Genome-wide effects of the antimicrobial peptide apidaecin on translation termination in bacteria
Figures
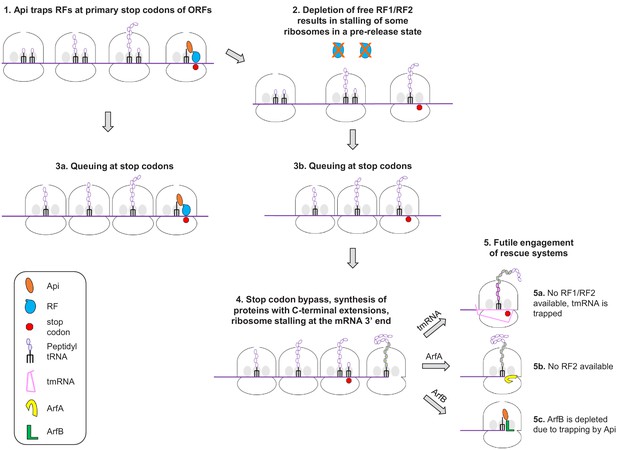
Api redistributes ribosomes toward the ends of mRNA ORFs.
(A) Comparison of ribosome footprint density in the tktA and dptA ORFs in untreated E. coli cells with that in cells treated with either Api or the translation initiation inhibitor retapamulin (Ret). The position of the ribosome footprint was assigned to the first nucleotide of the codon positioned in the P site, presumed to be at a distance of 16 nt from the 3’ end of the read (Woolstenhulme et al., 2015). (B) Ribosome footprint density within genes of the his operon in cells treated with no antibiotic, Api or Ret. The Ret Ribo-seq data are from Meydan et al., 2019.
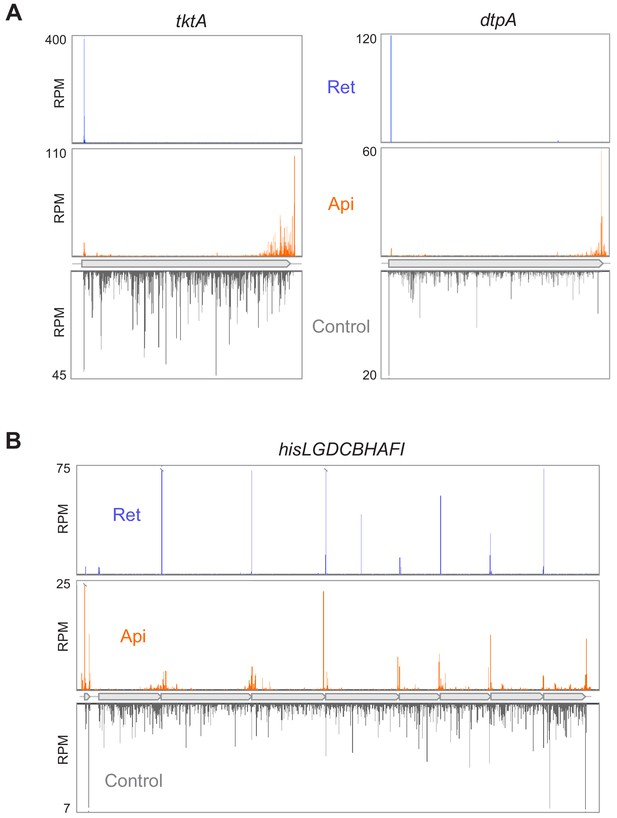
Api inhibits global protein synthesis in E. coli cells.
Api inhibits global protein synthesis. (A) Residual protein synthesis in E. coli cells (strain BL21 ΔtolC) exposed for 1 min to varying concentrations of Api137, as estimated by L-[35S]-methionine incorporation into TCA-precipitable proteins. Incorporation of L-[35S]-methionine in a sample devoid of Api was set as 100%. (B) Time course of inhibition of protein synthesis in cells exposed to 6.25 µM (4× MIC) of Api137. The shown data is the average of two independent experiments. Error bars indicate s.e.m.
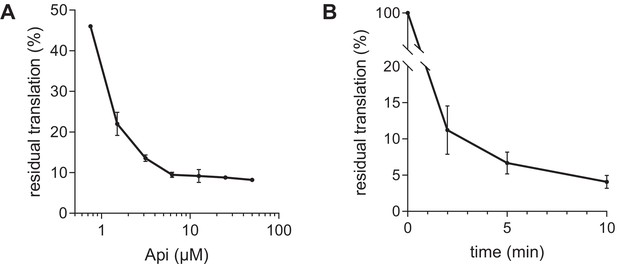
Api treatment leads to the accumulation of translating ribosomes near stop codons and depletion of free RFs in the cell.
