Starvation-induced regulation of carbohydrate transport at the blood–brain barrier is TGF-β-signaling dependent
Figures
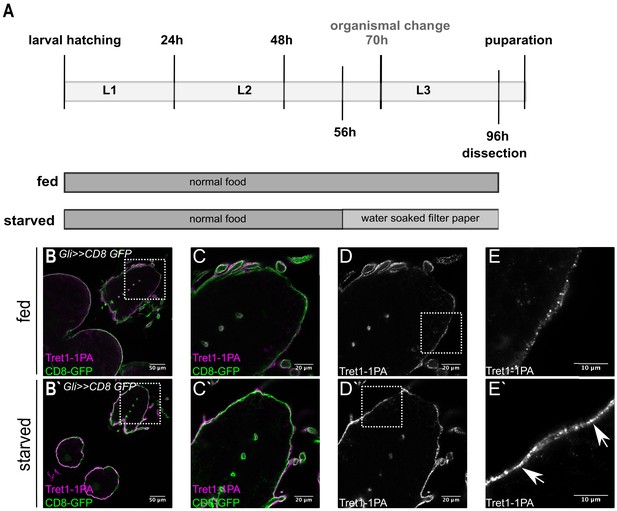
Tret1-1 expression is upregulated upon starvation.
(A) Scheme of the starvation paradigm. Fed animals were kept for 96 hr on normal food before dissecting. Starved animals were transferred onto a water-soaked filter paper 56 hr after larval hatching. Forty hour later these animals were also dissected and immunohistochemistry was performed. (B–E) Brains of fed larvae expressing CD8-GFP in the subperineurial glial cells (Gli>>CD8-GFP) stained for GFP (green) and Tret1-1 (magenta/gray). (B`–E`) Brains of starved larvae with the same genotype. (D, D`) Tret1-1 expression in the perineurial glial cells is induced upon starvation. (E, E`) Close up of the BBB. Tret1-1 is localized in vesicles and its expression is elevated upon starvation. Tret1-1 is localized to the plasma membrane (arrows).
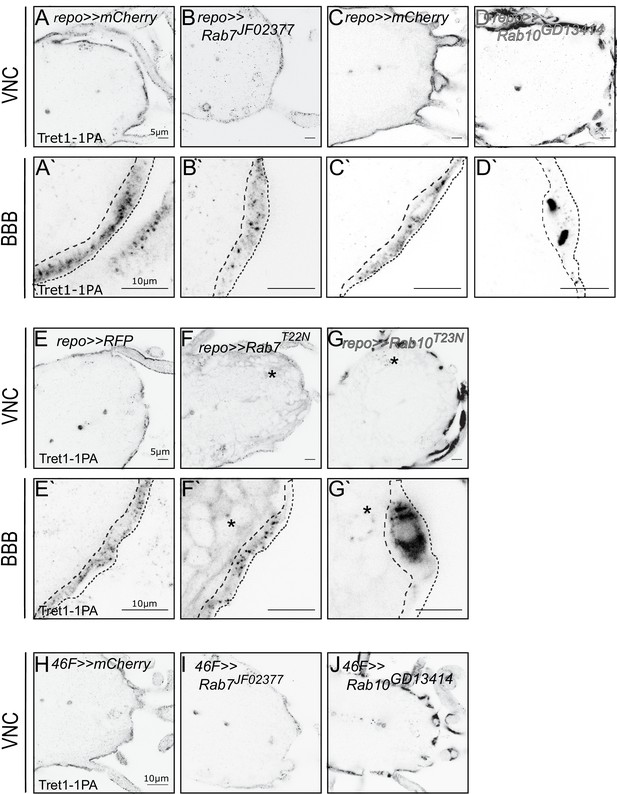
Tret1-1 intracellular trafficking depends on Rab7 and Rab10.
