5'-UTR SNP of FGF13 causes translational defect and intellectual disability
Figures
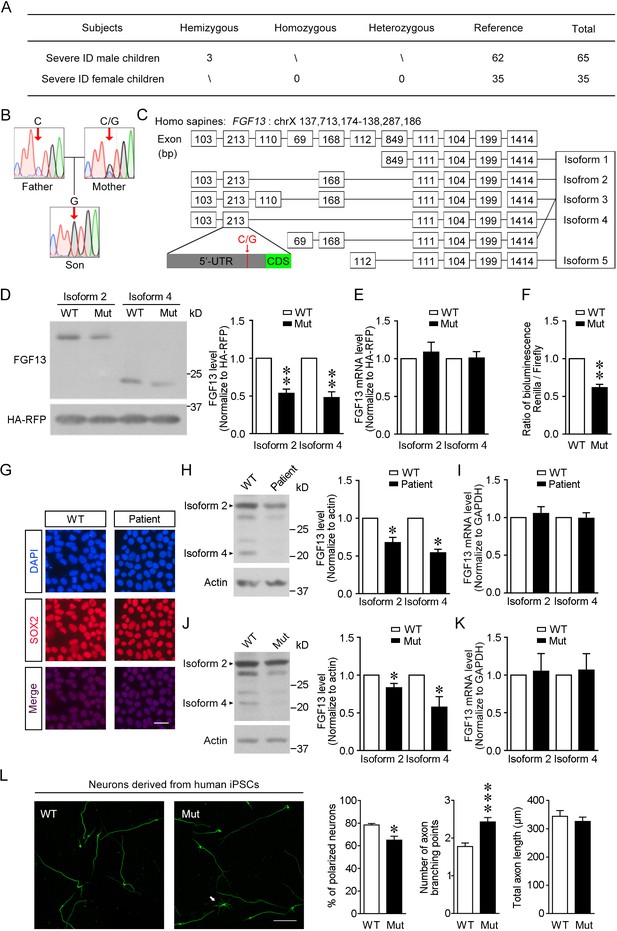
The newly identified single-nucleotide polymorphism (SNP) in FGF13 impairs the protein translation.
(A) The number of subjects involved in the present study is listed. A sum of 100 severe intellectual disability (ID) children (intelligence quotient [IQ] < 40) with detailed clinical histories were tested for the SNP of FGF13. (B) Sanger sequencing results of 1 ID boy with his parents revealed X-linked recessive inheritance model of the 5′-untranslated region (5′-UTR) SNP of FGF13. One of all three families is shown here, and in each family the son carried the SNP, while the mother showed heterozygous base in this site and the father showed normal base. (C) The point mutation (indicated by red arrow) is located at the 5′-UTR of FGF13 exon 2 and is involved in the expression of three FGF13 isoforms. (D, E) The 5′-UTR SNP of FGF13 reduced the protein levels of isoforms 2 and 4 of FGF13 in HEK293 cells. Immunoblotting results of FGF13 and HA-RFP are shown, and HA-RFP was used as a normalized control for the transfection of plasmids. The protein (D) and mRNA (E) levels of FGF13 isoforms 2 and 4 were examined, and the mutant FGF13 5′-UTR (Mut) reduced the protein levels of FGF13 isoform 2 (n = 4, p=0.0037) and isoform 4 (n = 4, p=0.0067), but did not alter their mRNA levels, compared to wildtype (WT) (isoform 2: n = 6, p=0.5364; isoform 4: n = 6, p=0.9306). (F) The activity of 5′-UTR was assessed in the dual-luciferase reporter assay, which was indicated by the luminescence ratio of Renilla and Firefly. The Mut 5′-UTR attenuated the reporter Renilla translation (n = 4, p=0.0032). (G) Representative images of DAPI and SOX2 immunostaining show the induced pluripotent stem cells (iPSCs) derived from the renal epithelial cells of ID patients and controls. Scale bar, 100 μm. (H) Immunoblotting showed that the expression of FGF13 isoforms was downregulated in the ID patient-derived iPSCs compared to WT iPSCs (isoform 2: n = 3, p=0.0437; isoform 4: n = 3, p=0.0101). Arrowheads indicate bands of FGF13 isoforms 2 and 4. (I) Quantitative real-time PCR shows that the FGF13 mRNA level was not decreased in the ID patient-derived iPSCs (isoform 2: n = 3, p=0.6136; isoform 4: n = 3, p=0.8951). (J) Immunoblotting shows that the introduction of the mutant site (C>G) in FGF13 5′-UTR in WT iPSCs could reduce the expression of FGF13 isoforms 2 and 4 (isoform 2: n = 3, p=0.0497; isoform 4: n = 3, p=0.0464). Arrowheads indicate bands of FGF13 isoforms 2 and 4. (K) Quantitative real-time PCR shows that the FGF13 mRNA level was not altered when WT iPSCs were transformed to Mut ones (isoform 2: n = 3, p=0.8527; isoform 4: n = 3, p=0.7903). (L) Immunostaining of Tuj1 (green) shows that the induced neurons derived from iPSCs carrying the 5′-UTR SNP of FGF13 displayed a lower ratio of polarized neurons (n = 3 experiments, p=0.0500) and increased axon branching points with unchanged total axon length (n = 120 and 131 for WT and Mut neurons, respectively, from three experiments, p=5.45E-05 for axon branching points and p=0.4409 for total axon length). Unpolarized neuron is labeled by white arrow in the Mut group. Scale bar, 100 μm. *p<0.05 and ***p<0.001 versus WT by Mann–Whitney U test. Data are represented as mean ± SEM (D–F, H–L). *p<0.05 and **p<0.01 versus WT by Student’s t-test (D, F, H, J).
-
Figure 1—source data 1
Source data for Figure 1.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig1-data1-v1.xlsx
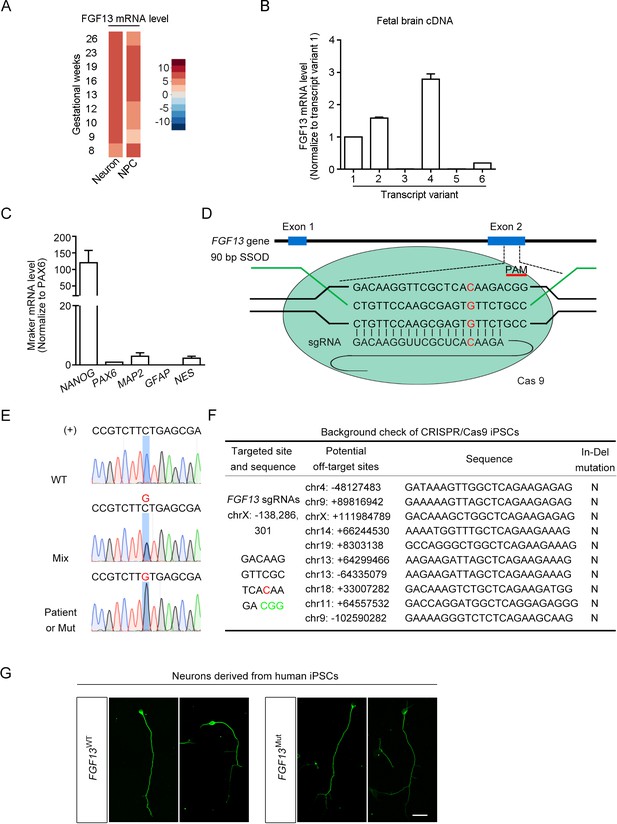
Manipulation of the FGF13 5′-untranslated region (5′-UTR) in induced pluripotent stem cells (iPSCs) with the CRISPR/Cas9 editing.
