PIE-1 SUMOylation promotes germline fates and piRNA-dependent silencing in C. elegans
Figures
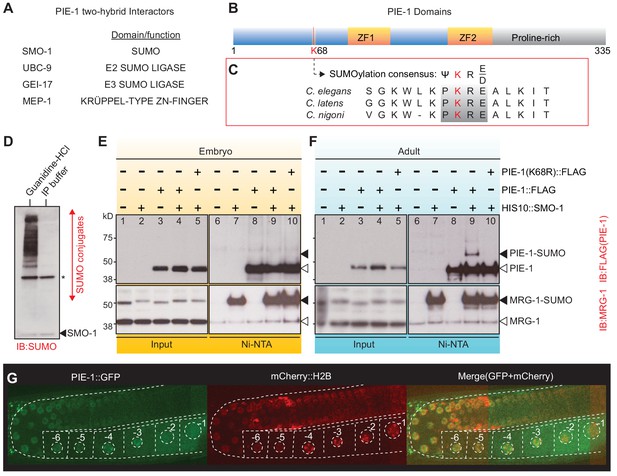
PIE-1 is SUMOylated on K68 residue in the Caenorhabditis elegans germline.
(A) Summary of PIE-1 interactors identified by yeast two-hybrid screen (see Supplementary file 1 for complete list). (B and C) Domain structure of PIE-1 containing two zinc fingers (ZF1 and ZF2) and proline-rich region, and location (red bar) of a consensus small ubiquitin-like modifier (SUMO) acceptor motif (ψKXE, where ψ represents a hydrophobic amino acid, K is the acceptor lysine, and X is any amino acid) conserved in PIE-1 from other Caenorhabditis species. (D) Western blot analysis of SUMO-conjugated proteins in total worm lysates prepared with guanidine-HCl denaturing buffer or IP buffer. The resistant band (asterisk) migrates with the expected size of the E1 enzyme AOS-1, which attaches to SUMO by a thioester bond and may therefore resist SUMO proteases, which cleave isopeptide bonds. The black triangle indicates free SMO-1. (E and F) Western blot analyses of SUMOylated proteins enriched from (E) early embryo or (F) adult lysates from wild-type pie-1::flag or pie-1(K68R)::flag worms. SUMOylated proteins were enriched from worms expressing HIS10::SMO-1 by Ni-NTA chromatography. Black triangles indicate SUMOylated forms of PIE-1 or MRG-1. White triangles indicate unmodified PIE-1 or MRG-1. MRG-1 is a robustly SUMOylated protein (Supplementary files 2 and 3; Drabikowski et al., 2018; Kaminsky et al., 2009) and thus serves as a positive control. (G) Confocal images of PIE-1::GFP and mCherry::H2B in adult germline of live pie-1::gfp; pie-1p::mCherry::his-58 worms. Oocyte nuclei are indicated by white circles and numbered.
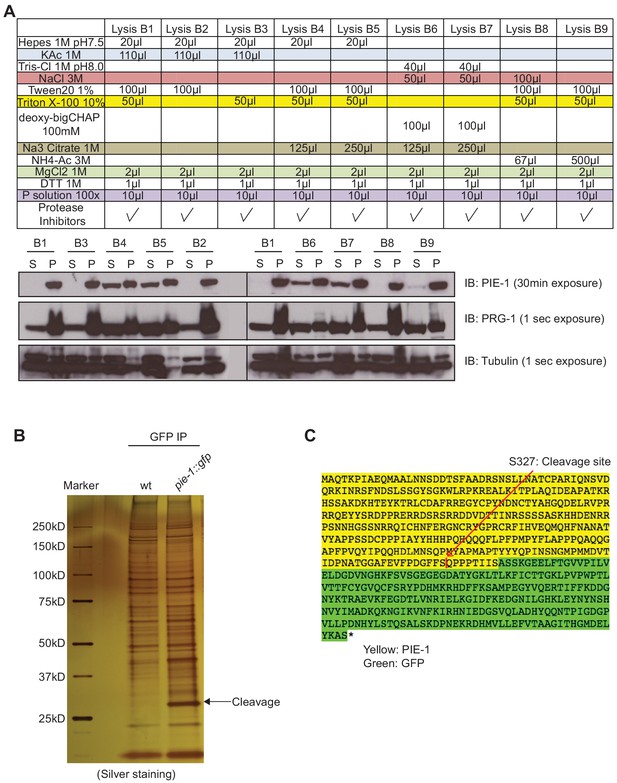
PIE-1 is insoluble and unstable.