(A, C) Tret1-1 staining of the ventral nerve cord of glia-specific (repo-Gal4) knockdowns (A`, B`, C`, D`) shows a close up of the BBB. Dotted lines show the outline of the perineurial glia. (B, B`) Tret1-1 expression is strongly reduced by a glial Rab7 (Rab7JF02377) knockdown. (D, D`) Disrupting Rab10 expression in glia (Rab10GD13414) induces accumulation of Tret1-1 in the perineurial glia cytosol. (E–G`) Glial expression of the dominant-negative constructs Rab7T22N and Rab10T23N induce similar phenotypes as the RNA interference mediated knockdowns. (F, F`) Expressing Rab7T22N reduces Tret1-1 staining. (G, G`) Glia expression of Rab10T23N induces transporter mislocalization and a strong accumulation in the perineurial cytosol. (F–G`) The dominant-negative Rab-constructs are Rab-YFP fusions. Panglial overexpression thus leads to a weak background staining in the green channel (asterisks). (C) Tret1-1 staining of surface and cortex glia-specific knockdown using 46 F-Gal4 and Rab7JF02377 and Rab10GD13414. Loss of Rab7 reduces Tret1-1 staining, while Rab10 disruption induces transporter mislocalization.
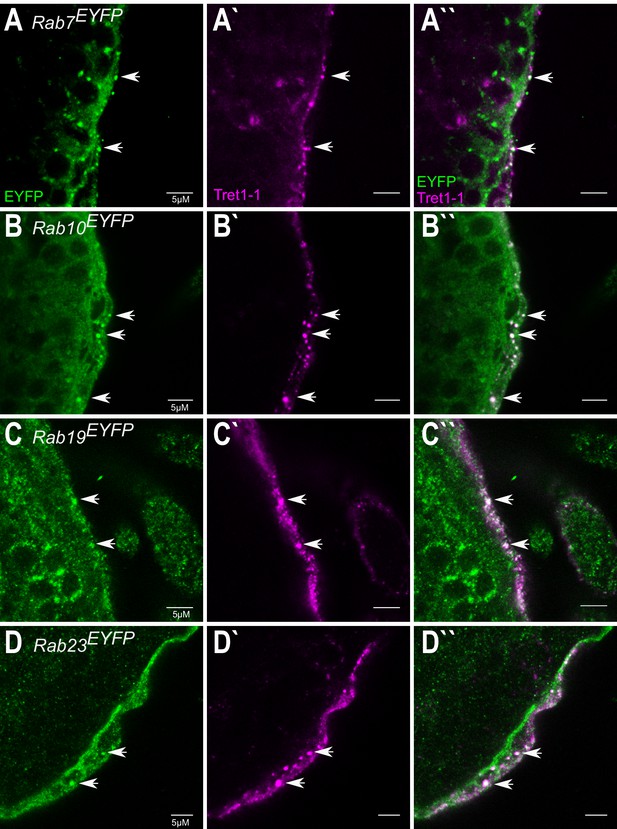
Rab7, Rab10, Rab19, and Rab23 colocalize with Tret1-1 vesicles.
Co-staining of endogenous EYFP-tagged Rab-GTPases (green) and Tret1-1 (magenta) in the surface glia of third-instar larval brains. (A–D) GFP-stained Rab-GTPases. (A`–D`) Tret1-1 is stained in magenta. (A``–D``) Merge of GFP and Tret1-1 staining. Representative overlapping stainings are indicated by an arrow. All Drosophila Rab-GTPases endogenously labeled with EYFP were tested. Tret1-1-positive vesicles show overlapping staining with Rab7EYFP, Rab10EYFP, Rab19EYFP, and Rab23EYFP.
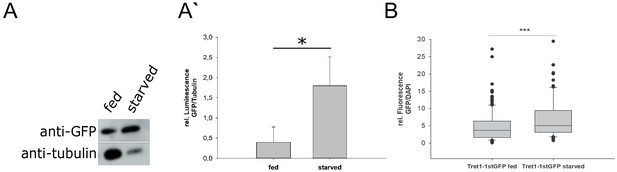
Tret1-1 is transcriptionally upregulated upon starvation.