(A) The expression of FGF13 mRNA is shown through reanalysis of single-cell data from the human fetal prefrontal cortex (Zhong et al., 2018). The abundance of FGF13 mRNA in the neuron and neural progenitor cells (NPC) is shown in the heatmap. FGF13 mRNA was expressed at relatively high levels through various gestational periods in both neurons and NPCs. (B) FGF13 transcripts 2 and 4 were more enriched in the human fetal brain. Quantitative real-time PCR was used to detect all six transcript variants of FGF13 in the human fetal brain cDNAs. Data were normalized to the transcript 1 (n = 4). Notably, the newly found single-nucleotide polymorphism (SNP) located in the 5′-UTR of FGF13 was included in the transcripts 2 and 4 (see Figure 1C). (C) Detection of the iPSC marker in the iPSCs cultured from the intellectual disability (ID) patients. The iPSC marker NANOG was highly expressed, while the progenitor cell marker PAX6, the neural cell marker MAP2 and NES, and the glial marker GFAP were expressed at low levels (n = 4). (D) Schematic diagram of editing the FGF13 SNP by CRISPR/Cas9 system. The Cas9 nuclease was targeted to the FGF13 exon 2 by the 20-nt sgRNA that covered the mutant (Mut) site upstream of the CGG PAM. After the induction of DNA double-stranded break, a repair template in the form of single-stranded DNA oligonucleotide (SSOD) was used to mediate the homology-directed repair and introduce the G substitution. (E) Sanger sequencing verified the insertion of the wildtype (WT) or Mut nucleotide in FGF13. Sense strand (labeled as '+') of genomic DNA had a C or G (highlighted in red color) base at the SNP site, which indicated the genomes of WT cells from normal children, mixed cells (Mix, middle), and cells from the ID patient or Mut cells, respectively. (F) Examination of the top 10 ranked off-target sites in edited iPSCs showed no undesired insertions and deletions (In-Del) (G) Representative images of Tuj1 immunostaining show the induced neurons derived from WT and Mut iPSCs. Scale bar, 100 μm. Data are represented as mean ± SEM (B, C).
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig1-figsupp1-data1-v1.xlsx
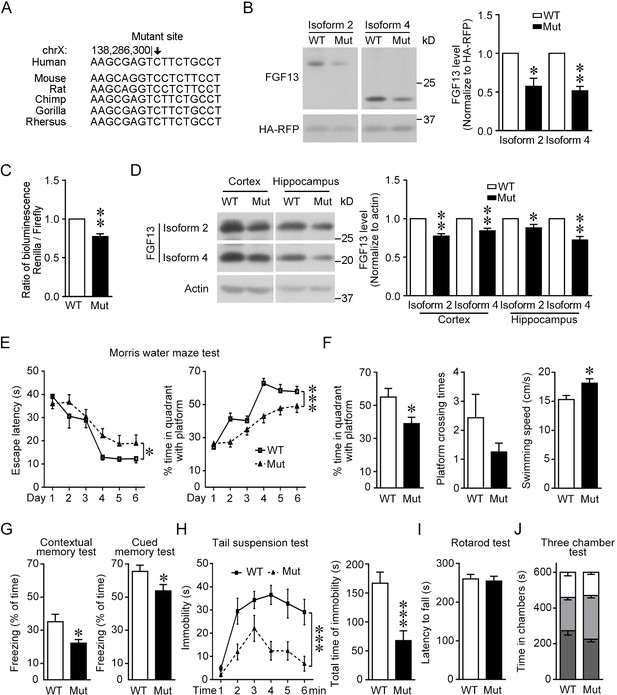
Mice with the point mutation in Fgf13 5′-untranslated region (5′-UTR) reduce protein translation and exhibit impaired learning and memory.
(A) The region around the mutation site is highly conserved among species. The mutation site is highlighted with the arrow and remains constant in various species. (B) The homology point mutation in the 5′-UTR of Fgf13 in the mutant (Mut) mouse could reduce the translation of FGF13 isoforms in HEK293 cells (isoform 2: n = 4, p=0.0263; isoform 4: n = 4, p=0.0038). The two FGF13 isoforms were immunoblotted in the same membrane but not adjacent. (C) Dual-luciferase reporter assay verified that the homologous mutation in Fgf13 5′-UTR impaired the reporter Renilla translation (n = 6, p=0.0013). (D) The expression of FGF13 isoforms was impaired in the cerebral cortex and the hippocampus of Fgf13 5′-UTR Mut mice compared to that of the wildtype (WT) mice at P0 (n = 5, p=0.0017 and p=0.0098 for cortex; n = 5, p=0.0286 and p=0.0021 for hippocampus). Bands of FGF13 isoforms 2 and 4 were immunoblotted in the same membrane. (E) The escape latency and performance of the Fgf13 5′-UTR Mut mice in Morris water maze were impaired compared to that of the WT mice during 6-day training period. The performance was indicated by the time spent in the quadrant with the platform and the escape latency to reach the platform (WT: n = 7; Mut: n = 12; left, p=0.0332; right, p=1.23E-05). *p<0.05 and ***p<0.001 versus WT by two-way ANOVA. (F) The Mut mice showed impaired spatial memory in the probe test day, which was indicated by the time spent in the quandrant with platform (p=0.0301) and the times crossing the platform position (p=0.2748 by Mann–Whitney U test). Moreover, the average swimming speed (p=0.0161) of Mut mice (n = 12) was higher than that of WT mice (n = 7). (G) Associate memory of Mut mice (n = 14) in short-term contextual or cued memory tests was impaired as compared to that of WT mice (n = 10), indicated by the freezing time in the chambers (p=0.0227 contextual and p=0.0405 for cued test). (H) Performance in the tail suspension test for Mut mice was quantitatively analyzed. The Mut mice (n = 14) showed less depression indicated by the reduced total time of immobility than WT mice (n = 10). Left, p=3.36E-10 (***) versus WT by two-way ANOVA; right, p=0.0009 (***) versus WT by Mann–Whitney U test. (I) Rotarod experiment showed that the motor coordinate function of Mut mice (n = 14) was not altered (p=0.8076 by Mann–Whitney U test) compared to that of WT mice (n = 10). (J) The results of three-chamber test showed that the social interaction of Mut mice was not altered compared to that of WT mice (WT: n = 10; Mut, n = 14; p>0.9999 by two-way ANOVA). White box stands for time in chamber of inanimate object, while gray box represents the time in center zone and dark gray box indicates the time sniffing another mouse in the chamber. Data are represented as mean ± SEM (B–J). *p<0.05 and **p<0.01 versus WT by Student’s t-test (B–D, F, G).
