Concerted conformational dynamics and water movements in the ghrelin G protein-coupled receptor
Figures
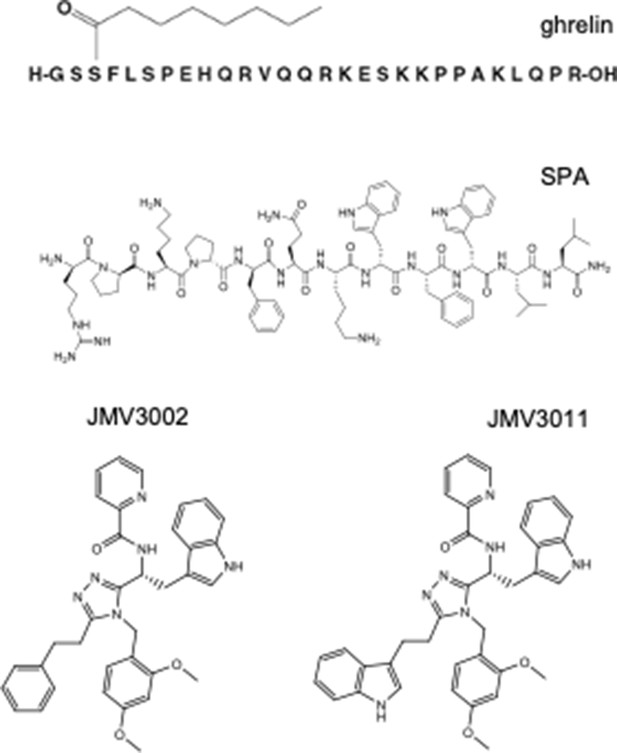
Growth hormone secretagogue receptor (GHSR) labeling.
(A) Position of the labeled residues in GHSR sequence. Red labeling indicates positions that were deleterious to GHSR expression and/or function. Green labeling indicates positions that did not markedly affect the pharmacological properties of the isolated receptor and were considered in the present work. (B) FRET-monitored competition assays of ghrelin for binding to GHSR assembled into nanodiscs. (C) GTP turnover for Gq catalyzed by GHSR and its labeled counterparts in the absence of ligand (apo) or in the presence of 10 µM of JMV3011, ghrelin, JMV3002, or SPA (substance-P analog). (D) Normalized emission spectrum of the apo wild-type and labeled GHSR with λexc set at 320 nm. Data in (B) and (C) is the mean value ± SD of three experiments. Statistical analyses for the data in (C) are provided in Figure 1—figure supplement 3.
-
Figure 1—source data 1
HTRF ratio for GHSR and its mutants.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Luminescence values for the GTP turnover assay.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig1-data2-v1.xlsx
-
Figure 1—source data 3
GHSR emission intensity.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig1-data3-v1.xlsx
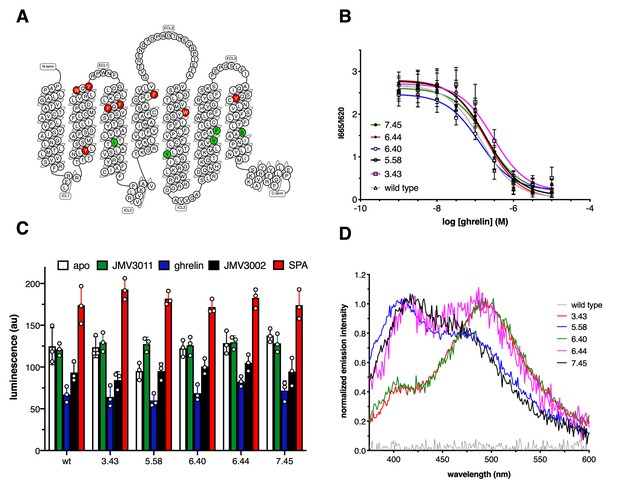
The growth hormone secretagogue receptor (GHSR)-containing nanodiscs.
Size-exclusion profile of the GHSR-containing nanodiscs. The nanodiscs were run on an S200 increase column (10×300) using a 25 mM HEPES, 150 mM NaCl, 0.5 mM EDTA, pH 7.5 buffer as the eluent and a 0.2 mL/min flow rate. Inset: SDS-PAGE profile of the nanodiscs. The fractions eluted from the main peak were pooled as indicated and run on a 15% polyacrylamide-0.1% SDS gel with Coomassie blue staining.
-
Figure 1—figure supplement 1—source data 1
SDS-PAGE of the GHSR-containing nanodiscs.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig1-figsupp1-data1-v1.xlsx
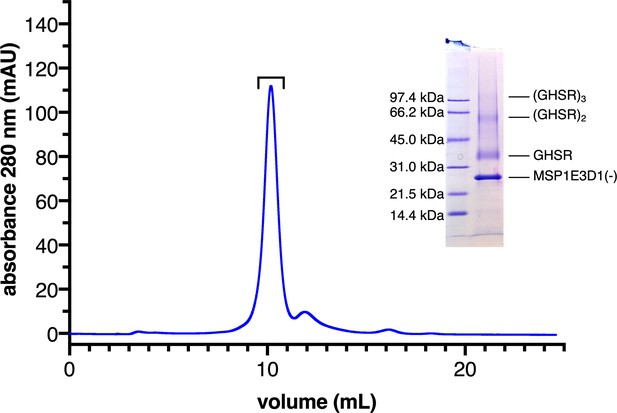
Control for the ligand-binding properties of the isolated growth hormone secretagogue receptor (GHSR) in nanodiscs.
The emission intensity of the dy647 acceptor at 665 nm and that of the Lumi4-Tb donor at 620 nm were measured after excitation of the donor at 337 nm. A 10−7 M concentration of fluorescent ghrelin was used, which is in the same range than that used in the competition plot in Figure 1B. For this assay, the Lumi4-Tb donor was attached to the N-terminus of either GHSR or the leukotriene B4 receptor BLT1 inserted into nanodiscs.