(A) Tret1-1 is transcriptionally upregulated upon starvation since the Tret1-1 reporter Tret1-1>stgGFP is significantly upregulated in brains of starved compared to fed larvae as seen in western blots. Shown are images of representative western blots for anti-GFP and anti-tubulin (loading control). n=3 (A`) Quantification of Western blots. N=3. (B) Tret1-1 protein is significantly upregulated upon starvation in Tret1-1>stgGFP animals. Quantified is the GFP fluorescence normalized to DAPI in individual nuclei. N=5 , n > 34.
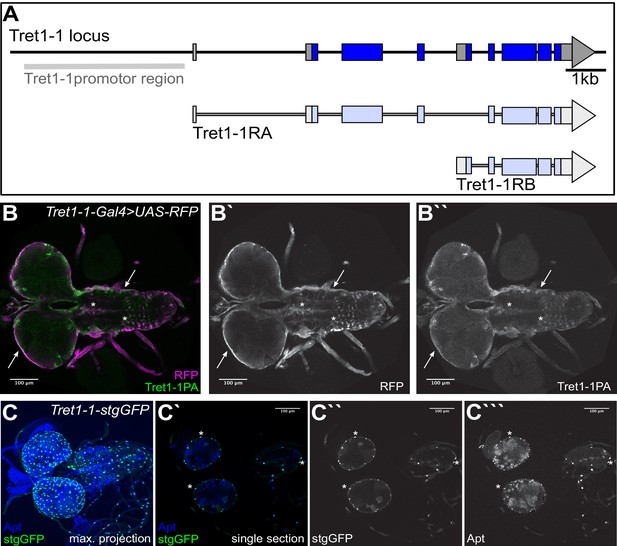
Tret1-1 promoter drives specific expression.
(A) Schematic of the Tret1-1 locus and the transcripts encoding the two Tret1-1 isoforms. The Tret1-1 promoter region used to generate Tret1-1-Gal4 and tret1-1-stgGFP is highlighted in gray. (B–B``) Tret1-1PA staining (green, gray) overlaps with Tret1-1-driven RFP (magenta, gray) (Tret1-1-Gal4 UAS-RFP), verifying the specificity of the Tret1-1 promotor region. Arrows indicate representative overlays. (C–C```) Co-staining of Apontic (blue, gray) and stgGFP (green, gray) that shows that Tret1-1-driven stgGFP is specifically expressed in perineurial glial nuclei. Arrows indicate representative overlay expressions.
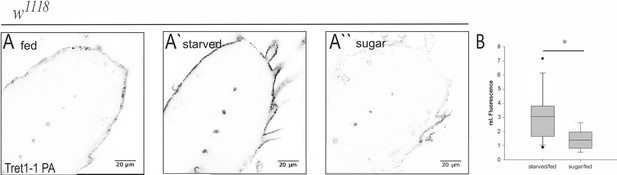
Tret1-1 upregulation upon starvation is sugar-dependent.
(A- A``) Brains of larvae kept on normal food, under starvation conditions or on sugar food (10% sucrose) were stained for Tret1-1. (A`) Tret1-1 expression is elevated upon starvation of the animal. (A``) Dietary sugar reverses Tret1-1 upregulation completely. (B) Quantification of Tret1-1 expression in starved wild type animals and wild type animals on sugar food. The quantification shows the ratio of relative Tret1-1 fluorescence intensity in the perineurial glial cells of starved versus fed and sugar-fed vs. fed animals. N≥4; n=9-15; *p<0,05.
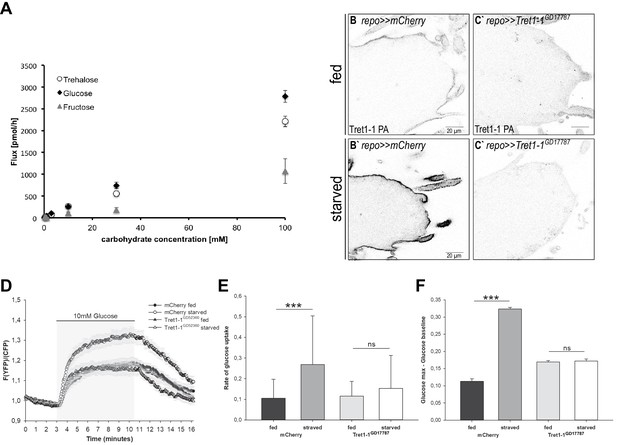
Carbohydrate uptake rate into the surface glia is elevated upon starvation.