-
Figure 2—source data 1
Source data for Figure 2.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig2-data1-v1.xlsx
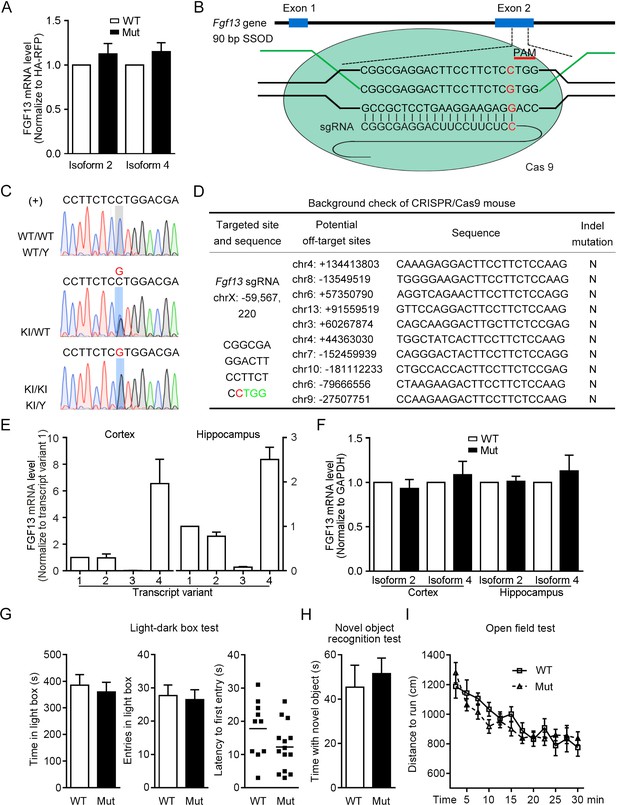
Construction of mice with the point mutation in Fgf13 5′-untranslated region (5′-UTR) using CRISPR/Cas9 system.
(A) The FGF13 mRNA level was not changed in HEK293 cells expressing the mutant (Mut) 5′-UTR of Fgf13 compared with that of the wildtype (WT) one (n = 5, p=0.3489 and p=0.2096 for isoforms 2 and 4). (B) Schematic diagram of the construction for Fgf13 point mutant mice with the CRISPR/Cas9 system. After the pSpCas9 plasmid was expressed in the mouse fertilized eggs, the Cas9 nuclease targeted to the gene by the 20-nt sgRNA that covered the mutant site upstream of the TGG PAM. A repair template in the form of single-stranded DNA oligonucleotide (SSOD) was used to mediate the homology-directed repair and introduce the mutation. (C) Sanger sequencing identification of F1 in the Fgf13 Mut mice verified the insertion of the mutant nucleotide (knock-in [KI]) in the mouse genome. Sense strand (labeled as '+') of genomic DNA had a G (highlight in red color) or C base at the point mutation site, which represents the genomes of WT mice (WT/WT and WT/Y, upper), heterozygous mice (KI/WT, middle), and Mut mice (KI/KI and KI/Y, lower). (D) Examination of the top 10 ranked off-target sites in the mutant mice showed no undesired insertions and deletions (In-Del) mutations. (E) The mRNA levels of various Fgf13 transcripts in the mouse cerebral cortex and hippocampus were examined at P0. Fgf13 transcripts 2 and 4 were highly expressed in both the cortex and the hippocampus, normalized to transcript 1 (cortex: n = 4, hippocampus: n = 6). (F) The level of FGF13 mRNA was not changed in the cerebral cortex and hippocampus from Mut mice at P0 (cortex: n = 10, p=0.5115 and 0.5810 for isoforms 2 and 4; hippocampus: n = 5, p=0.8375 and 0.5050 for isoforms 2 and 4). (G) The results of light-dark box test showed that the anxiety of Mut mice (n = 14) was not altered compared to that of WT mice (n = 10). The time staying in the light box (p=0.6494), entries in light box (p=0.7739), and the latency of first entry in light box (p=0.1178) was examined, respectively. (H) In the novel object recognition test, the time spent in sniffing new object was not significantly changed between WT mice (n = 10) and Mut mice (n = 14; p=0.6208). (I) Records of the traveling distance within 30 min in the open field test showed that the locomotion of Mut mice (n = 14) was similar to that of WT mice (n = 10; p=0.3273 by two-way ANOVA). Data are represented as mean ± SEM (A, E–I). p-values are calculated between Mut and WT by Student’s t-test (A, E–H).
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig2-figsupp1-data1-v1.xlsx
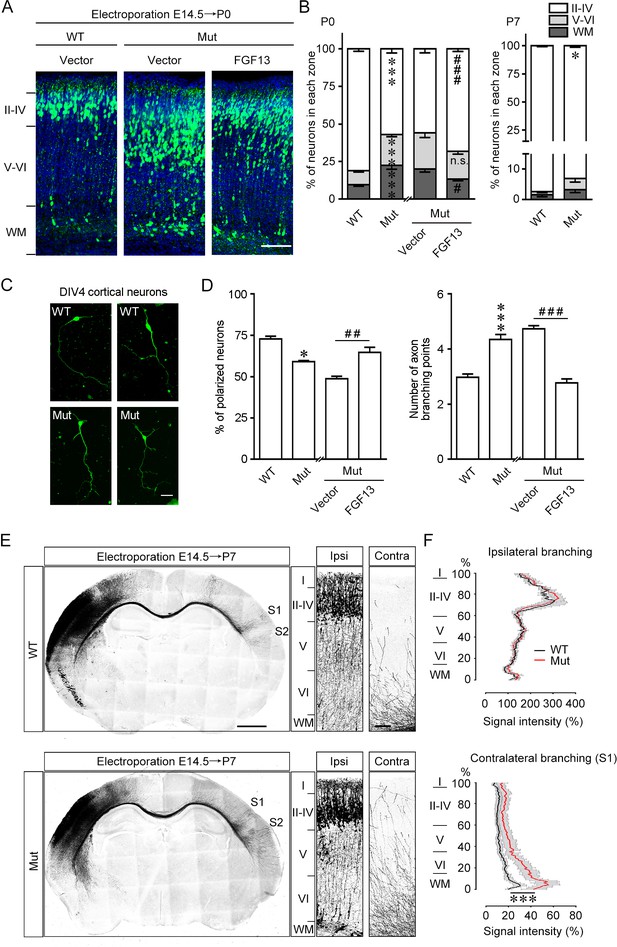
Fgf13 point mutation mice show delayed radial migration and increased axon branching.