(A) Metagene plot of the average normalized ribosome occupancy near the annotated stop codons of genes in cells treated (orange) or not (gray) with Api. The solid line indicates the mean value and the shadow plot reflects the standard error between two independent biological replicates. The metagene plot is based on 836 actively translated genes separated from neighboring genes by ≥50 nt. The cartoon illustrates ribosomes stalled at the stop codon in the post- or pre-release state, ribosomes queued upstream from those stalled at termination sites, and ribosomes present in mRNA regions downstream from the main ORF stop codon (readthrough). (B) Analysis of ribosome-associated and free RF2 in untreated cells and cells treated with Api. Ribosomes from lysates of untreated or Api-treated cells were pelleted through a sucrose cushion. The presence of RF2 in the ribosome pellet or in the post-ribosomal supernatant was tested by western blotting using the anti-RF2 serum. The purified E. coli His6-tagged RF2 was used as a control (lane marked ‘RF2’). An unknown protein (indicated with an asterisk) cross-reacting with the anti-RF2 serum served as the loading control. The cartoon illustrates how Api depletes free RFs by sequestering them on the ribosome.
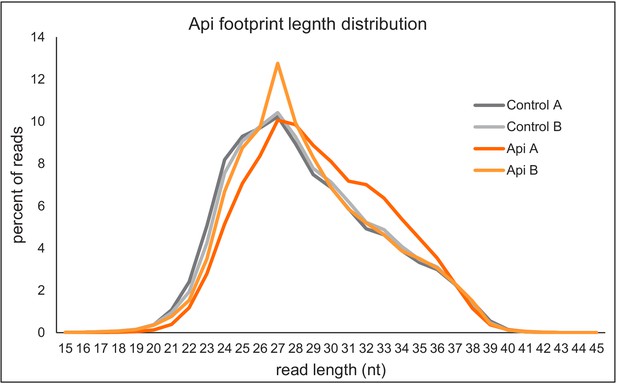
Footprint length distributions for Api and control samples.
Length distribution of ribosome footprint reads aligned to the genome after filtering out non-coding RNA reads.
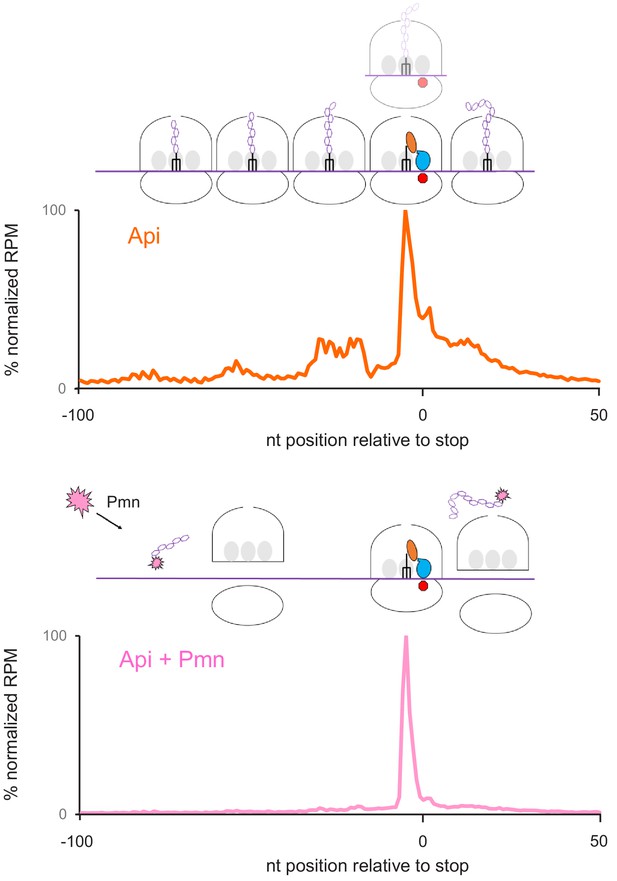
Puromycin treatment simplifies the ribosome distribution pattern in Api-treated cells.
Metagene plots of average normalized ribosome density in the vicinity of stop codons of genes in cells exposed to Api without (top) or with (bottom) puromycin (Pmn) treatment. The Pmn treatment removes peptidyl-tRNA and facilitates the dissociation of ribosomes from mRNA. Ribosomes arrested at the stop codon in the post-release state are expected to be refractory to Pmn action. Note that the metagene plots show the relative (normalized) distribution of ribosomes rather than the absolute number of ribosomes associated with each mRNA nucleotide.