(A) Carbohydrate uptake capacity of Xenopus laevis oocytes heterologously expressing Tret1-1PA-3xHA. Tret1-1PA facilitates uptake of glucose and trehalose. In contrast, fructose uptake rate is minor. (B–C`) Expressing Tret1-1GD17787 in glial cells induces a loss of the specific Tret1-1 staining in perineurial glia. (C`) No increase upon starvation can be detected. (D) Glucose uptake was measured in ex vivo brains of fed or starved larvae using the genetically encoded glucose sensor FLII12Pglu-700µδ6. Shown are mean traces (n = 10). Error bars are standard error. (E) The glucose uptake rate is significantly higher in brains of starved control larvae compared to fed control larvae. Knocking down Tret1-1 prohibits the increased glucose uptake upon starvation. (F) In addition, the maximum intracellular glucose concentration is significantly higher in starved control larvae than in fed control larvae, suggesting that the uptake rate exceeds the rate of metabolism. This effect is also abolished when Tret1-1 is impaired in BBB-glia. N = 3, n ≥ 10.
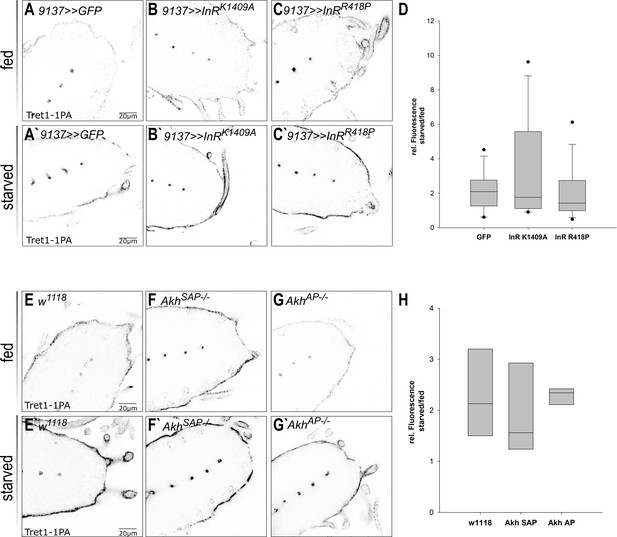
Tret1-1 regulation upon starvation is insulin and AKH independent.
(A–C`) Tret1-1 staining of the ventral nerve cord of starved and fed larvae expressing dominant-negative forms (InRK1409A or InRR418P) of the insulin receptor (InR) in the perineurial and subperineurial glial cells. (B, B`, C, C`) Tret1-1 levels in fed and starved animals expressing InR dominant negative are indistinguishable from wild type. (B`, C`) Tret1-1 upregulation upon starvation is seen in all cases. (D) Quantification of Tret1-1 upregulation in animals expressing InRK1409A or InRR418P. Shown is the ratio of relative Tret1-1 fluorescence intensity in the perineurial glial cells of starved versus fed animals. No significant differences between the genotypes are observed. N = 4, n = 12–16. (E–G`) Tret1-1 staining of the ventral nerve cord of starved and fed wild-type and Akh−/− mutant animals (AkhSAP or AkhAP). Tret1-1 levels in fed and starved mutant animals are indistinguishable from wild type. (F`, G`) Tret1-1 upregulation upon starvation can be seen in all mutants. (H) Quantification of Tret1-1 intensities of AkhSAP or AkhAP. Shown is the ratio of relative Tret1-1 fluorescence intensity in the perineurial glial cells of starved versus fed animals. No significant differences are observed. N = 3, n = 5–8.