(A, B) Representative images and quantitative analysis of the cerebral cortex at P0 in Fgf13 point mutant (Mut) mice electroporated with pcDNA3.1 vector at E14.5 showed that the radial migration of cortical neurons was delayed, and such a delay of neuronal migration was sustained till P7, compared to that of wildtype (WT) mice. Re-expression of FGF13 could rescue the migratory defects. Left of (B), n = 15, 19, 26, and 31 brain slices from 3 to 5 WT or Mut littermate mice. Layers II–IV: p=3.23E-08 (***) versus WT, while p=0.0005 (###) versus Mut plus vector; layers V–VI: p=5.47E-07 (***) versus WT, while p=0.3193 (n.s.: not significant) versus Mut plus vector; WM: p=0.0006 (***) versus WT, while p=0.0121 (#) versus Mut plus vector. Right of (B), n = 9 and 12 brain slices from 3 to 4 WT or Mut littermate mice. Layers II–IV: p=0.0271 (*) versus WT; layers V–VI: p=0.2388; WM: p=0.2342. Scale bar, 100 μm (A). All statistical analyses were performed by Mann–Whitney U test. (C, D) DIV4 cortical neurons cultured from Mut mice showed lower ratio of polarized morphology and displayed more axon branches compared to that from WT mice (n = 179 neurons for WT and 160 neurons for Mut from three experiments). Neurons were immunostained with Tau. Such changes were rescued by re-expression of FGF13 (n = 295 neurons for Mut plus vector and 127 neurons for Mut plus FGF13 from five experiments). p=0.0500 (*) and p=8.78E-09 (***) versus WT, while p=0.0040 (##) and p<1.00E-15 (###) versus indicated group by Mann–Whitney U test. Scale bar, 50 μm (C). (E) Representative images of P7 brain slices in mice electroporated with green fluorescent protein (GFP) plasmid at E14.5 show the distribution of axon projection in the cerebral cortex. Right panels show magnification of bilateral primary somatosensory cortex. The Mut mice showed that the axon arbors were more complex, and the axons could project to both the contralateral S1 and S2 cortical regions, while in WT mice the axons mainly reached the contralateral S1 cortical region. Scale bars, 1000 (left) and 100 (right) μm. (F) Quantitative analysis of axon arbors of bilateral somatosensory cortex showed increased axon branching in the contralateral hemisphere in Mut mice. The axon intensity in the ipsilateral layer V was used for normalization. The Mut mice showed more complex and intense axon projections in the contralateral side (n = 5 brain slices from three WT mice, and n = 8 slices from three Mut littermate mice). p<1.00E-15 (***) versus WT in contralateral branching by two-way ANOVA. Data are represented as the mean ± SEM (B, D, F).
-
Figure 3—source data 1
Source data for Figure 3.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig3-data1-v1.xlsx
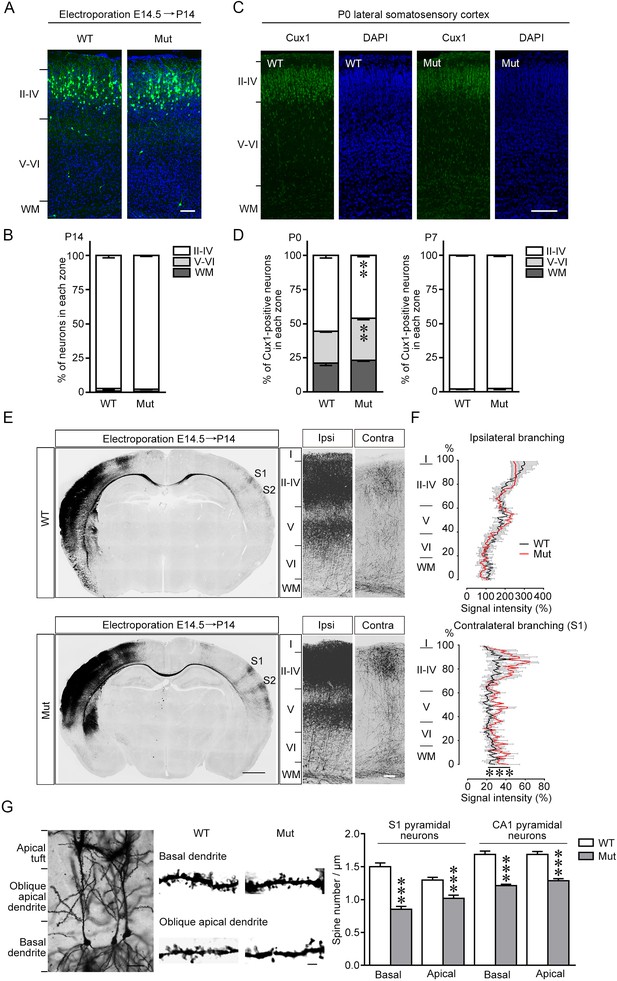
Alterations of neuronal migration, axon projection, and spine density in the brain of Fgf13 5′-untranslated region (5′-UTR) mutant (Mut) mice.
(A, B) The representative images of the cerebral cortex of Fgf13 5′-UTR Mut mice at P14, electroporated with the plasmid expressing the green fluorescent protein (GFP) at E14.5, show that the radial migration of cortical neurons was similar to that of wildtype (WT) mice, suggesting that the migratory defects at P0 (Figure 4A) were compensated during late phase (n = 4 brain slices from four WT and four Mut littermate mice). Scale bar, 100 μm. (C, D) Images of the somatosensory cortex of Mut mice at P0 show the disorganized cortical layers. Brain slices were immunostained for Cux1 (green) and DAPI (blue), and many Cux1-positive neurons remained in the deep cortical layers in Mut mice at P0, while the cortical lamination of Mut mice was similar to that of WT mice at P7 (n = 5 and 6 brain slices from five WT and six Mut littermate mice for P0, and n = 4 brain slices from four WT and four Mut littermate mice for P7). For P0: p=0.0043 (**), p=0.0043 (**), and p=0.1775 for layers II–IV, V–VI, and WM by Mann–Whitney U test. For P7: p=0.8857, p=0.8857, and p>0.9999 for layers II–IV, V–VI, and WM by Mann–Whitney U test. Scale bar, 100 μm. (E, F) Representative images of the slices of mouse brain at P14, electroporated with the GFP plasmid at E14.5, show the distribution of axon projection in the somatosensory cortex. Right panel of (E) shows the higher magnification of bilateral primary somatosensory cortex. Mut mice showed more complex axon arbors that could project to more superficial cortical layers on the contralateral side compared to WT mice (n = 3 brain slices from three WT and four Mut littermate mice). p<1.00E-15 (***) by two-way ANOVA. Scale bars, 1000 (left) and 100 (right) μm. (G) Representative images of adult mouse CA1 pyramidal neurons after Golgi staining show typical dendritic structure. Dendritic spine density from apical oblique and basal dendrites were quantified, and Mut mice had decreased spine density in both basal and apical dendrites of primary somatosensory cortex (S1) layer V and CA1 pyramidal neurons (n = 21–31 dendrites from two WT and 35–51 dendrites from three Mut littermate mice for each group). p=9.61E-12, p=5.65E-05, p=8.61E-10, and p=1.23E-08, respectively, by Student’s t-test. Scale bars, 30 (left) and 2 (right) μm. Data are represented as mean ± SEM (B, D, F, G).