(A) Analysis of lysis buffers (upper) used to test PIE-1 solubility in lysates (lower). PIE-1 is insoluble in most buffers, whereas PRG-1 and tubulin are soluble. S, supernatant; P, pellet. (B) Silver stain gel of GFP immunoprecipitation experiment from wild-type or pie-1::gfp worms. Cleavage product of PIE-1 is indicated with black arrow. (C) Sequence of the expected PIE-1::GFP fusion protein showing the location of the cleavage site at S327 identified by analysis (red arrow), which removes the last eight residues of PIE-1 and GFP.
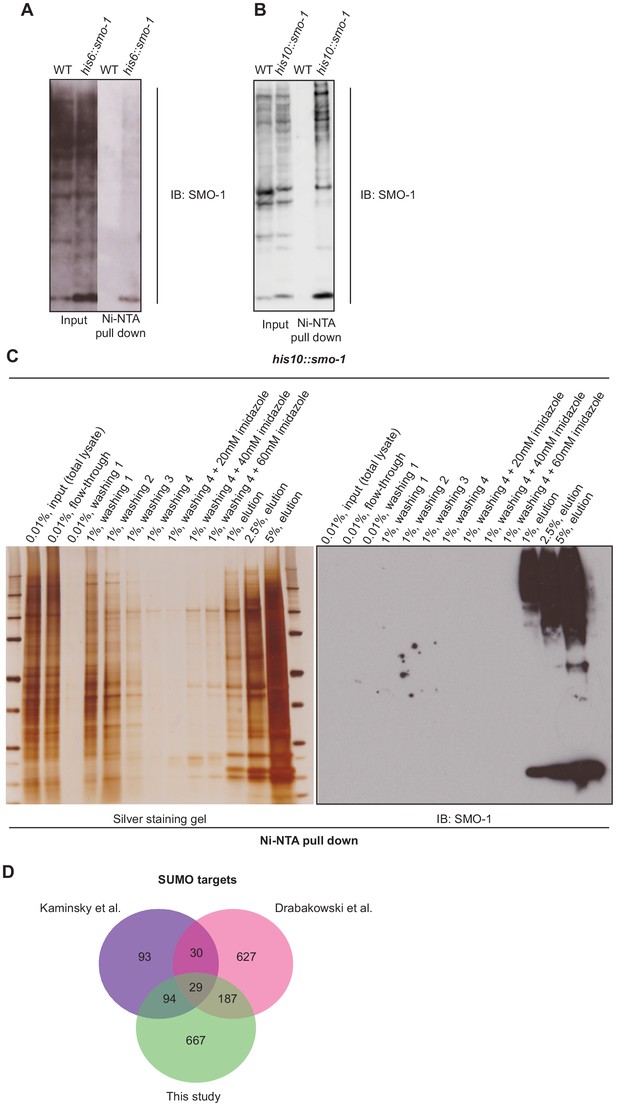
Enrichment of SUMOylated proteins from worms expressing HIS-tagged SMO-1.
(A and B) Western blots comparing Ni-affinity enrichment of SUMOylated proteins from (A) his6::smo-1 or (B) his10::smo-1 worms. (C) Silver staining gel (left) and western blot (right) showing the results of Ni-affinity chromatography (binding, washes, and elution) and enrichment of SUMOylated proteins. (D) Venn diagram showing the overlap of Caenorhabditis elegans SUMO targets identified in this study and by Kaminsky et al., 2009 and Drabikowski et al., 2018. Our list of potential SUMOylated proteins included approximately half of the 146 proteins identified by Kaminsky et al., and one-quarter of the 873 proteins identified by Drabikowski et al. Nearly 70% of the proteins we identified were not identified by these studies. Only 29 proteins (3%) were identified by all three studies. These findings suggest that the full SUMO proteome in C. elegans is far from complete.