Tret1-1 regulation upon starvation is ALK-independent.
(A, B`) Tret1-1 staining of the ventral nerve cord of starved and fed control (repo>>mCherry-dsRNA, A, A`) and alk knockdown (repo>>alkGD42, B, B`) animals. (B`) Tret1-1 upregulation is still induced in starved alk knockdown animals.
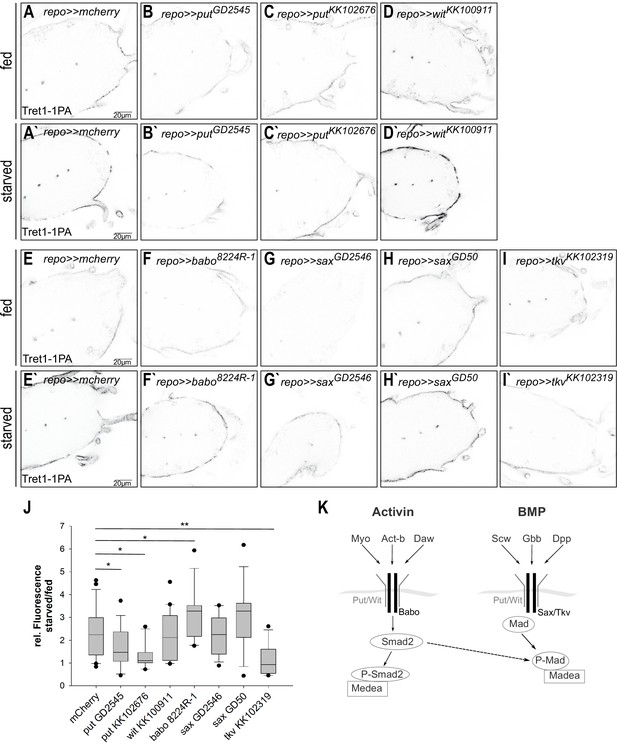
Tret1-1 upregulation upon starvation is BMP-mediated TGF-β signaling dependent.
(A, A`, I, I`) Tret1-1 staining of the ventral nerve cord of starved and fed control (repo>>mCherry-dsRNA) animals and animals with a glial TGF-β knockdown. (B, B`, C, C`) Knockdown of the type I receptor Put in glial cells using two different dsRNA constructs (putGD2545 and putKK102676) abolished Tret1-1 upregulation upon starvation. (D, D`) Glia-specific knockdown of wit using witKK100911 does not affect Tret1-1 upregulation upon starvation. (F, F`) Glia-specific knockdown of the activin-branch-specific type II receptor Babo (using baboNIG8224R-1) does not have any influence on Tret1-1 upregulation upon starvation (compare to control in E, E`). (G, G`, H, H`) The glia-specific knockdown of the BMP-branch-specific type II receptor Sax (using saxGD2546 and saxGD50) does not influence Tret1-1 expression (compare to control in E, E`). (I, I`) In contrast, glia-specific knockdown of the main BMP-branch-specific type II receptor Tkv (using tkvKK1023019) abolishes Tret1-1 upregulation upon starvation (compare to control in E, E`). This indicates that signaling via the BMP branch of TGF-β signaling regulates Tret1-1 induction upon starvation. (J) Quantification of Tret1-1 upregulation upon starvation. Shown is the ratio of relative Tret1-1 fluorescence intensity in the perineurial glial cells of starved versus fed animals. N ≥ 4, n = 10–22. (K) Schematic representation of the two branches of the TGF-β signaling pathway.