-
Figure 1—figure supplement 2—source data 1
HTRF ratio for GHSR- and BLT-containing discs.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig1-figsupp2-data1-v1.xlsx
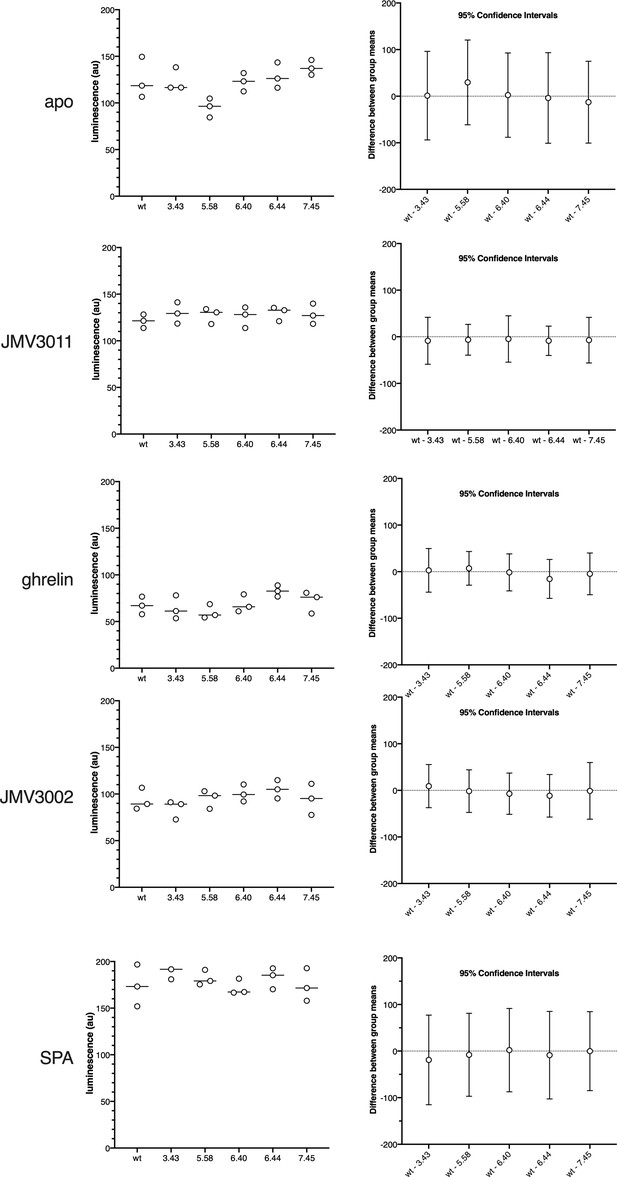
Statistical analysis of the GTP turnover assay.
The left panels show free GTP for the wild-type receptor and each of the mutants used in the present work in the absence or in the presence of ligands (ligands indicated in each panel). All data is from Figure 1. The right panels show the mean difference between the apo and each of the mutant for each condition with multiplicity-adjusted 95% confidence intervals.
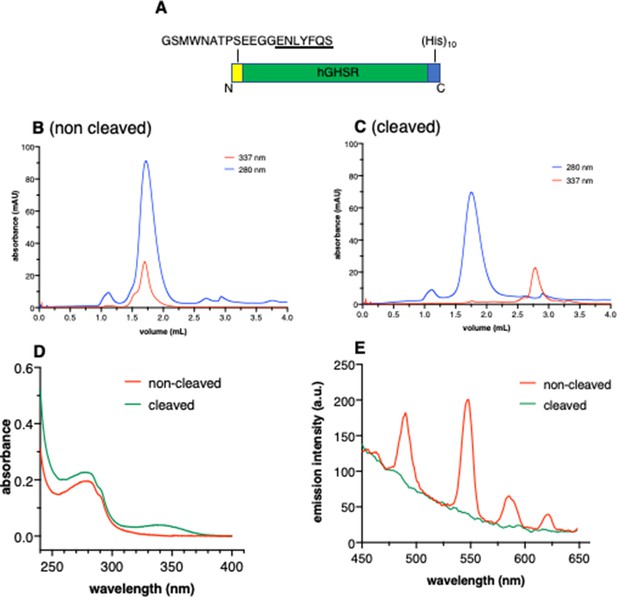
N-terminal labeling of growth hormone secretagogue receptor (GHSR).
(A) Schematic representation of the construct used. This construct includes a TEV cleavage site and two additional glycines 11 residues after the receptor N-terminus. (B,C) Size-exclusion chromatography (SEC) profile of amphipol (APol)-stabilized GHSR labeled with Lumi-4 Tb before (B) and after (C) cleavage with the TEV protease. The receptor in A8-35 after labeling and desalting was run on an S200 increase (5×150) GL column (GE Healthcare) using a 50 mM potassium phosphate, 100 M KCl, pH 7.7 buffer as the eluent and a 0.2 mL/min flow rate. UV absorbance (D) and fluorescence (E) of the labeled receptor before and after cleavage with TEV protease. In both cases, the receptor used corresponds to the main peak of the SEC. The fluorescence emission spectra were recorded with an excitation wavelength set at 337 nm, that is, at the maximum emission wavelength of the caged Tb.