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig3-figsupp1-data1-v1.xlsx
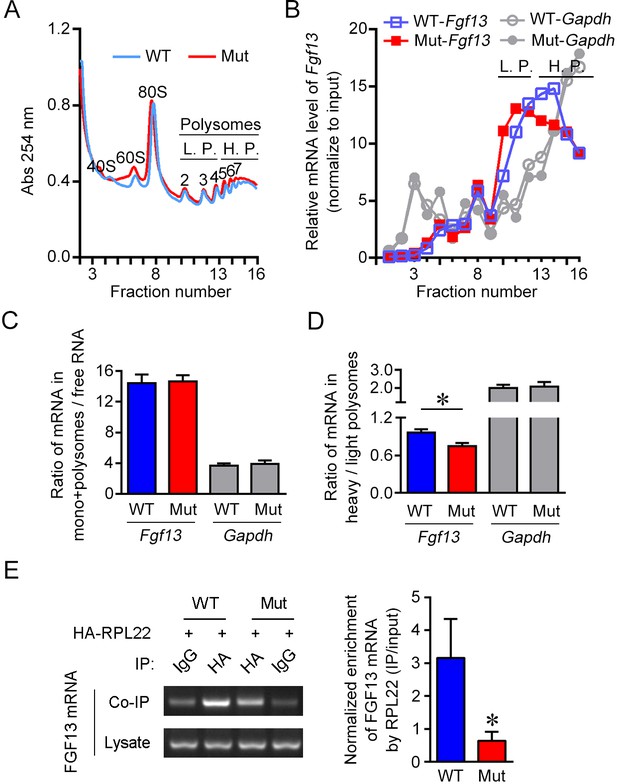
The 5′-untranslated region (5′-UTR) point mutation impairs association of FGF13 mRNA with ribosomes.
(A) The overall mRNA levels in the brain of wildtype (WT) or mutant (Mut) mouse revealed by UV absorption profiles (254 nm). A total of 16 fractions were collected from the sucrose density gradients. Components of ribosome subunits (40S, 60S), monosome (80S) and polysomes are indicated in the curve. The light polysomes (L.P.) represent 2–4 polysomes, and heavy polysomes (H.P.) represent five and more polysomes. (B) Polysome profiles show the association level of FGF13 mRNA and GAPDH mRNA with ribosomes in the Mut mouse compared with the WT mouse. The distribution of FGF13 mRNA and GAPDH mRNA in different fractions after normalization to the input is shown here. The level of FGF13 mRNA in the heavier polysome fractions of Mut mouse was lower than that of WT mouse, while the level of GAPDH mRNA was not altered. (C, D) The ratio of FGF13 or GAPDH mRNA in monosomes plus polysomes to free RNA in Mut mice was not changed (WT: n = 6 mouse brains; Mut: n = 6 mouse brains; p=0.9372 for FGF13 and p=0.8182 for GAPDH by Mann–Whitney U test). The ratio of FGF13 mRNA, but not GAPDH mRNA, in the heavy polysome versus that in the light polysome of Mut mice was lower than that of WT mice (WT: n = 6 mouse brains; Mut: n = 6 mouse brains; p=0.0260 [*] for FGF13 and p=0.9372 for GAPDH by Mann–Whitney U test). (E) The 5′-UTR point mutation impaired the binding of FGF13 mRNA with translating complex. RNA immunoprecipitation (RIP) assay was used to examine the ribosome-binding mRNAs, in which FGF13 mRNA enriched in ribosomes was detected through IP with antibody against HA in HEK293 cells expressing HA-RPL22 with WT or Mut mouse FGF13 isoform 2. Representative PCR results are shown in the left panel, and quantitative real-time PCR shows a reduced enrichment of Mut FGF13 mRNAs with HA-RPL22 (n = 5, p=0.0355). Data are represented as the mean ± SEM (C–E). *p<0.05 versus WT by Student’s t-test (D, E).
-
Figure 4—source data 1
Source data for Figure 4.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig4-data1-v1.xlsx
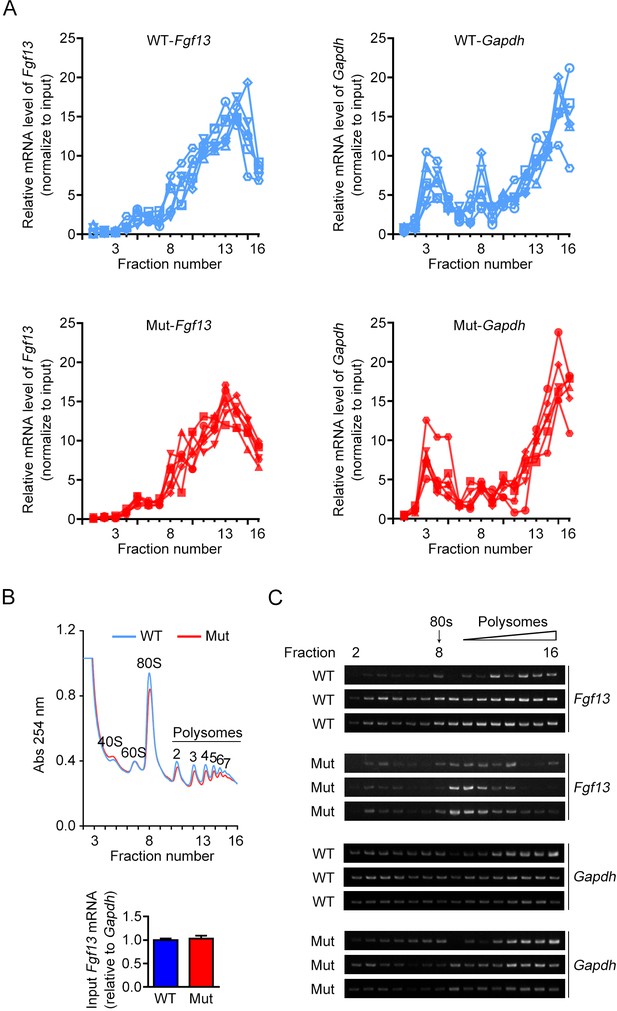
The association of FGF13 mRNA with heavy polysomes is decreased in Fgf13 5′-untranslated region (5′UTR) mutant mice.