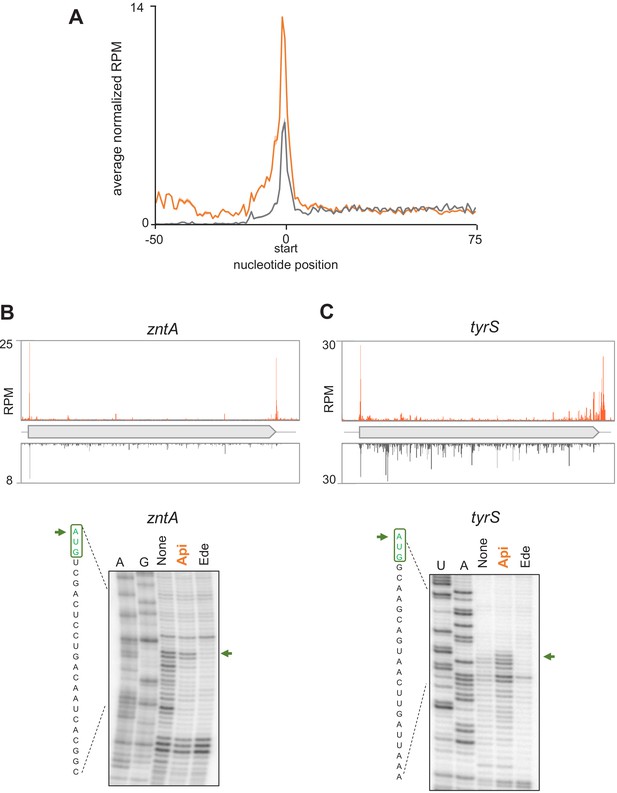
Api moderately increases ribosome density at start codons.
(A) Metagene plot of the average normalized ribosome density in the vicinity of the start codons in E. coli cells treated (orange) or not (gray) with Api. (B) and (C) Examples of genes where Api causes ribosome stalling at the start codons during in vivo and in vitro translation. Top: Ribosome footprint density observed in Ribo-seq experiments. Bottom: toeprinting analysis showing ribosome stalling at the start codons (green arrows) during cell-free translation in the presence of 2 mM of Api. The control antibiotic edein (Ede), which prevents ribosome binding to mRNA, was used to distinguish the toeprint bands originating from translating ribosomes.
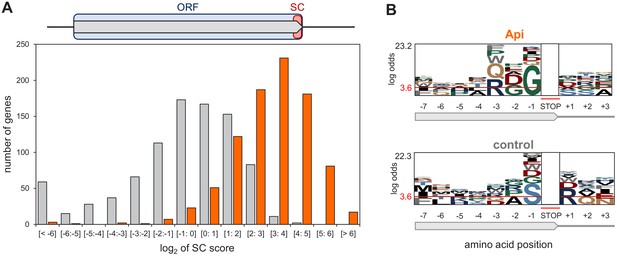
Api arrests ribosomes at translation termination sites.
(A) Top: Stop codon (SC) score was calculated for actively translated genes separated by ≥50 nt as the ratio of ribosome density near the stop codon (‘SC’, orange rectangle) to the ribosome density within the entire ORF (light blue rectangle) (see Materials and methods for details). Bottom: Histogram of the SC scores of genes from cells treated (orange) or not (gray) with Api. (B) pLogo analyses of amino acid sequence bias around the termination regions of the top 50% SC-scoring versus the bottom 50% SC-scoring genes from Api-treated cells (top) or control cells (bottom).
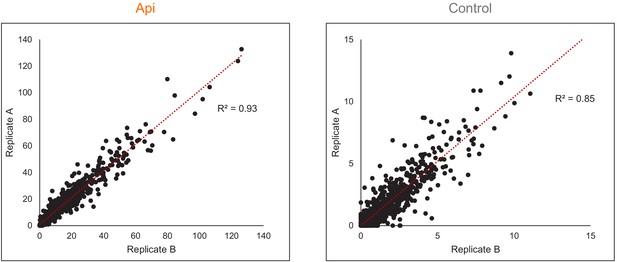
Reproducibility of stop codon effects.
Correlation of the stop codon (SC) scores in Api-treated or untreated (control) cells estimated from the Ribo-seq data obtained in two independent biological replicates.
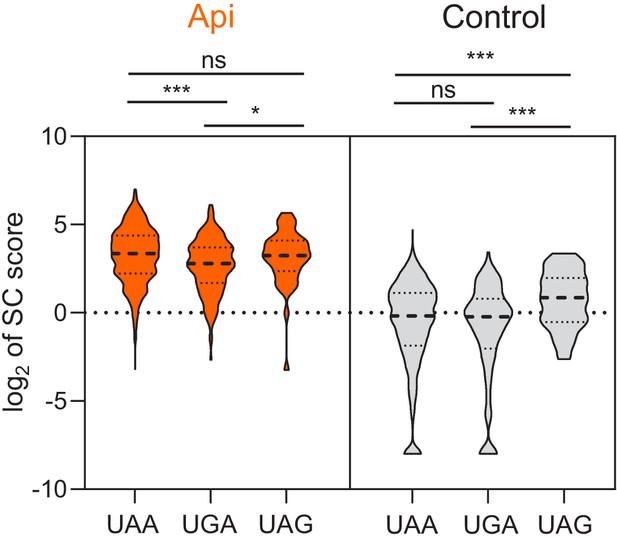
Stop codon scores at different stop codons.