-
Figure 1—figure supplement 4—source data 1
Absorbance of GHSR-containing nanodiscs before and after TEV cleavage.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig1-figsupp4-data1-v1.xlsx
-
Figure 1—figure supplement 4—source data 2
Emission intensity of GHSR-containing nanodiscs before and after TEV cleavage.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig1-figsupp4-data2-v1.xlsx
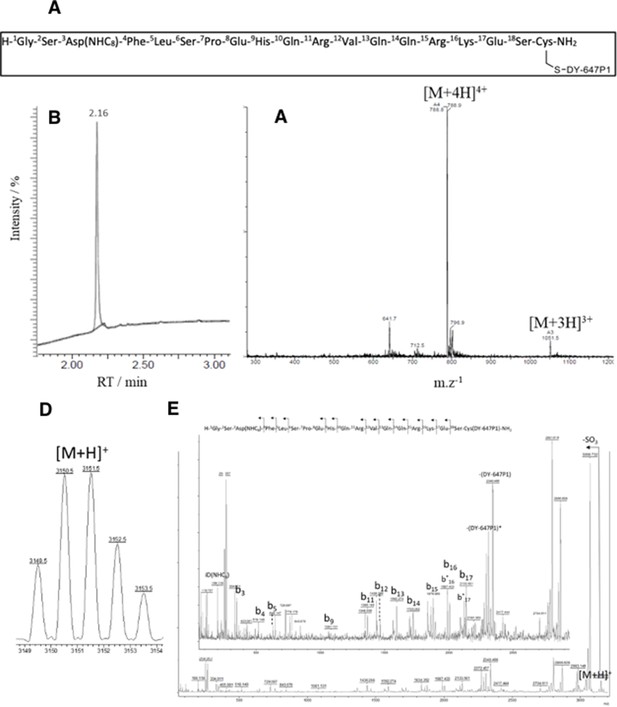
Fluorescent ghrelin characterization.
(A) Structure of the fluorescent ghrelin peptide used in the ligand-binding experiments. (B) RP-HPLC chromatogram; RT = 2.16 min, UV purity (214 nm) = 99%; (C) MS-ESI(+) spectrum: Mcalculated = 3150.6 g/mol, observed m/z 1051.2 [M+3H]3+, m/z 788.6 [M+4H]4+. (D) Isotopic pattern of [M+H]+, calculated Mmonoisotopic = 3148.5 Da; observed [M+H]+ m/z 3149.5; (E) MS/MS spectrum of parent ion m/z=3149.5.
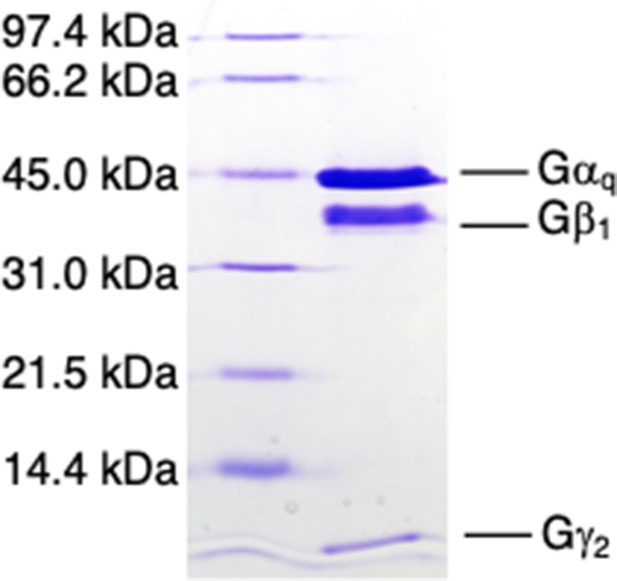
The G protein trimer.
SDS-PAGE profile of the Gαqβ1γ2 trimer used in the functional assays. The G protein trimer was run on a 15% polyacrylamide-0.1% SDS gel, with Coomassie blue staining.
-
Figure 1—figure supplement 6—source data 1
SDS-PAGE profile of the G protein used in the GTP-binding assays.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig1-figsupp6-data1-v1.xlsx
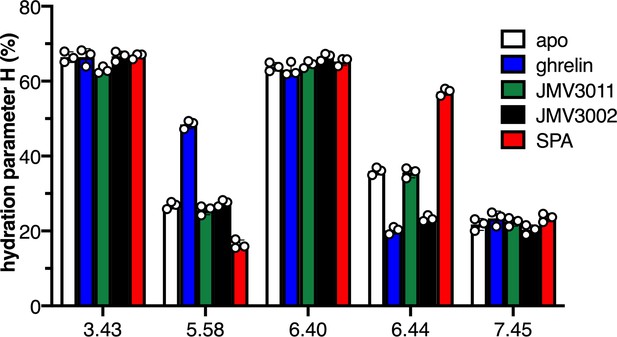
Local hydration of growth hormone secretagogue receptor (GHSR) as a function of ligands.
H parameter for the 7H4MC-labeled GHSR in the absence of ligand (apo) and in the presence of JMV3011, ghrelin, JMV3002, or SPA (substance-P analog). All ligands were used at a 10 µM concentration. In all cases, the data represents the mean value ± SD of three experiments. Statistical analyses are provided in Figure 1—figure supplement 2 and 4.
-
Figure 2—source data 1
H parameter for GHSR and its mutants.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig2-data1-v1.xlsx
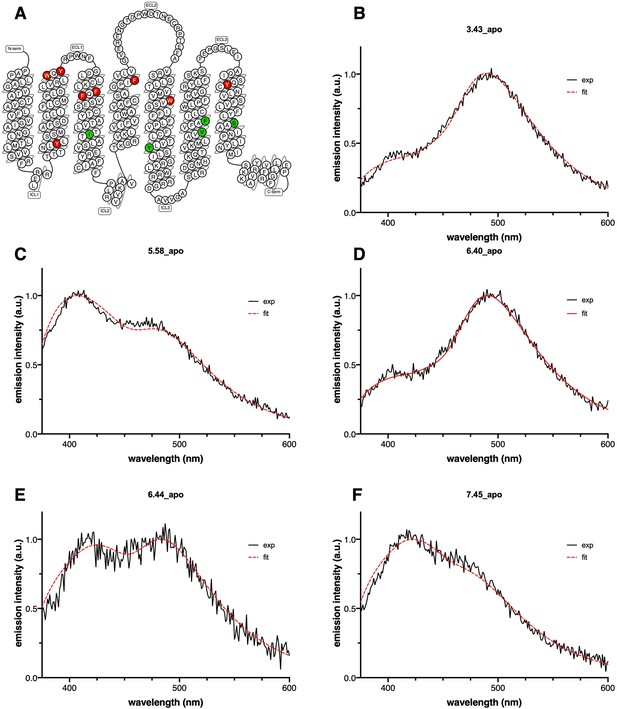
Emission spectra of L-(7-hydroxycoumarin-4-yl) (7H4MC)-ethylglycine incorporated into isolated growth hormone secretagogue receptor (GHSR).