(A) Polysome profiles from brains of six wildtype (WT) and six mutant (Mut) mice (for analysis in Figure 4C, D) are shown. The distribution of FGF13 mRNA and GAPDH mRNA in different fractions was normalized to the input. (B, C) The brains of WT or Mut mice were collected and used for polysome profiling analysis. Overall mRNA levels revealed by UV absorption profiles (254 nm) in different fractions of the sucrose density gradients are shown, and different components are indicated in the curve. The mRNA levels of Fgf13 in the input of Mut mice were not changed by quantitative real-time PCR (WT: n = 3 mouse brains; Mut: n = 3 mouse brains; p>0.9999 by Mann–Whitney U test). Polysomal distribution of indicated mRNAs was determined by semi-quantitative PCR, and each replicate is shown. Fgf13 was preferentially distributed in lighter polysomal fractions in Mut mice compared to WT mice, while the positions of Gapdh were not affected.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig4-figsupp1-data1-v1.xlsx
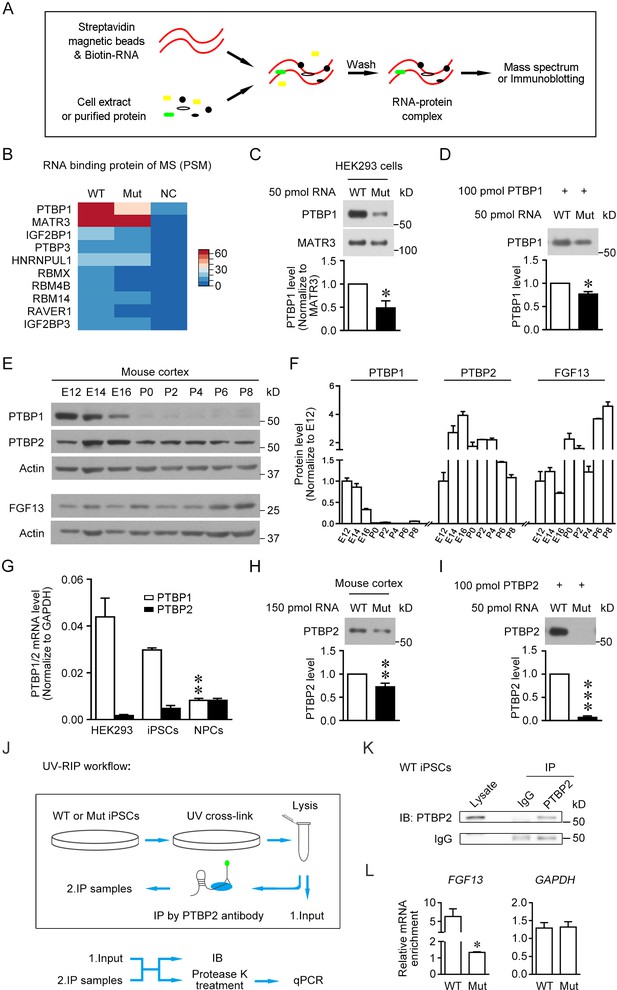
The 5′-untranslated region (5'-UTR) point mutation impairs the binding of PTBP1/2 with FGF13 5′-UTR.
(A) Schematic graph shows the procedure of RNA pull-down assay. Biotin-labeled synthesized RNA was perfused with cell extracts or purified protein and enriched in streptavidin magnetic beads that were further eluted for mass spectrum or immunoblotting detection. (B) Top 10 ranked liquid chromatography tandem-mass spectrometry (LC-MS/MS) results of pull-downed proteins with synthesized RNAs of wildtype (WT) FGF13 5′-UTR and mutant (Mut) FGF13 5′-UTR are shown in heatmap. PTBP1 and MATR3 had fundamental enrichment with the WT RNA, but the Mut RNA selectively abolished its binding with PTBP1. NC indicates negative control without RNA in pull-down experiment. (C) Immunoblotting confirmed the decreased binding affinity of synthesized Mut RNA with endogenous PTBP1 but not MATR3 in HEK293 cells (n = 3, p=0.0399). (D) Immunoblotting shows the direct binding between purified PTBP1 (100 pmol) and synthesized FGF13 RNA (50 pmol). The mutation decreased its binding level with PTBP1 (n = 3, p=0.0107). (E, F) Immunoblotting shows the change of FGF13, PTBP1, and PTBP2 in the mouse cerebral cortex at various developmental stages. FGF13 (isoform 2) was increased persistently from E14 to P8. PTBP1 was continuously decreased from E12 to P0, and largely absent at P0–P8. The PTBP2 expression reached the peak at E14–E16, then decreased and kept at certain levels at P0–P8 (n = 2–4 mice at each stage). (G) The mRNA level of PTBP1 and its neuronal paralog PTBP2 were examined in HEK293 cells, human induced pluripotent stem cells (iPSCs), and induced neural progenitor cells (NPCs). PTBP2 was specifically enriched in neural cells in which PTBP1 was decreased (n = 3 for iPSCs and n = 4 for HEK293 and NPCs; NPCs versus iPSCs: p=0.0034 [**] for PTBP1, p>0.9999 for PTBP2, by two-way ANOVA followed with Bonferroni's multiple comparisons test). (H) Immunoblotting showed that the homologous point mutation in Fgf13 5′-UTR hampered the binding of mouse PTBP2 with synthesized Mut RNA of the Fgf13 5′-UTR. Biotin-labeled synthesized RNAs were incubated with the mouse cerebral cortex extract to pull-down binding proteins (n = 10, p=0.0027). (I) Immunoblotting shows the decreased binding affinity between purified PTBP2 (100 pmol) and synthesized Mut FGF13 RNA (50 pmol). The point mutation almost totally abolished their interaction (n = 3, p=0.0006). (J) Flowchart of ultraviolet -RNA immunoprecipitation (UV-RIP) experiments in WT or Mut iPSCs. (K) Immunoblotting showed that PTBP2 was immunoprecipitated by PTBP2 antibody. (L) Quantitative real-time PCR shows that the abundant FGF13 in PTBP2 immunoprecipitates of WT iPSCs was largely decreased compared with that of Mut iPSCs (n = 3, p=0.0204). GAPDH was not apparently enriched and used as control (n = 3, p=0.4581). The mRNA enrichment was defined as the ratio of immunoprecipitated sample to input sample and normalized to corresponding IgG group. Data are represented as mean ± SEM (C, D, F, G–I, L). *p<0.05, ***p<0.01, and ***p<0.001 versus WT by Student’s t-test (C, D, H, I, L).
-
Figure 5—source data 1
Source data for Figure 5.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig5-data1-v1.xlsx
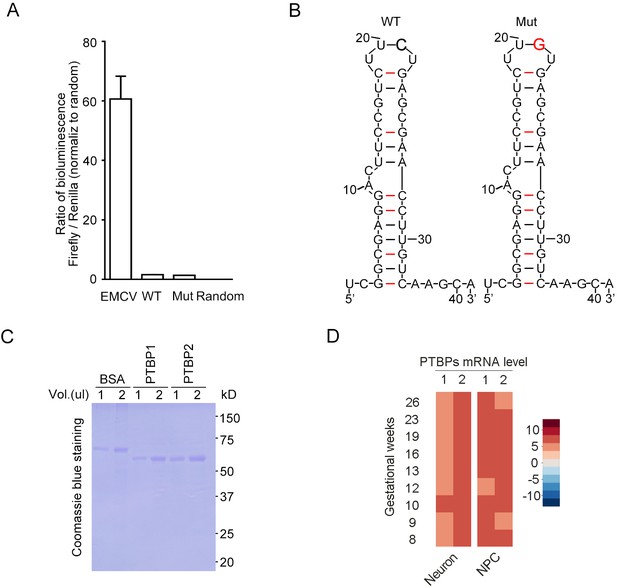
The FGF13 5′-untranslated region (5′-UTR) single-nucleotide polymorphism (SNP) does not induce the alteration of internal ribosome entry site (IRES) entry and mRNA structure.