Violin plots of genome-wide stop codon (SC) scores binned by stop codon identity. Two-sided Mann-Whitney U tests were performed to assess the significance of difference between genes grouped by stop codons: Api UAA versus UGA p=1.2E-5, Api UGA versus UAG p=0.032, control UAA versus UAG p=5.2E-4, control UGA versus UAG p=1.5E-4.
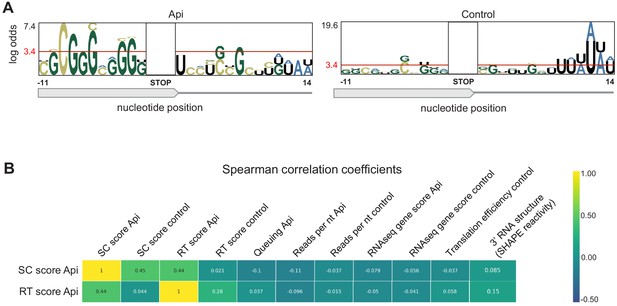
Evaluation of cellular factors that could potentially contribute to Api-mediated ribosome arrest at stop codons.
(A) The pLogo analysis of nucleotide sequences in the vicinity of the termination sites of the 50% high-SC score vs. 50% low SC-score genes in Api treated (left) and control (right) cells. (B) Estimated correlation between stop codon (SC) scores and readthrough (RT) scores of genes from Api-treated cells with various parameters calculated for the 905 well-separated (50 nt away from the nearest gene) and actively translated (ribosome footprint [rfp] density per nt ≥ 0.1) genes. The following parameters were used: Queuing score: a parameter reflecting the difference in the number of rfps in the first half of the gene relative to the number of rfps in the second half of the gene (Mohammad et al., 2019); Reads per nt: the total number of rfps within an ORF (excluding the first and last nine nts) divided by the length of an ORF; RNAseq gene score: total number of reads per million mapped to an ORF; Translation efficiency: Ribo-seq gene score dividied by RNA-seq gene score; 3’ RNA structure: the mean SHAPE reactivity of mRNA segment that includes 40 nt upstream and 10 nt downstream of the stop codon; (Huang et al., 2019). The latter was calculated for 114 out of the aforementioned 905 genes, for which SHAPE-seq data were available.
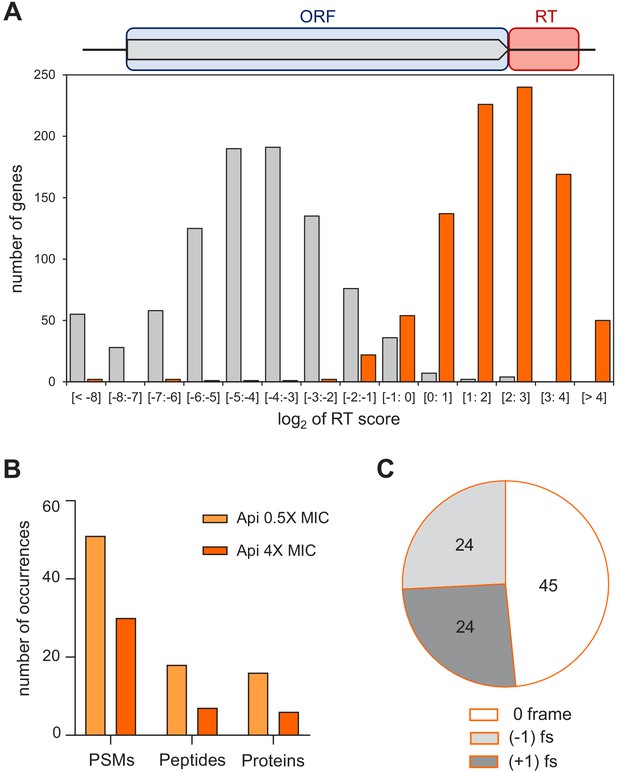
Api leads to stop codon bypass and generation of proteins with unintended C-terminal extensions.
