co-localization, expression, and distribution of HIV-1 RNA-associated host proteins during infection.
(A) Summary of cellular co-localization phenotypes observed by fluorescence in situ hybridization/immunofluorescence (FISH/IF) analysis of HIV-1 unspliced (US) RNA and select host proteins. Host proteins represented include both positive and negative effectors of HIV gene expression as was determined by siRNA knockdown. (B) Models of host protein changes in expression and cellular distribution from early to late HIV gene expression. Host protein quantities represented here show the general, but not exact, scale of changes in host protein abundance in the nucleus and cytoplasm over the course of HIV replication. The extent of shading in the nucleus or cytoplasm correlates with that same change in abundance. Figure created with BioRender.com.
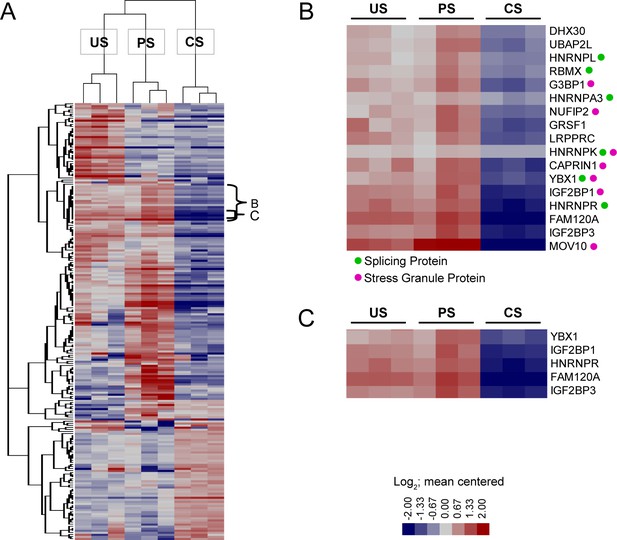
Heatmap summary of closely clustered proteins of interest.
(A) Hybridization Purification of RNA-Protein Complexes Followed by Mass Spectrometry (HyPR-MS) heatmap displaying the HIV splice variant classes differential protein interactomes. (B) A cluster containing a high concentration of stress granule proteins and splicing proteins. (C) Cluster of proteins with closely related interaction profiles. Related to Figures 2, 3 and 6 and Supplementary file 4.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | Jurkat, Clone E6-1 | NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH | Cat#177; RRID:CVCL_0367 | Male |
| Cell line (Homo sapiens) | HEK293T | ATCC | CRL-11268; RRID:CVCL_1926 | Fetus |
| Cell line (Homo sapiens) | HEK293T YFP-ACT | ATCC | CRL-11268; RRID:CVCL_1926 | HEK293T cells stably expressing YFP-ACT fusion protein |
| Cell line (Homo sapiens) | HeLa | ATCC | CCL-2; RRID:CVCL_0030 | Female |
| Recombinant DNA reagent | HIV-1 E-R- Gag-3x CFP mCherry/nef (plasmid) | Knoener et al., 2017 | NL4-3 | |
| Recombinant DNA reagent | HIV-1 E-R-CFP (plasmid) | Knoener et al., 2017 | NL4-3 | |
| Recombinant DNA reagent | psPAX2 (plasmid) | RRID:Addgene_12660 | A gift from Didier Trono | |
| Sequence-based reagent | Intron-1 capture oligonucleotide | This paper, Supplementary file 1 | Biotinylated; Supplementary file 1 | |
| Sequence-based reagent | Intron-2 capture oligonucleotide | This paper, | Biotinylated; Supplementary file 1 | |
| Sequence-based reagent | 3’-Exon capture oligonucleotide | This paper, Supplementary file 1 | Biotinylated; Supplementary file 1 | |
| Sequence-based reagent | Intron-1 release oligonucleotide | This paper, Supplementary file 1 | Supplementary file 1 | |
| Sequence-based reagent | Intron-2 release oligonucleotide | This paper, Supplementary file 1 | Supplementary file 1 | |
| Sequence-based reagent | 3’-Exon release oligonucleotide | This paper, Supplementary file 1 | Supplementary file 1 | |
| Sequence-based reagent | Intron-1 qPCR assay | This paper, Supplementary file 1 | Supplementary file 1 | |
| Sequence-based reagent | Intron-2 qPCR assay | This paper, Supplementary file 1 | Supplementary file 1 | |
| Sequence-based reagent | Intron-3 qPCR assay | This paper, Supplementary file 1 | Supplementary file 1 | |
| Sequence-based reagent | RNA FISH probes | Biosearch Technologies; This paper, Supplementary file 7 | Supplementary file 7 | |
| Transfected construct (Homo sapiens) | siRNAs for knockdown (KD) screen | This paper, Supplementary file 3 | Supplementary file 3 | |