The spectra were recorded in the absence of ligand. The labeling positions are given in a snake-like plot of GHSR (Shiimura et al., 2020) (A) (red: positions deleterious to GHSR function; green: positions considered in this work), and then indicated in each panel (B–F). The excitation wavelength was set to 320 nm with an emission intensity recorded between 340 and 600 nm. The gray curve represents the experimental emission spectrum (exp). The red, blue, green, and magenta spectra represent the decomposition of the experimental spectrum into its neutral (N), complexed (C), anionic (A), and tautomeric (T) components, respectively. The orange curve (fit) corresponds to sum of the individual N, C, A, and T curves.
-
Figure 2—figure supplement 1—source data 1
7-H4MC emission intensity of the apo GHSR mutants.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig2-figsupp1-data1-v1.xlsx
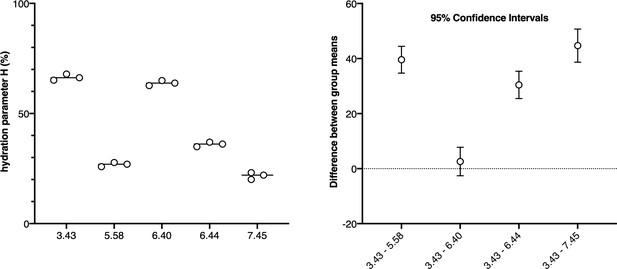
Statistical analysis of the data in Figure 2.
The left panel shows the hydration parameter for each of the mutants in the absence of ligand. Data is from Figure 2. The right panel shows the mean difference between the apo state of each of the mutants taking the modified receptor at position 3.43 as a reference, with multiplicity-adjusted 95% confidence intervals.
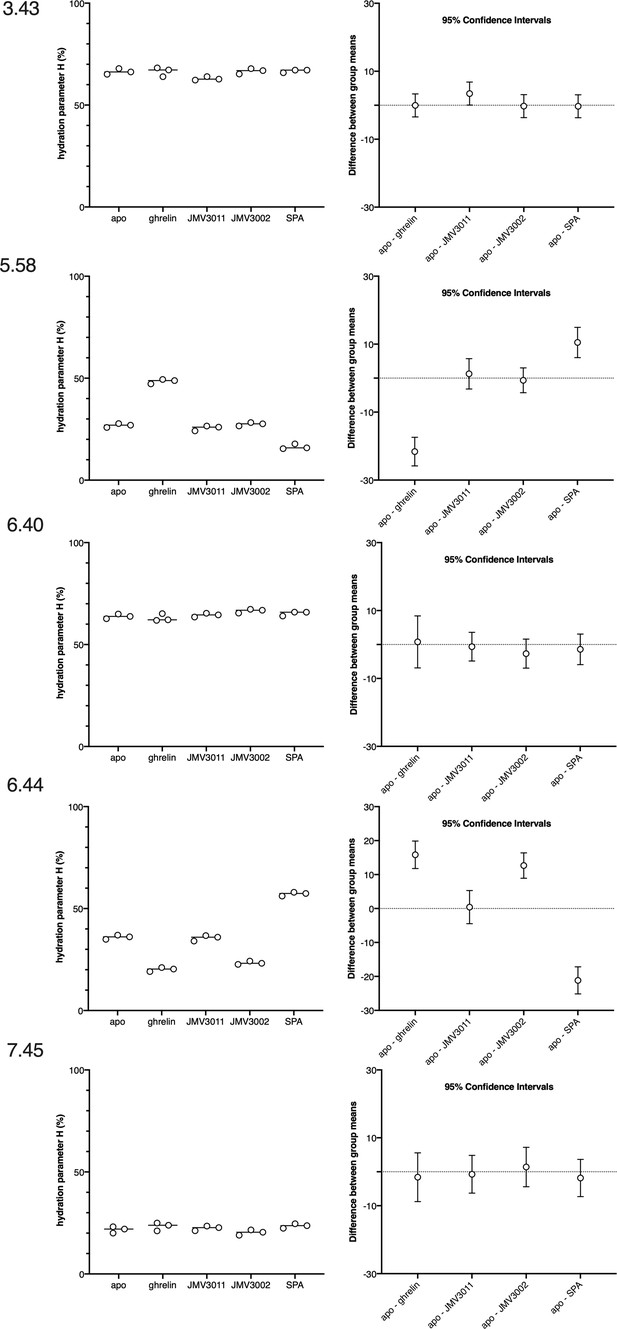
Statistical analysis of the data in Figure 2.
The left panels show the hydration parameter for each of the mutants in the absence or in the presence of ligands. Data are from Figure 2. The right panels show the mean difference between the apo and the ligand-loaded states for each of the mutants with multiplicity-adjusted 95% confidence intervals.