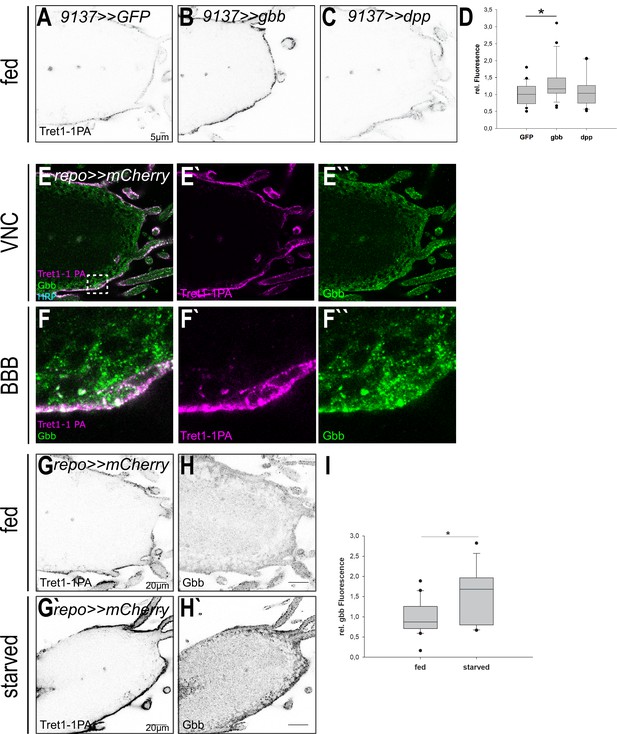
Tret1-1 upregulation depends on Gbb-mediated signaling.
(A–C) Tret1-1 staining of the ventral nerve cord of fed control (9137>>GFP) animals or animals overexpressing either Gbb or Dpp in the perineurial and subperineurial glial cells (using 9137-Gal4). (B) Overexpression of Gbb induces increased Tret1-1 expression in fed animals. (C) Differently, overexpression of Dpp does not have any effect on the Tret1-1 expression. (D) Quantification of Tret1-1 upregulation upon overexpression of Gbb or Dpp in fed animals. The quantification shows Tret1-1 expression levels in the perineurial glial cells normalized to those in controls (9137>>GFP). N = 4; n = 20–25; (E–F``) Gbb is expressed in perineurial glial cells (coexpression with Tret1-1) and in other glial cell types, most likely subperineurial glial cells and cortex glial cells. (G–H`) Upon starvation expression of Tret1-1 and Gbb is increased in the VNC. (I) Quantification of Gbb upregulation upon starvation in the brain. N = 5; n > 17.
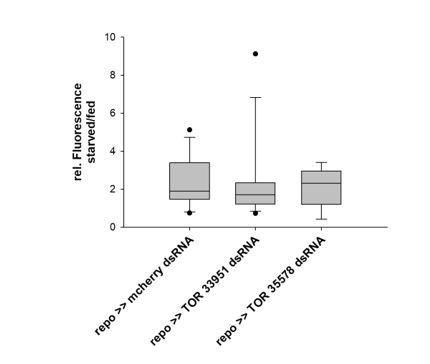
Tret1-1 upregulation upon starvation is independent of TOR signaling.
Quantified is the ratio of Tret1-1 fluorescence intensities in starved larval VNCs vs. VNCs of fed animals. Knockdown of TOR expressing using either TOR33951 or TOR35578 in glial cells shows a comparable upregulation of Tret1-1 to the control. N=5 n >9.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | Tret1-1 | FBgn0050035 | ||
| Genetic reagent (D. melanogaster) | jebKK111857 | Vienna Drosophila Resource Center | v103047; FBgn0086677; FBst0474909 | |
| Genetic reagent (D. melanogaster) | jebGD5472 | Vienna Drosophila Resource Center | v30800; FBgn0086677; FBst0458662 | |
| Genetic reagent (D. melanogaster) | AlkGD42 | Vienna Drosophila Resource Center | v11446; FBgn0040505; FBst0450267 | |
| Genetic reagent (D. melanogaster) | putKK102676 | Vienna Drosophila Resource Center | v107071; FBgn0003169; FBst0478894 | |
| Genetic reagent (D. melanogaster) | putGD2545 | Vienna Drosophila Resource Center | v37279; FBgn0003169; FBst0461929 | |
| Genetic reagent (D. melanogaster) | witKK100911 | Vienna Drosophila Resource Center | v103808; FBgn0024179; FBst0475666 | |
| Genetic reagent (D. melanogaster) | saxGD50 | Vienna Drosophila Resource Center | v42457; FBgn0003317; FBst0464598 | |
| Genetic reagent (D. melanogaster) | saxGD2546 | Vienna Drosophila Resource Center | FBgn0003317 | |
| Genetic reagent (D. melanogaster) | tkvKK102319 | Vienna Drosophila Resource Center | v105834; FBgn0003716; FBst0477660 | |
| Genetic reagent (D. melanogaster) | Rab10GD13414 | Vienna Drosophila Resource Center | v28758; FBgn0015789; FBst0457628 | |