(A) Top: Readthrough (RT) score was calculated for actively translated genes separated by ≥50 nt as the ratio of ribosome density within the 40 nt downstream from the stop codon (‘RT’, orange rectangle) to the ribosome density within the associated ORF (light blue rectangle). Bottom: Histogram of the RT scores of genes from cells treated (orange) or not (gray) with Api. (B) The number of proteins with C-terminal extensions, peptides belonging to the C-terminal extensions of proteins, and peptide-spectrum matches (PSMs) identified by mass spectrometry in cells treated with 0.5× MIC (light orange bars) or 4× MIC (orange bars) of Api. With the same data-filtering criteria, no such products were detected in the control cells. (C) The number of peptides identified using relaxed criteria (see Figure 4—figure supplement 3B) in cells treated with 0.5× MIC of Api belonging to C-terminal extensions of proteins generated via bypassing the stop codon of the main ORF by amino acid misincorporation (0-frame), or as a result of −1 or +1 ribosomal frameshifting (fs).
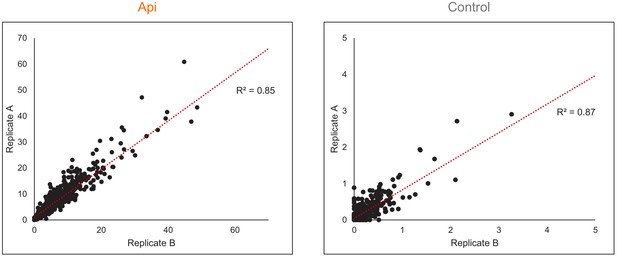
Reproducibility of stop codon bypass effects.
Correlation of the readthrough (RT) scores in Api-treated or control cells estimated from the Ribo-seq data obtained in two independent biological replicates.
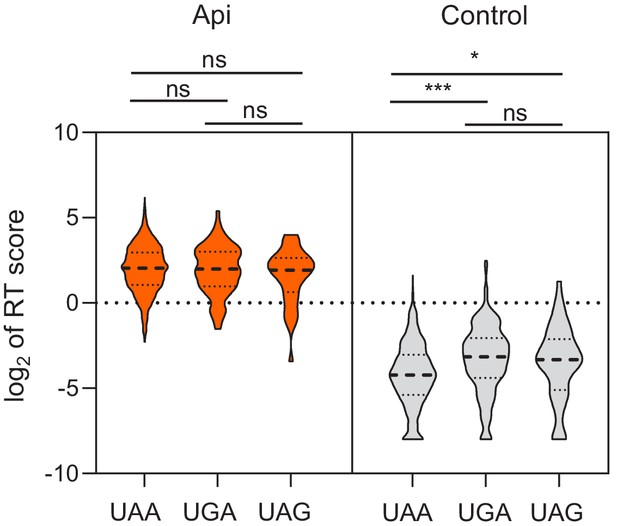
Readthrough scores at different stop codons.
Violin plots of genome-wide readthrough (RT) scores binned by stop codon identity. Two-sided Mann-Whitney U tests were performed to assess the significant difference between genes grouped by stop codons: Control UAA vs UGA p=5.91E-9, control UAA vs UAG p=0.012.
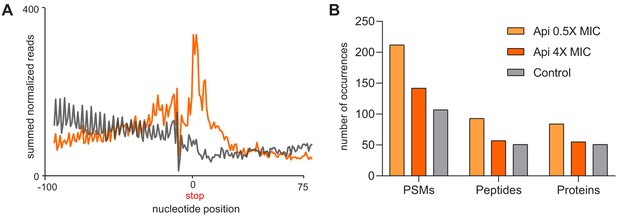
In the Api-treated cells, ribosomes translate mRNA segments downstream from the stop codon of the main ORF.
(A) Metagene plot of the ribosome footprints density in the vicinity of the first in-frame stop codon downstream from the main ORF termination codon in cells treated (orange) or not (gray) with Api. (B) The number of proteins with C-terminal extensions or PSMs and peptides counts belonging to the C-terminal extensions of proteins in the control and Api-treated cells identified by shotgun proteomics (more relaxed criteria compared to those used for generation of the Figure 4B plots, see Materials and methods).
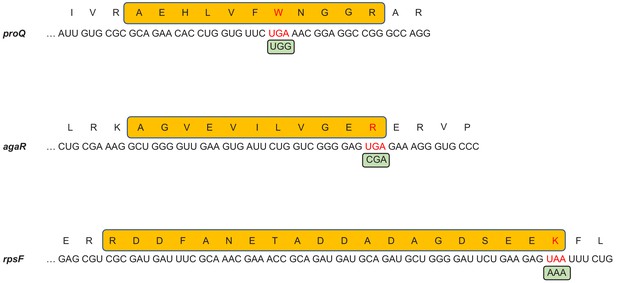
Stop codon bypass by near-cognate aminoacyl-tRNA misincorporation.