| Antibody | Anti-BUB3 (mouse polyclonal) | ThermoFisher | PA5-20388; RRID:AB_11154661 | IF (1:200) WB (1:1000) |
| Antibody | Anti-CSDE1 (rabbit polyclonal) | ThermoFisher | PA5-22394; RRID:AB_11154127 | IF (1:200) WB (1:1000) |
| Antibody | Anti-DHX30 (rabbit polyclonal) | ThermoFisher | PA5-41298; RRID:AB_2607114 | IF (1:200) WB (1:1000) |
| Antibody | Anti-DLD (rabbit polyclonal) | ThermoFisher | PA5-27367; RRID:AB_2544843 | IF (1:200) WB (1:1000) |
| Antibody | Anti-DNM2 (rabbit polyclonal) | ThermoFisher | PA1-661; RRID:AB_2293040 | IF (1:200) WB (1:1000) |
| Antibody | Anti-DYNC1H1 (rabbit polyclonal) | ThermoFisher | PA5-49451; RRID:AB_2634905 | IF (1:200) WB (1:1000) |
| Antibody | Anti-FAM120A (rabbit polyclonal) | ThermoFisher | PA5-54069; RRID:AB_2641236 | IF (1:200) WB (1:1000) |
| Antibody | Anti-G3BP1 (mouse monoclonal) | Santa Cruz | SC-98561; RRID:AB_2294329 | IF (1:200) WB (1:1000) |
| Antibody | Anti-GSDMA (rabbit polyclonal) | ThermoFisher | PA5-24813; RRID:AB_2542313 | IF (1:200) WB (1:1000) |
| Antibody | Anti-HNRNPR (rabbit polyclonal) | ThermoFisher | PA5-55290; RRID:AB_2642500 | IF (1:200) WB (1:1000) |
| Antibody | Anti-IGF2BP3 (rabbit polyclonal) | ThermoFisher | PA5-51672; RRID:AB_2642656 | IF (1:200) WB (1:1000) |
| Antibody | Anti-LRPPRC (rabbit polyclonal) | ThermoFisher | PA5-22034; RRID:AB_11153345 | IF (1:200) WB (1:1000) |
| Antibody | Anti-MBOAT7 (rabbit polyclonal) | Abcam | ab105643; RRID:AB_10862084 | a.k.a. Anti-LENG4 IF (1:200) WB (1:1000) |
| Antibody | Anti-NCLN (rabbit polyclonal) | ThermoFisher | PA5-34356; RRID:AB_2551708 | IF (1:200) WB (1:1000) |
| Antibody | Anti-RBM4 (rabbit polyclonal) | ThermoFisher | PA5-21755; RRID:AB_11153613 | IF (1:200) WB (1:1000) |
| Antibody | Anti-RBMX (rabbit polyclonal) | ThermoFisher | PA5-49468; AB_2634922 | IF (1:200) WB (1:1000) |
| Antibody | Anti-RPL15 (rabbit polyclonal) | ThermoFisher | PA5-48446; RRID:AB_2633903 | IF (1:200) WB (1:1000) |
| Antibody | Anti-SRRM2 (rabbit polyclonal) | ThermoFisher | PA5-59559; RRID:AB_2647934 | IF (1:200) WB (1:1000) |
| Antibody | Anti-SRSF6 (rabbit polyclonal) | ThermoFisher | PA5-56034; RRID:AB_2647943 | IF (1:200) WB (1:1000) |
| Antibody | Anti-TRIM56 (mouse monoclonal) | ThermoFisher | MA5-27066; RRID:AB_2725573 | IF (1:200) WB (1:1000) |
| Antibody | Anti-HIV-1 Gag/p24 | NIH AIDS Reagent Program | 183-H12-5C; RRID:AB_2819250 | IF (1:200) WB (1:1000) |
| Antibody | Goat anti-Rabbit secondary antibody | ThermoFisher | A11008; RRID:AB_143165 | Conjugate Alexa Fluor 488 IF (1:200) WB (1:10000) |
| Antibody | Goat anti-Mouse secondary antibody | ThermoFisher | A21235; RRID:AB_2535804 | Conjugate Alexa Fluor 647 IF (1:200) WB (1:10000) |
| Antibody | Goat anti-Rabbit secondary antibody | LiCor Biosciences | 926–32211; RRID:AB_621843 | LiCor IRDye800 WB (1:10000) |
| Antibody | Goat anti-Mouse secondary antibody | LiCor Biosciences | 926–68020; RRID:AB_10706161 | LiCor 680LT WB (1:10000) |
| Commercial assay or kit | High Capacity cDNA Reverse Transcription Kit | ThermoFisher | 4368814 | |
| Peptide, recombinant protein | Trypsin | Promega | V5111 | |
| Peptide, recombinant protein | Proteinase K | Sigma | 3115828001 | |
| Peptide, recombinant protein | RNasin Plus | Promega | N2611 | |
| Chemical compound, drug | Halt Protease Inhibitors | ThermoFisher | 78430 | |
| Chemical compound, drug | Tri Reagent | Sigma | T9424 | |
| Other | GlycoBlue | ThermoFisher | AM9516 | |
| Other | OMIX C18 solid-phase extraction pipette tip | Agilent | A57009100 | |