| Genetic reagent (D. melanogaster) | Rab10GD16778 | Vienna Drosophila Resource Center | v46792; FBgn0015789; FBst0466897 | |
| genetic reagent (D. melanogaster) | Rab10KK109210 | Vienna Drosophila Resource Center | v101454; FBgn0015789; FBst0473327 | |
| Genetic reagent (D. melanogaster) | Tret1-1GD17787 | Vienna Drosophila Resource Center | v52360; FBgn0050035; FBst0469787 | |
| Genetic reagent (D. melanogaster) | Rab7T22N | Bloomington Drosophila Stock Center | 9778; FBgn0015795 | |
| Genetic reagent (D. melanogaster) | Rab10T23N | Bloomington Drosophila Stock Center | 9778; FBgn0015795; FBst0009778 | |
| Genetic reagent (D. melanogaster) | Rab7EYFP | Bloomington Drosophila Stock Center | 62545; FBgn0015795; FBst0062545 | |
| Genetic reagent (D. melanogaster) | Rab10EYFP | Bloomington Drosophila Stock Center | 62548; FBgn0015789; FBst0062548 | |
| Genetic reagent (D. melanogaster) | Rab19EYFP | Bloomington Drosophila Stock Center | 62552; FBgn0015793; FBst0062552 | |
| Genetic reagent (D. melanogaster) | Rab23EYFP | Bloomington Drosophila Stock Center | 62554; FBgn0037364; FBst0062554 | |
| Genetic reagent (D. melanogaster) | Rab7TRIP.JF02377 | Bloomington Drosophila Stock Center | 27051; FBgn0015795; FBst0027051 | |
| Genetic reagent (D. melanogaster) | InRK1409A | Bloomington Drosophila Stock Center | FBgn0283499 | |
| Genetic reagent (D. melanogaster) | InRR418P | Bloomington Drosophila Stock Center | FBgn0283499 | |
| Genetic reagent (D. melanogaster) | UAS-dpp | Bloomington Drosophila Stock Center | 1486; FBgn0000490 | |
| Genetic reagent (D. melanogaster) | CherrydsRNA | Bloomington Drosophila Stock Center | 35785; FBti0143385 | |
| Genetic reagent (D. melanogaster) | UAS-CD8-GFP | Bloomington Drosophila Stock Center | 30002 or 30003 | |
| Genetic reagent (D. melanogaster) | AkhAP | Gáliková et al., 2015 doi: 10.1534/genetics.115.178897 | FBal0319564 | |
| Genetic reagent (D. melanogaster) | AkhSAP | Gáliková et al., 2015 doi: 10.1534/genetics.115.178897 | FBal0319565 | |
| Genetic reagent (D. melanogaster) | baboNIG8224R | Japanese National Institute of Genetics | FBal0275907 | |
| Genetic reagent (D. melanogaster) | gliotactin-Gal4 | Sepp et al., 2001 Doi: 10.1006/dbio.2001.0411 | – | |
| Genetic reagent (D. melanogaster) | repo-Gal4 | Sepp et al., 2001 Doi: 10.1006/dbio.2001.0411 | – | |
| Genetic reagent (D. melanogaster) | 46 F-Gal4 | Xie and Auld, 2011 Doi: 10.1242/dev.064816 | – | |
| Genetic reagent (D. melanogaster) | 9137-Gal4 | DeSalvo et al., 2014 Doi: 10.3389/fnins.2014.00346 | – | |
| Genetic reagent (D. melanogaster) | UAS-FLII12Pglu-700µδ6 | Volkenhoff et al., 2018 Doi: 10.1016/j.jinsphys.2017.07.010 | Maintained at S. Schirmeier lab | |