Api-induced 0-frame stop codon bypass occurs via decoding of the stop codon by a near-cognate aminoacyl-tRNA. Three peptides (yellow boxes), encoded by the mRNA sequences spanning the stop codons were identified by mass spectrometry in the cells exposed to 0.5× MIC of Api. The nucleotide sequences of the 3’-proximal segments of the corresponding ORFs and amino acid sequences of the encoded proteins are shown. Stop codons and amino acids erroneously incorporated at stop codons are shown in red. The codon recognized by aminoacyl-tRNA that likely decoded the stop codon is shown in green boxes.
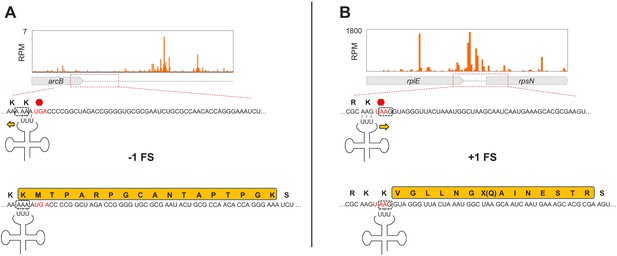
Possible scenarios for stop codon bypass via frameshifting in Api-treated cells.
(A) Top: Ribosome footprints density downstream of the arcB ORF in Api-treated cells. Bottom: Inability of the ribosome to rapidly terminate translation at the arcB UGA stop codon (red), due to Api-induced RF2 depletion, may allow for to a backward shift of the peptidyl-tRNALys occupying the last arcB sense codon AAA (boxed) within the slippery sequence, landing in an identical (−1) frame codon (boxed). (B) Top: Ribosome footprints density in the region downstream of the rplE ORF in Api-treated cells. Bottom: Slow termination of translation at the rplE UAA stop codon (red), may lead to repairing of the peptidyl-tRNALys occupying the last rplE sense codon AAG to the nearby identical codon in a (+1) frame (boxed). In (A) and (B), the amino acid sequences of the tryptic peptide identified by shotgun proteomics in cells treated with Api are highlighted in yellow. Note that the stop codon within the (+1) frame downstream of rplE is bypassed by the misincorporation of a near-cognate Gln-tRNA(UUG). Also of note, the same tryptic peptide as the one encoded in the mRNA segment downstream of the arcB gene could be also identified in untreated cells.
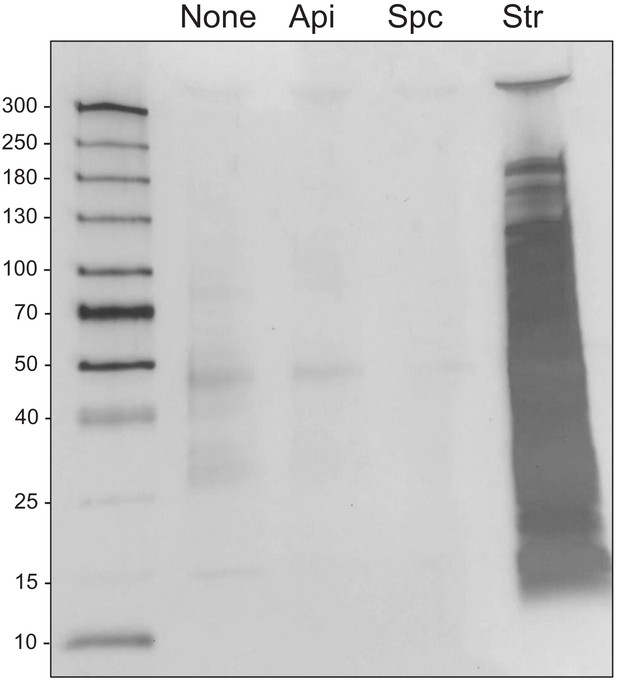
Api treatment does not lead to pronounced protein aggregation in E.
coli cells. Cells were treated or not with apidaecin (Api), spectinomycin (Spc), or streptomycin (Str) at twofold the respective MICs for 30 min. Protein aggregates were isolated, separated by SDS-PAGE, and visualized by silver staining. Spc and Str are verified positive and negative controls in these conditions (Ling et al., 2012).