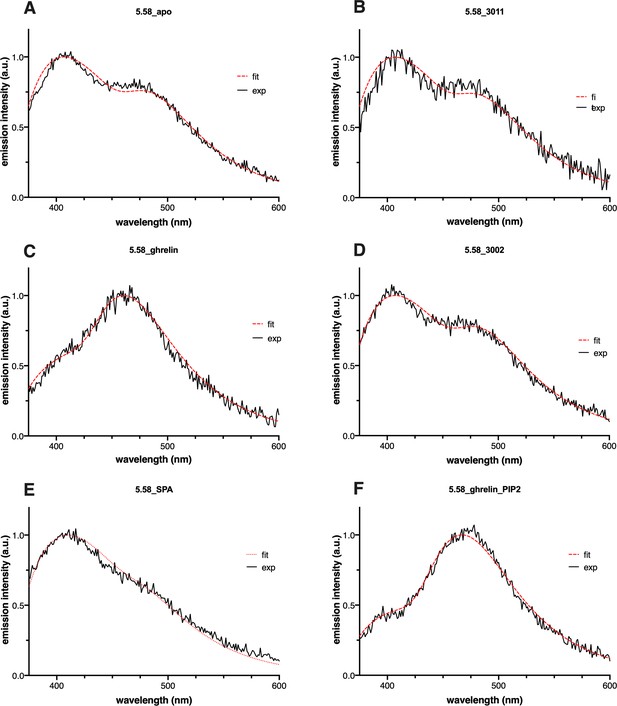
Emission spectra of L-(7-hydroxycoumarin-4-yl) (7H4MC)-ethylglycine incorporated at position 5.58 of growth hormone secretagogue receptor (GHSR) in the presence of pharmacologically distinct ligands or phosphatidylinositol-4,5-bisphosphate (PIP2).
All ligands were used at a 10 µM concentration (JMV3011: antagonist, ghrelin: full agonist, JMV3002: Gq-biased agonist, substance-P analog [SPA]: inverse agonist). The excitation wavelength was set to 320 nm with an emission intensity recorded between 340 and 600 nm. The black curve represents the experimental emission spectrum (exp). The red one (fit) corresponds to sum of the individual curves after decomposition of the experimental spectrum into its neutral, complexed, anionic, and tautomeric components.
-
Figure 2—figure supplement 5—source data 1
7-H4MC emission intensity of the GHSR 5.58 mutant in the presence of ligands.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig2-figsupp5-data1-v1.xlsx
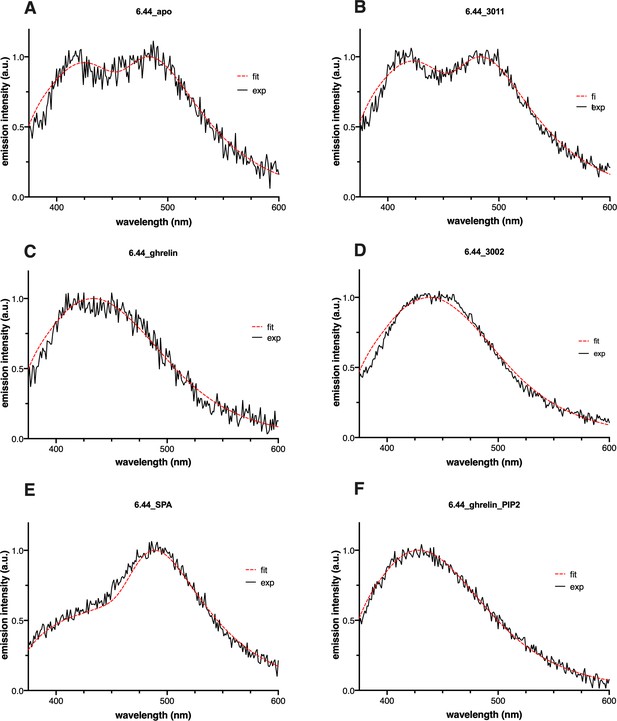
Emission spectra of L-(7-hydroxycoumarin-4-yl) (7H4MC)-ethylglycine incorporated at position 6.44 of growth hormone secretagogue receptor (GHSR) in the presence of pharmacologically distinct ligands or phosphatidylinositol-4,5-bisphosphate (PIP2) in the nanodiscs.
All ligands were used at a 10 µM concentration (JMV3011: antagonist, ghrelin: full agonist, JMV3002: Gq-biased agonist, substance-P analog [SPA]: inverse agonist). The excitation wavelength was set to 320 nm with an emission intensity recorded between 340 and 600 nm. The black curve represents the experimental emission spectrum (exp). The red one (fit) corresponds to sum of the individual curves after decomposition of the experimental spectrum into its neutral, complexed, anionic, and tautomeric components.
-
Figure 2—figure supplement 6—source data 1
pa 7-H4MC emission intensity of the GHSR 6.44 mutant in the presence of ligands.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig2-figsupp6-data1-v1.xlsx
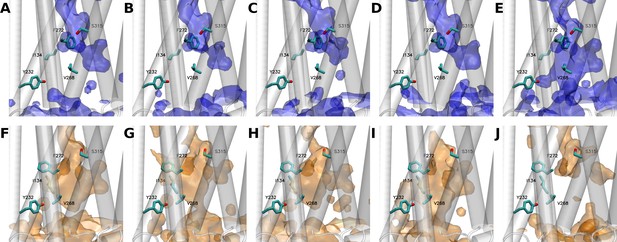
Water distribution in growth hormone secretagogue receptor (GHSR) as observed along the five independent 5 µs molecular dynamics (MD) simulations starting from either the inactive (A to E) or the active (F to J) states of the receptor.
(E and J) panels show simulations where GHSR transited from inactive to active (E) or from active to inactive (J) states, respectively. The backbone of the protein is represented as a transparent-white cartoon, while the five positions at which the L-(7-hydroxycoumarin-4-yl) (7H4MC)-ethylglycine residue was inserted are shown in licorice. Blue or orange surfaces respectively describe the hydration of the receptor using a probability of 0.3. Volumetric maps were computed using the volmap tool of visual molecular dynamics (VMD).