(A) Dual-luciferase assay was used to test the IRES activity of FGF13 5′-UTR. The known IRES sequence (EMCV) in the pRIF plasmid was used as a positive control, and the wildtype (WT) or mutant (Mut) sequence of FGF13 5′-UTR was inserted to replace EMCV sequence. Both WT and Mut 5′-UTR could not exhibit a robust reporter expression (luminescence ratio of Firefly to Renilla, which is opposite to traditional Renilla/Firefly due to the plasmid construct sequence). Random indicates the random reverse complement sequence of EMCV sequence (n = 4). (B) The 5′-UTR SNP of FGF13 did not alter the secondary structure of mRNA by prediction. The 41 bp RNA sequence from the FGF13 5′-UTR was used to predict the secondary structure on website (see Materials and methods), and Mut 5′-UTR had the same structure to the WT one. The C>G substitution is highlighted with red color and bold text in the upper part. (C) Coomassie blue staining shows the purified protein of PTBP1 and PTBP2 used in Figure 5D/H. First two lanes were loaded with 1 and 2 μg of BSA (1 μg/μL) as the quantification standard. (D) The mRNA levels of PTBP1 and PTBP2 are shown in the heatmap generated from the single-cell sequencing data of the fetal prefrontal cortex in human brain (Zhong et al., 2018). The expression level of PTBP2 was higher than that of PTBP1 in the neurons of human brain. Data are represented as mean ± SEM (A).
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig5-figsupp1-data1-v1.xlsx
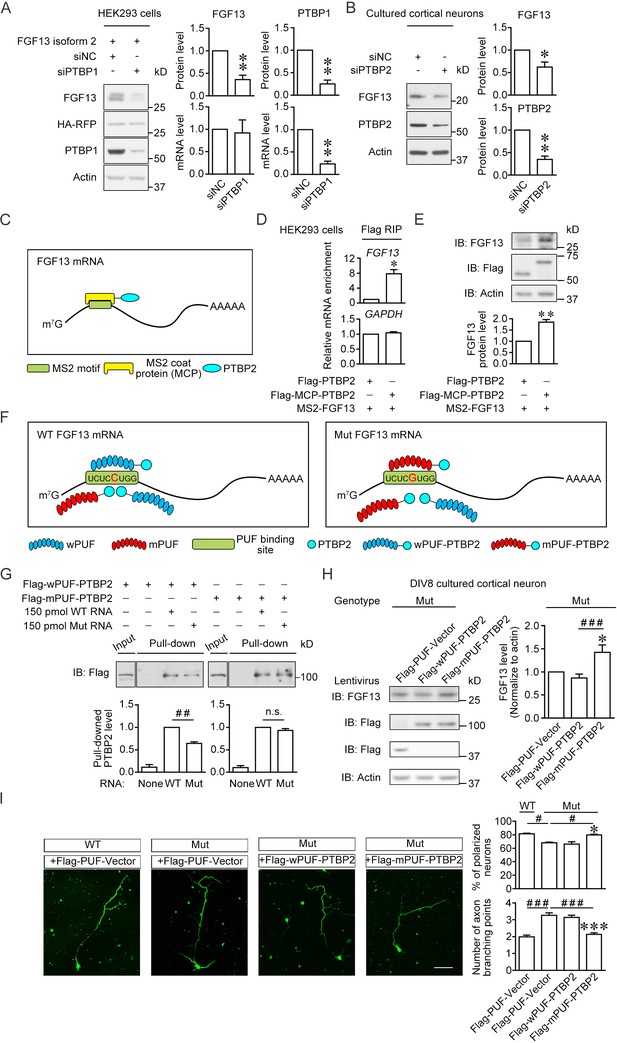
Enhancement of the PTBP2 binding with mutant (Mut) Fgf13 5′-untranslated region (5′-UTR) rescues the impairment of neuronal morphogenesis.
(A) Immunoblotting showed that the PTBP1 knockdown reduced the translation of co-expressed human FGF13 isoform 2 in HEK293 cells (n = 4, p=0.0077 for FGF13, p=0.0032 for PTBP1). The mRNA level of FGF13 remained unchanged when PTBP1 was silenced (n = 3, p=0.8100 for FGF13, p=0.0066 for PTBP1). (B) Immunoblotting showed that the PTBP2 knockdown reduced the FGF13 (isoform 4) expression in cultured mouse neurons (n = 4, p=0.0425 for FGF13, p=0.0018 for PTBP2). Three different small interference RNAs (siRNAs) targeting PTBP2 were mixed to enhance the knockdown efficiency in neurons. (C) Schematic graph shows that MCP-fused PTBP2 is recruited to the MS2-inserted FGF13 5′-UTR. (D) Quantitative real-time PCR showed that exogenous MS2-FGF13 mRNA exhibited an increased binding to Flag-MCP-PTBP2, after RNA immunoprecipitation (RIP) by Flag beads in HEK293 cells (n = 3, p=0.0262), while GAPDH mRNA was not enriched (n = 3, p=0.3807). (E) Immunoblotting showed that MS2-PTBP2 overexpression increased the protein level of FGF13 in HEK293 cells compared to PTBP2 (n = 4, p=0.0054). (F) Schematic graph shows that engineered PTBP2 protein with PUF scaffold recognizes 8 nt RNA sequence and is recruited to the Fgf13 5′-UTR. Due to the difference in designed recognition RNA sequence (the mutation site of Fgf13 is highlighted in red color), wPUF recognizes and binds to wildtype (WT) 5′-UTR, while mPUF binds to mutant (Mut) 5′-UTR. However, the PTBP2 within fused proteins normally binds to WT Fgf13 5′-UTR but has a lower binding affinity with Mut Fgf13 5′-UTR. (G) Immunoblotting showed that wPUF-PTBP2 with Flag tag at the N-terminus had lower binding level with synthesized Mut RNA compared to synthesized WT RNA (n = 3, p=0.0099), but mPUF-PTBP2 reversed the binding difference between synthesized WT and Mut RNAs (n = 3, p=0.2386). Proteins of input were simultaneously immunoblotted. ##p<0.01 and not significant (n.s.) versus indicated group by Student’s t-test. (H) The engineered PTBP2 with PUF scaffold could increase the FGF13 expression in cultured neurons of Mut mice. Lentivirus expressing PUF-Vector, wPUF-PTBP2, and mPUF-PTBP2 were used to transfect cortical neurons cultured from Mut mice. Immunoblotting showed that the FGF13 expression was increased by mPUF-PTBP2, but not by wPUF-PTBP2 (n = 8). p=0.0357 (*) versus Flag-PUF-Vector and p=0.0008 (###) versus indicated group by Student’s t-test. (I) Neurons from the Mut mice exhibited a lower ratio of polarized neurons with single axon and an increase in axon branching compared to that of WT mice. Introducing wPUF-PTBP2 did not alter the aberrant neuronal morphogenesis, but mPUF-PTBP2 could increase the ratio of polarization and decrease the axon branching. Neurons were immunostained with axon marker SMI-312. n = 100, 92, 78, and 95 neurons for four groups, respectively, from three experiments. For polarization ratio: p=0.0500 (*) and p=0.0500 (*) versus Flag-wPUF-PTBP2, while p=0.0500 (#) versus indicated group by Mann–Whitney U test. For axon branching: p=1.67E-10 (***) and p=1.88E-08 (***) versus Flag-wPUF-PTBP2, while p=3.65E-09 (###) versus indicated group by Mann–Whitney U test. Scale bar, 50 μm. Data are represented as mean ± SEM (A, B, D, E, G–I). *p<0.05 and **p<0.01 by Student’s t-test (A, B, D and E).