| Other | DarmaFECT | Horizon Discovery | T-2001–02 | |
| Other | DAPI | Sigma | D9542-10MG | |
| Other | FISH wash buffer A | Biosearch Technologies | SMF-WA1-60 | |
| Other | FISH wash buffer B | Biosearch Technologies | SMF-WB1-20 | |
| Other | FISH hybridization buffer | Biosearch Technologies | SMF-HB1-10 | |
| Other | TaqMan Fast Advanced Master Mix | Fisher Scientific | 4444963 | |
| Other | Ribonucleoside Vanadyl Complex | Sigma | R3380-5ML | |
| Other | Sera-Mag Streptavidin Coated Magnetic Speedbeads | Fisher Scientific | 09981140 | |
| Software, algorithm | MaxQuant software | https://www.maxquant.org/ | RRID:SCR_014485 | |
| Software, algorithm | Perseus software | https://www.maxquant.org/perseus/ | RRID:SCR_015753 | |
| Software, algorithm | Cluster software | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm | ||
| Software, algorithm | TreeView software | https://sourceforge.net/projects/jtreeview/ | RRID:SCR_016916 | |
| Software, algorithm | Gene Ontology software | http://geneontology.org/ | RRID:SCR_002811 | |
| Software, algorithm | FIJI/ImageJ2 software | https://imagej.nih.gov/ij/ | RRID:SCR_003070 |
Additional files
-
Supplementary file 1
Human immunodeficiency virus type 1 (HIV-1) splice variant capture oligonucleotide and qPCR assay sequences and genomic locations.
Related to Figure 1 and Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/62470/elife-62470-supp1-v1.docx
-
Supplementary file 2
Proteins identified by Hybridization Purification of RNA-Protein Complexes Followed by Mass Spectrometry (HyPR-MS) to be ‘common protein interactors,’ or proteins identified in at least two of three biological replicates of each splice variant class capture, and proteins identified to be ‘differential protein interactors.’.
Includes comparison of common protein interactors to a meta-analysis of studies identifying general mRNA interactors. Also includes comparison of proteins previously determined to interact with human immunodeficiency virus type 1 (HIV-1) RNAs.
- https://cdn.elifesciences.org/articles/62470/elife-62470-supp2-v1.xlsx
-
Supplementary file 3
siRNA sequences used for gene-specific knockdown screen.
Related to Figure 3.
- https://cdn.elifesciences.org/articles/62470/elife-62470-supp3-v1.docx
-
Supplementary file 4
Summary of human immunodeficiency virus type 1 (HIV-1) splice variant interactome MS data and siRNA knockdown (KD) expression changes data.
- https://cdn.elifesciences.org/articles/62470/elife-62470-supp4-v1.xlsx
-
Supplementary file 5
Antibodies used for immunoblots and immunofluorescence.
Related to Figures 4 and 5 and Figure 4—figure supplements 1–3.
- https://cdn.elifesciences.org/articles/62470/elife-62470-supp5-v1.docx
-
Supplementary file 6
Western blot quantitation, with and without siRNA knockdown (KD), with and without human immunodeficiency virus type 1 (HIV-1) infection.
Related to Figure 3 and Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/62470/elife-62470-supp6-v1.xlsx
-
Supplementary file 7
Stellaris-designed fluorescence in situ hybridization (FISH) probes specific to unspliced (US) human immunodeficiency virus (HIV) RNA.
Related to Figures 4 and 5, Figure 4—figure supplements 1–3.
- https://cdn.elifesciences.org/articles/62470/elife-62470-supp7-v1.docx
-
Supplementary file 8
Host protein immunofluorescence in cells with and without siRNA knockdown (KD).
Related to Figure 4 and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/62470/elife-62470-supp8-v1.xlsx