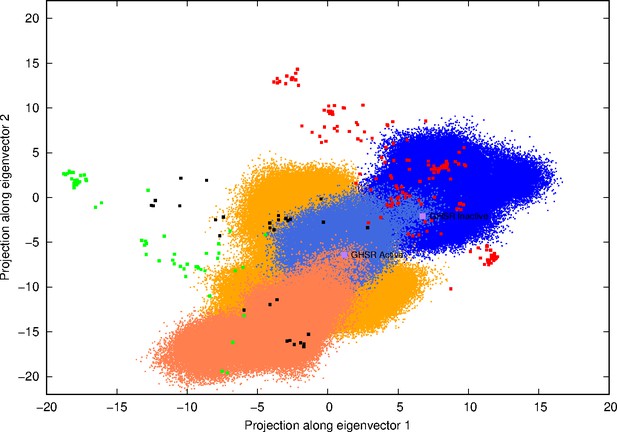
Projection of structures and molecular dynamics (MD) conformers on the first two eigenvectors obtained from the principal component analysis (PCA) of experimental structures.
Red and green squares represent inactive and active experimental structures, respectively, whereas the black squares stand for structures describing intermediates. Both purple squares represent either the experimental-inactive structure or the model-active conformer of growth hormone secretagogue receptor (GHSR). Blue dots represent the projections of all conformers obtained along our MD simulations starting from the inactive state, lighter-blue dots resulting from the simulation which transited from the inactive to the active conformations. In the same way, orange dots resulted from the projection of all conformers obtained from MD simulations starting from the active-like state, while dark-orange dots stand for the simulation where TM7 closed the intracellular side of the receptor.
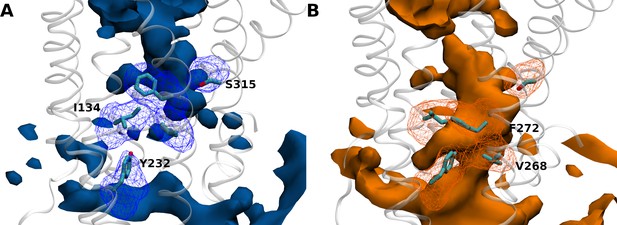
Amino acid positions and hydration patterns of inactive growth hormone secretagogue receptor (GHSR) (A) and active GHSR (B) explored by molecular dynamics (MD) simulations.
GHSR is represented in white ribbons. Volumetric maps in solid surface represent the water distribution with a probability of presence of 0.3. Meshes represent the most probable (probability of 0.3) positions of residues I1343.43, Y2325.58, V2686.40, F2726.44, and S3157.45 in both states. Snapshots representing the residues in their respective shapes are drawn in licorice for visualization. Volumetric maps were computed using the volmap tool of visual molecular dynamics (VMD).
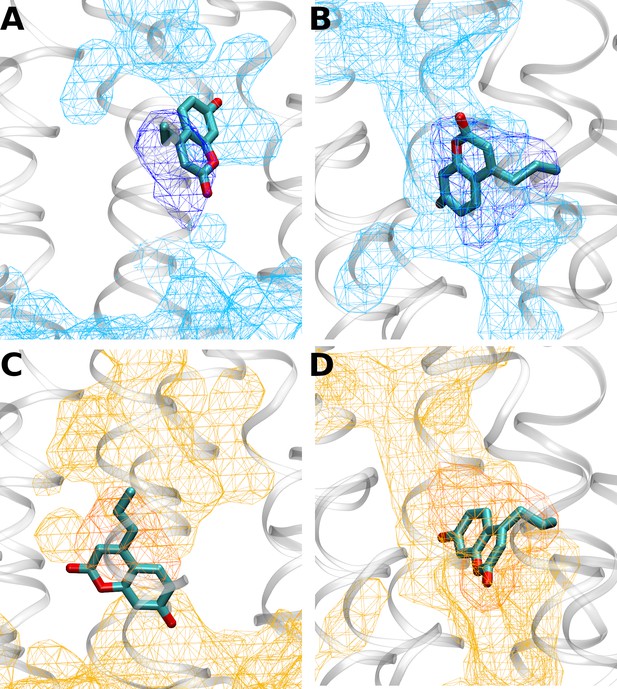
Intersections between water distributions and predicted L-(7-hydroxycoumarin-4-yl) (7H4MC) orientations in the inactive growth hormone secretagogue receptor (GHSR) (A and B) and the active GHSR (C and D) at positions 268 (A and C) and 315 (B and D).
The dark isocontours in mesh represent a probability of presence of the mutated residue of 0.3, while the light isocontours in mesh represent a probability of presence of water of 0.3. The protein backbone is represented in white tubes and the most probable orientation(s) of 7H4MC are represented in licorice. Volumetric maps were computed using the volmap tool of visual molecular dynamics (VMD).
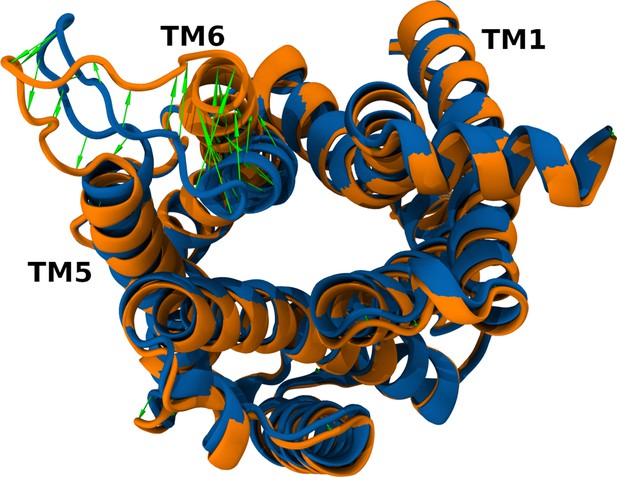
Structural differences between both starting conformers: inactive (blue) and active (orange).
Protein backbone is represented in cartoon; the green arrows emphasize the difference between the two models.