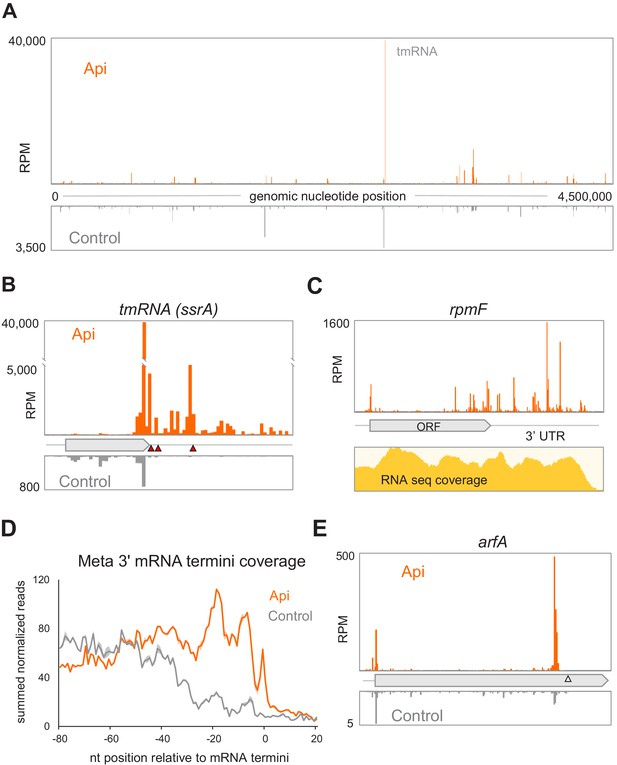
Alteration of the proteome and activation of the ribosome rescue systems in response to Api.
(A) The density of ribosome footprints throughout the E. coli genome in cells treated or not with Api. (B) The density of ribosomal footprints at the termination region of the tmRNA ORF encoding the degron tag. The position of the main stop codon and two in-frame downstream stop codons of the tmRNA are indicated with red triangles. (C) Ribosome footprints near the 3’-ends of the rpmF transcript in Api-treated cells. The RNA-seq coverage is shown below. (D) The Metagene plot of summed normalized reads toward the 3’ termini of 1006 mRNAs (Dar and Sorek, 2018; see Materials and methods). (E) The density of ribosome footprints in the arfA gene in Api-treated or control cells. The gray triangle marks the position of the RNase III cleavage site involved in the tmRNA- and ArfA-dependent regulation of arfA expression.
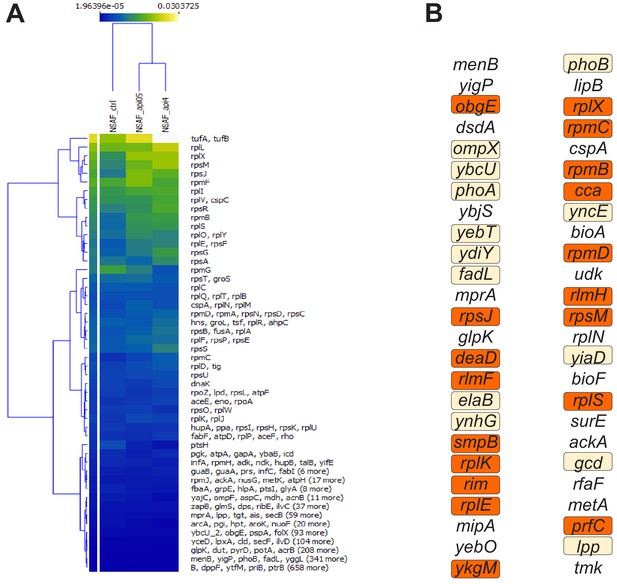
Api-induced changes in protein abundance.
(A) Api-induced changes in protein abundance. Changes in relative protein abundance in E. coli cells exposed to 0.5× MIC and 4× MIC of Api relative to the untreated control cells. The heatmap scale reflects length-adjusted fractional abundance of the protein in the total protein sample (blue – less abundant, yellow – more abundant). Genes are clustered using a hierarchical k-mean clustering algorithm. The color represents the relative abundance of each protein or protein cluster using a normalized spectral abundance factor (NSAF). (B) Top 50 proteins showing significant increase in abundance in the cells treated with 0.5× MIC of Api relative to the untreated control. Proteins related to translation are highlighted in orange. Periplasmic, inner-, and outer-membrane proteins are highlighted in beige.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21 (ΔtolC) | Meydan et al., 2019 | ||
| Strain, strain background (Escherichia coli) | BW25113 (ΔarfA) | Baba et al., 2006 | ||
| Strain, strain background (Escherichia coli) | BW25113 (ΔarfB) | Baba et al., 2006 | ||
| Strain, strain background (Escherichia coli) | BW25113 (ΔsmpB) | Baba et al., 2006 | ||
| Strain, strain background (Escherichia coli) | BW25113 | Baba et al., 2006 | ||
| Antibody | Anti-RF2 (rabbit serum polyclonal) | Cruz-Vera laboratory | (1:500) | |
| Sequence-based reagent | zntA_F | Integrated DNA Technologies | PCR primers | TAATACGACTCACTATAGGGAAACTTAACCGGAGGATGCCATGTCG |
| Sequence-based reagent | zntA_R | Integrated DNA Technologies | PCR primers | GGTTATAATGAATTTTGCTTATTAACTTTGAACGCAGCAAATTGAGGGGC |
| Sequence-based reagent | tyrS_F | Integrated DNA Technologies | PCR primers | TAATACGACTCACTATAGGGTTATATACATGGAGATTTTGATGGCAAGC |
| Sequence-based reagent | tyrS_R | Integrated DNA Technologies | PCR primers | GGTTATAATGAATTTTGCTTATTAA CGACTGGGCTACCAGCCCCCG |
| Sequence-based reagent | Linker oligonucleotide: NI-810-NI817 | McGlincy and Ingolia, 2017 | /5Phos/NNNNNATCGTAGA TCGGAAGAGCACACGTCTGAA/3ddC/ | |
| Sequence-based reagent | RT primer | Integrated DNA Technologies | /5Phos/RNAGATCGGAAGAGCGTCGTGTAGGGAAAGAG/iSp18/GTGACTGGAGTTCAGACGTGTGCTC | |
| Peptide, recombinant protein | Api137 | NovoPro Biosciences Inc | ||
| Commercial assay or kit | Oligo Clean and Concentrator kit | Zymo Research | D4060 | |