| Genetic reagent (D. melanogaster) | UAS-Gbb | P. Soba | – | |
| Genetic reagent (D. melanogaster) | UAS-RFP | S. Heuser | – | |
| Genetic reagent (D. melanogaster) | w1118 | Lindsley and Zimm, 1992 ISBN 9780124509900 | – | |
| Genetic reagent (D. melanogaster) | Tret1-1-stGFP | This paper | Maintained at S. Schirmeier | Tret1-1 promoter fusion to a nuclei-targeted GFP |
| Genetic reagent (D. melanogaster) | Tret1-1-Gal4 | This paper | Maintained at S. Schirmeier | Tret1-1 promoter induced Gal4 expression |
| Antibody | anti-Tret1-1 guinea pig polyclonal | Volkenhoff et al., 2015 | Maintained at S. Schirmeier Lab | (1:50) |
| Antibody | anti-Laminin rabbit polyclonal | Abcam | ab11575 | (1:1000) |
| Antibody | anti-Repo mouse monoclonal | Developmental Studies Hybridoma Bank | 8D12 anti-Repo | (1:2) |
| Antibody | anti-GFP mouse monoclonal | Molecular Probes | A11120 | (1:1000) |
| Antibody | anti-GFP chicken polyclonal | Abcam | Ab92456 | (1:1000) |
| Antibody | anti-GFP JL-8 mouse monoclonal | Clontech | Cat. 632381 | (1:10000) WB |
| Antibody | anti-Tubulin mouse monoclonal | Developmental Studies Hybridoma Bank | 12G10 anti-alpha-tubulin | (1:80) WB |
| Antibody | anti-Apontic rabbit polyclonal | Eulenberg and Schuh, 1997 | Gifted from Reinhard Schuh | (1:150) |
| Antibody | anti-Gbb mouse monoclonal | Developmental Studies Hybridoma Bank | GBB 3D6-24 | (1:20) |
| Recombinant DNA reagent | pBPGuw-stingerGFP (vector) | C. Klämbt | ||
| Recombinant DNA reagent | pBPGuwGal4 (vector) | Addgene | 17575 | |
| Recombinant DNA reagent | pGEM-He-Juel (vector) | S. Bröer | ||
| Sequence-based reagent | Forward primer_Tret1-1prom | This paper | PCR primers | CACCGGTCTCAAGCTCTCTTTTTTGCCTTACATATTTT |
| Sequence-based reagent | Reverse primer_Tret1-1prom | This paper | PCR primers | TGGGTAAGTTGGAGAGAGAG |
| Sequence-based reagent | Forward primer Tret1-1 PA | This paper | PCR primers | CGTCTAGAATGAGTGGACGCGAC |
| Peptide, recombinant protein | Reverse primer Tret1-1 PA | This paper | PCR primers | CGAAGCTTCTAGCTTACGTCACGT |
| Commercial assay or kit | pENTR/D- TOPO Cloning Kit | Thermo Fisher Scientific | K240020 | |
| Commercial assay or kit | mMESSAGE mMACHINE T7 Kit | Thermo Fisher Scientific | AM1344 | |
| Chemical compound, drug | 14C12-trehalose | Hartmann Analytic, Braunschweig | #1249 | |
| Chemical compound, drug | 14C6-glucose | Biotrend, Köln | #MC144-50 | |
| Chemical compound, drug | 14C6-fructose | Biotrend, Köln | #MC1459-50 | |
| Chemical compound, drug | Rotiszint eco plus scintillation cocktail | Carl Roth | Art. No. 0016.3 | |
| Software, algorithm | SigmaPlot | Jadel | SPSS Inc | |
| Software, algorithm | Fiji | NIH |