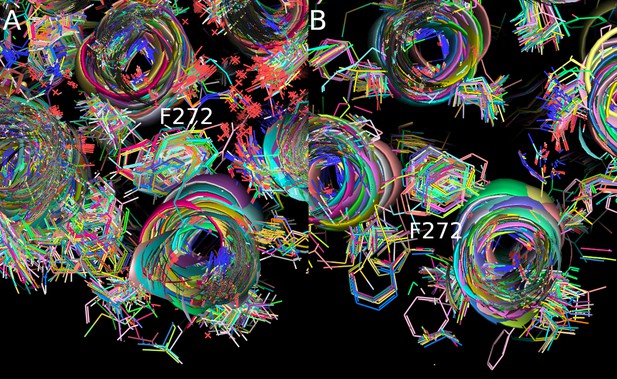
Orientation of F2726.44 in all experimental-inactive (A) and experimental-active (B) structures.
Protein atoms are represented as lines and cartoon, each receptor has been colored differently.
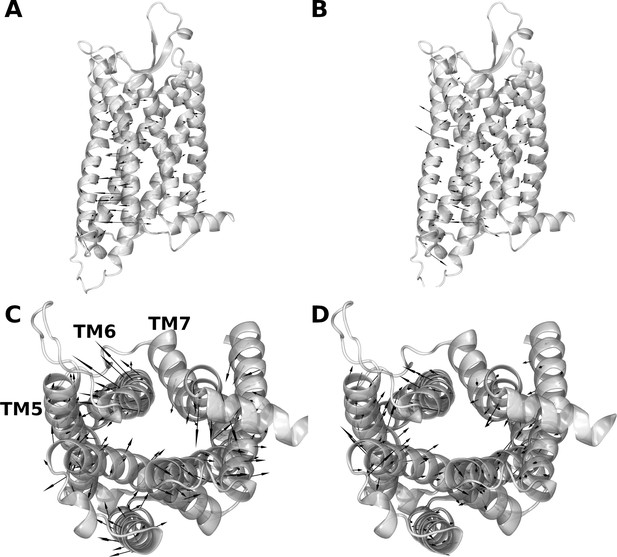
Motions encoded by the first two eigenvectors resulting from the principal component analysis (PCA) of the conserved Cα coordinates from all experimental structures.
The receptor is represented as cartoon and black arrows represent the direction and amplitude of eigenvectors. (A and C) represent a side and an intracellular view of the first eigenvector respectively, while (B and D) represent a side and an intracellular view of the second eigenvector, respectively.
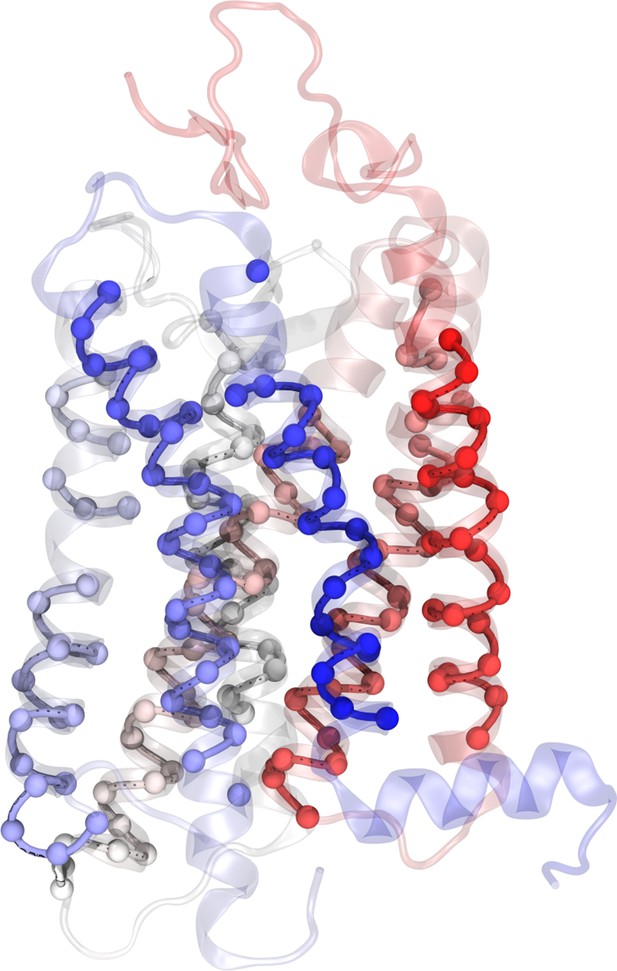
Residue selection for principal component analysis (PCA).
The receptor is represented in transparent cartoon colored by residue number from red (N-terminus) to blue (C-terminus). The Cα atoms kept for the PCA are represented in spheres.
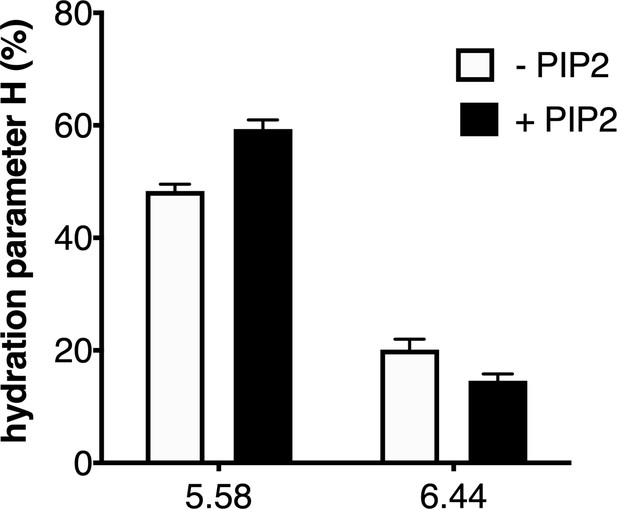
Impact of lipids on the local hydration of growth hormone secretagogue receptor (GHSR).
H parameter for L-(7-hydroxycoumarin-4-yl) (7H4MC)-labeled GHSR assembled into nanodiscs containing or not phosphatidylinositol-4,5-bisphosphate (PIP2) (2.5% PIP2-to-total lipids molar ratio), in the presence of 10 µM ghrelin. The data represents the mean value ± SD of three experiments.