| Commercial assay or kit | MEGAclear | Thermo Fisher | AM1908 | |
| Commercial assay or kit | MicrobEXPRESS | Thermo Fisher | AM1905 | |
| Commercial assay or kit | High pH reverse phase peptide fractionation | Thermo Fisher | 84868 | |
| Chemical compound, drug | GMPPNP | Sigma | G0635 | |
| Chemical compound, drug | L-[35S]-methionine | Perkin Elmer | ||
| Peptide, recombinant protein | MNase (S7 Microccocal Nuclease) | Roche | 10107921001 | |
| Peptide, recombinant protein | DNase I, RNase free | Roche | 04716728001 | |
| Peptide, recombinant protein | SUPERase*In | Thermo Fisher | AM2696 | |
| Peptide, recombinant protein | T4 PNK | New England Biolabs | M0201L | |
| Peptide, recombinant protein | Mth RNA Ligase | New England Biolabs | E2610S | |
| Peptide, recombinant protein | 5’ Deadenylase | New England Biolabs | M0331S | |
| Peptide, recombinant protein | RecJf | New England Biolabs | M0264S | |
| Peptide, recombinant protein | Superscript III | Invitrogen | 18080–093 | |
| Peptide, recombinant protein | CircLigase ssDNA Ligase | Lucigen | CL4115K | |
| Peptide, recombinant protein | Phusion high-fidelity polymerase | New England Biolabs | M0530S | |
| Peptide, recombinant protein | MS grade modified Trypsin | Promega | V5113 | |
| Software, algorithm | Scripts for RNA-seq and Ribo-seq metagene analysis | https://github.com/adamhockenberry/ribo-t-sequencing | ||
| Software, algorithm | Scripts for Ribo-seq processing | https://github.com/mmaiensc/RiboSeq | ||
| Software, algorithm | Bedtools | https://github.com/arq5x/bedtools2 | ||
| Software, algorithm | pLogo | https://plogo.uconn.edu/ | ||
| Software, algorithm | Bowtie2 | https://github.com/BenLangmead/bowtie2 | v2.2.9 | |
| Software, algorithm | cutadapt | https://github.com/marcelm/cutadapt/ | ||
| Software, algorithm | Mass-spectrometry raw data conversion | Chambers et al., 2012 | ||
| Software, algorithm | Peptide database searching | Xu et al., 2015 | ||
| Software, algorithm | Peptide database searching | Kong et al., 2017 | ||
| Software, algorithm | Custom databases used for peptide searching | http://git.pepchem.org/gaolab/api_ribo_ext | ||
| Software, algorithm | Data analysis scripts in Python | http://git.pepchem.org/gaolab/api_ribo_ext |
Additional files
-
Source data 1
Source data from ribosome profiling analysis used in Figure 2A.
- https://cdn.elifesciences.org/articles/62655/elife-62655-data1-v1.xlsx
-
Source data 2
SC score correlations.
- https://cdn.elifesciences.org/articles/62655/elife-62655-data2-v1.csv.zip
-
Source data 3
Peptides encoded in the downstream mRNA regions found by proteomics.
- https://cdn.elifesciences.org/articles/62655/elife-62655-data3-v1.xlsx
-
Source data 4
Protein abundance in the untreated and Api-exposed cells.
- https://cdn.elifesciences.org/articles/62655/elife-62655-data4-v1.xlsx
-
Source data 5
RNA-seq gene scores.
- https://cdn.elifesciences.org/articles/62655/elife-62655-data5-v1.xlsx
-
Source data 6
Ribo-seq gene scores.
- https://cdn.elifesciences.org/articles/62655/elife-62655-data6-v1.csv.zip
-
Source data 7
Api MIC in E coli strains.
- https://cdn.elifesciences.org/articles/62655/elife-62655-data7-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62655/elife-62655-transrepform-v1.docx