-
Figure 6—source data 1
Source data for Figure 6.
- https://cdn.elifesciences.org/articles/63021/elife-63021-fig6-data1-v1.xlsx
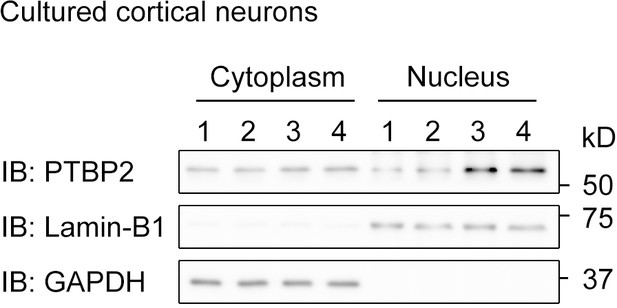
The subcellular distribution of PTBP2 in mouse cortical neurons at DIV4.
Immunoblotting with antibody against PTBP2 detected bands in both nuclear and cytoplasmic components, which are marked by Lamin-B1 and GAPDH, respectively.
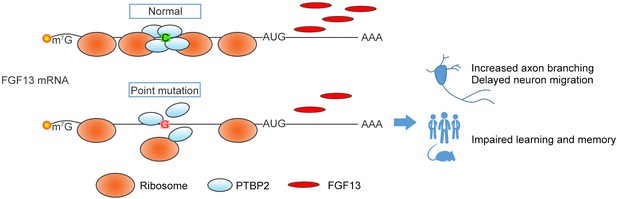
A proposed model of the FGF13 5′-untranslated region (5′-UTR) single-nucleotide polymorphism (SNP) in attenuating protein translation and impairing brain development.
In normal situation, PTBP2 binds to the 5′-UTR of FGF13 and recruits the ribosomes to initiate the protein translation. However, the C>G mutation in the FGF13 5′-UTR of intellectual disability (ID) patient impairs the PTBP2 binding, attenuates the ribosome association and translating complex recruitment, and therefore causes the decrease in FGF13 protein synthesis. During brain development, the FGF13 reduction results in increased axon branching and delayed neuronal migration, and finally leads to the cognitive impairment in both ID patients and corresponding mouse models.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) male and female | Fgf13 mutant | This paper | Home-made | Obtained by contacting the correspondence author |
| Antibody | Anti-FGF13(goat polyclonal) | Santa Cruz | Cat# sc-16811 RRID:AB_2104044 | IB (1:500) |
| Antibody | Anti-FGF13(rabbit polyclonal) | Wu et al., 2012 | Home-made by immunization with FGF13B 182–192 aa | IB (1:5000) |
| Antibody | Anti-HA(mouse monoclonal) | Sigma | Cat# H3663 RRID:AB_262051 | IB (1:2000); IP (1 μg) |
| Antibody | Anti-Flag(mouse monoclonal) | Sigma | Cat# F3165 RRID:AB_259529 | IB (1:10,000) |
| Antibody | Anti-actin(mouse monoclonal) | Chemicon | Cat# MAB1501 RRID:AB_2223041 | IB (1:400,000) |
| Antibody | Anti-MATR3(rabbit monoclonal) | Abcam | Cat# ab151739 RRID:AB_2885091 | IB (1:2000) |
| Antibody | Anti-PTBP1(mouse monoclonal) | This paper | Home-made from ATCC Cat# CRL-2501 | Source: RRID:CVCL_L596; IB (1:2000) |
| Antibody | Anti-PTBP2(mouse monoclonal) | Abcam | Cat# ab57619 RRID:AB_2284865 | IB (1:500); IP (4 μg) |
| Antibody | Anti-Lamin B1(mouse monoclonal) | Proteintech | Cat# 66095-1-Ig RRID:AB_2721256 | IB (1:5000) |
| Antibody | Anti-SOX2(rabbit polyclonal) | Stem Cell Technologies | Cat# 60055 RRID:N/A | ICC (1:100) |
| Antibody | Anti-Tuj1(mouse monoclonal) | Chemicon | Cat# CBL412 RRID:AB_11205398 | ICC (1:1000) |
| Antibody | Anti-Tuj1(chicken polyclonal) | Abcam | Cat# ab107216 RRID:AB_10899689 | ICC (1:1000) |
| Antibody | Anti-GFP(chicken polyclonal) | Abcam | Cat# ab13970 RRID:AB_300798 | ICC/IHC (1:2000) |
| Antibody | Anti-SMI-312(mouse monoclonal) | BioLegend | Cat# 837904 RRID:AB_2566782 | ICC (1:1000) |
| Antibody | Anti-Cux1(rabbit polyclonal) | Santa Cruz | Cat# sc-13024 RRID:AB_2261231 | IHC (1:1000) |
| Recombinant DNA reagent | Plasmids | This paper | Supplementary file 6 | |
| Sequence-based reagent | Oligonucleotides, primers | This paper | Supplementary file 5 | |
| IB: immunoblotting; IP: immunoprecipitation; IHC: immunohistochemistry; ICC: immunocytochemistry. | ||||
Additional files
-
Supplementary file 1
List of 262 reported intellectual disability-related genes from database and literature screening.
- https://cdn.elifesciences.org/articles/63021/elife-63021-supp1-v1.docx
-
Supplementary file 2
List of single-nucleotide polymorphisms and insertions and deletions of three families from exon-capture-sequencing.
- https://cdn.elifesciences.org/articles/63021/elife-63021-supp2-v1.docx
-
Supplementary file 3
List of single-nucleotide polymorphisms, insertions and deletions, and copy number variations of three children from whole-genome sequencing.
- https://cdn.elifesciences.org/articles/63021/elife-63021-supp3-v1.docx
-
Supplementary file 4
Clinical records of three intellectual disability individuals.
- https://cdn.elifesciences.org/articles/63021/elife-63021-supp4-v1.docx
-
Supplementary file 5
Primers and oligonucleotides used for real-time PCR, Cas9 targeting, sequencing, plasmid construction, and biotin probe.
- https://cdn.elifesciences.org/articles/63021/elife-63021-supp5-v1.docx
-
Supplementary file 6
Antibodies, reagents, cell lines, plasmid constructs, software, and other resources used in this study.
- https://cdn.elifesciences.org/articles/63021/elife-63021-supp6-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63021/elife-63021-transrepform-v1.docx