-
Figure 5—source data 1
H parameter as a function of PIP2 in the nanodiscs.
- https://cdn.elifesciences.org/articles/63201/elife-63201-fig5-data1-v1.xlsx
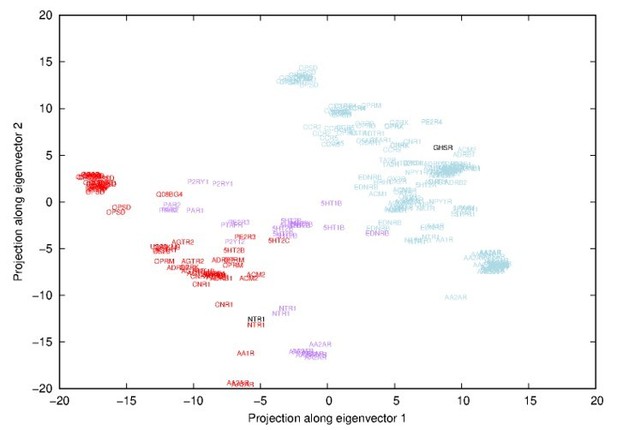
Projections along the first-two eigenvectors inferred from a Principal Component Analysis (PCA) of all experimental structures.
Names correspond to uniprot names of GPCR and have been placed in respect to the projection. Red names correspond to activated receptors, purple to intermediate conformations and blue names to inactivated receptors (according to the classification of the GPCRdb).
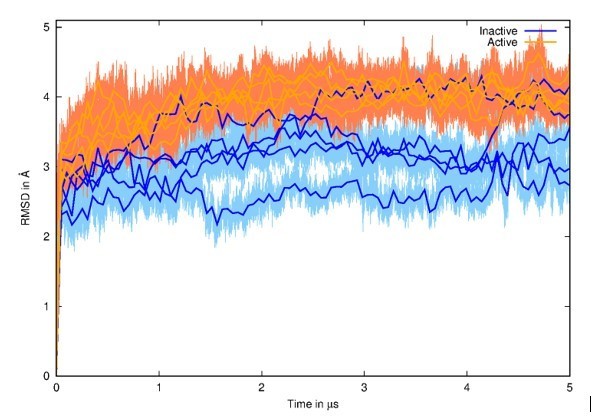
Root Mean Square Deviation (RMSD) of all MD simulations with initial structures as references.
The 5 MD simulations from the inactive state are represented in blue, and the 5 MD simulations from the active state are represented in orange. Running averages on top of curves are shown for clarity.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background | BL21(DE3) Escherichia coli | Sigma-Aldrich | CMC0014 | Chemically competent cells |
| Recombinant DNA reagent | pEvol-aaRS | doi: 10.1021/ja062666k | ||
| Recombinant DNA reagent | pMSP1E3D1 | Addgene | #20066 | |
| Recombinant DNA reagent | pET21a-α5-GHSR (transfected construct; Homo sapiens) | doi: 10.1074/jbc.M111.288324 | ||
| Peptide, recombinant protein | Ghrelin | This work | Synthesis is described in the Materials and methods section | |
| Peptide, recombinant protein | Fluorescent ghrelin | This work | Labeling is described in the Materials and methods section | |
| Peptide, recombinant protein | Thrombin | Sigma | T7009 | |
| Commercial assay or kit | GTPase-GloTM assay | Promega | V7681 | |
| Chemical compound, drug | 7H4MC-ethylglycine | This work | Synthesis is described the Materials and methods section | |
| Chemical compound, drug | Ampicillin | Sigma | A9518 | |
| Chemical compound, drug | Chloramphenicol | Calbiochem | 220551 | |
| Chemical compound, drug | IPTG | Sigma | I6758 | |
| Chemical compound, drug | Amphipol A8-35 | Anatrace | A835 100 MG | |
| Chemical compound, drug | β-DDM | Anatrace | D310 | |
| Chemical compound, drug | Cholesteryl-hemisuccinate | Anatrace | CH210 | |
| Chemical compound, drug | POPC | Avanti Polar Lipids | 850457C | |
| Chemical compound, drug | POPG | Avanti Polar Lipids | 840457C | |
| Chemical compound, drug | PIP2 | Avanti Polar Lipids | 850155P | |
| Chemical compound, drug | Bio-Beads SM-2 | BIO-RAD | 1528920 | |
| Chemical compound, drug | Lumi4-Tb NHS | CisBio | 62TBSPEA | |
| Chemical compound, drug | DY647P1-maleimide | Dyomics | 647P1-03 | |
| Chemical compound, drug | Amine reactive Tb chelate | Fisher | 11563467 | |
| Chemical compound, drug | NiNTA Superflow | Qiagen | 30430 | |
| Chemical compound, drug | Streptavidin-agarose | Thermofisher | 20361 | |
| Chemical compound, drug | Superdex S200 increase 10×300 GL | GE Healthcare (Cytiva) | 28990944 | |
| Chemical compound, drug | Source 15Q 4.6×100 PE | GE Healthcare (Cytiva) | 17518101 | |
| Chemical compound, drug | ZebaSpin 40K MWCO column | Thermofisher | 87766 | |
| Software, algorithm | Prism | GraphPad | Version 8.4.3 | |
| Software, algorithm | VMD | doi: 10.1016/0263-7855(96)00018-5 | ||
| Software, algorithm | Bio3D | doi: 10.1093/bioinformatics/btl461 | ||
| Software, algorithm | Pymol | Schrodinger LLC | ||
| Software, algorithm | Gromacs 2020.3 | doi: 10.5281/zenodo.3923645 |
Additional files
-
Supplementary file 1
Equilibration procedure.
- https://cdn.elifesciences.org/articles/63201/elife-63201-supp1-v1.xlsx
-
Supplementary file 2
List of structures used for principal component analysis.
- https://cdn.elifesciences.org/articles/63201/elife-63201-supp2-v1.xls
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63201/elife-63201-transrepform-v1.docx