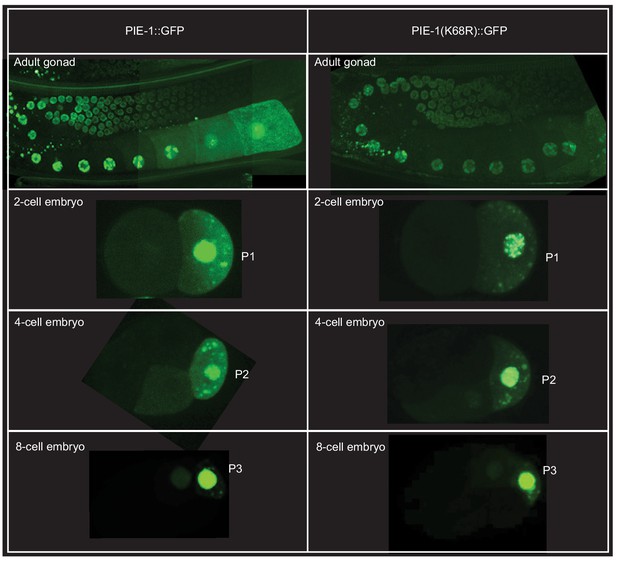
PIE-1 expression in the adult germline and early embryos.
Confocal images of PIE-1::GFP or PIE-1(K68R)::GFP fluorescence in adult germline (top) and in the germ cells (P1, P2, and P3) of early embryos.
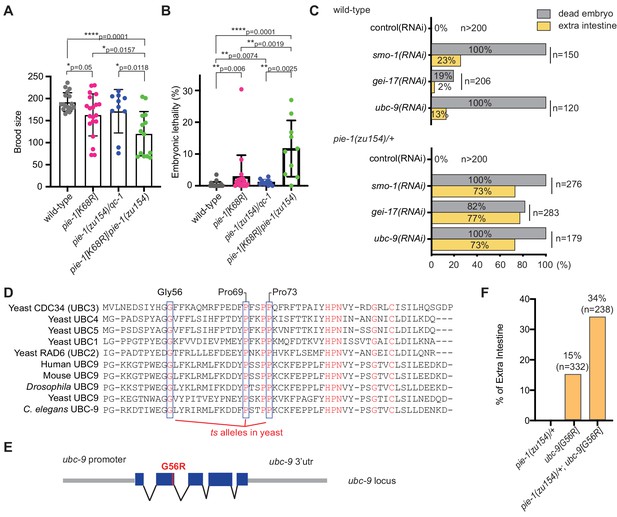
Genetic interactions between pie-1 and SUMO pathway.
(A) Brood size analysis and (B) embryonic lethality of wild-type (N2), pie-1(ne4303[K68R]), pie-1(zu154)/qC-1, and pie-1(ne4303[K68R])/pie-1(zu154). Statistical significance was determined by Wilcoxon-Mann-Whitney test: *p≤0.05; **p≤0.01; ****p≤0.0001. (C) Tests of genetic interactions between pie-1 and SUMO pathway mutants. Bar graphs show the percentage of dead embryos (gray) and percentage of dead embryos with extra intestine (yellow) among ‘n’ embryos scored. (D) Partial sequence alignment of UBC enzymes, including C. elegans UBC-9. Residues conserved in all UBC proteins are shown in red. Temperature-sensitive (ts) alleles of yeast Cdc34 result from mutations in highly conserved residues (blue boxes). Mutating the proline resides (P69S and P73S) resulted in non-conditional lethality in C. elegans. A G56R mutation in C. elegans UBC-9 caused a ts phenotype. (E) Location of the G56R mutation introduced into the endogenous ubc-9 gene by CRISPR genome editing. (F) Genetic interaction between pie-1 and ubc-9(ne4446[G56R]) allele at 25°C. Bar graphs show the percentage of embryos with extra intestine among ‘n’ embryos scored.
-
Figure 2—source data 1
Brood size and embryonic lethality.
- https://cdn.elifesciences.org/articles/63300/elife-63300-fig2-data1-v1.xlsx
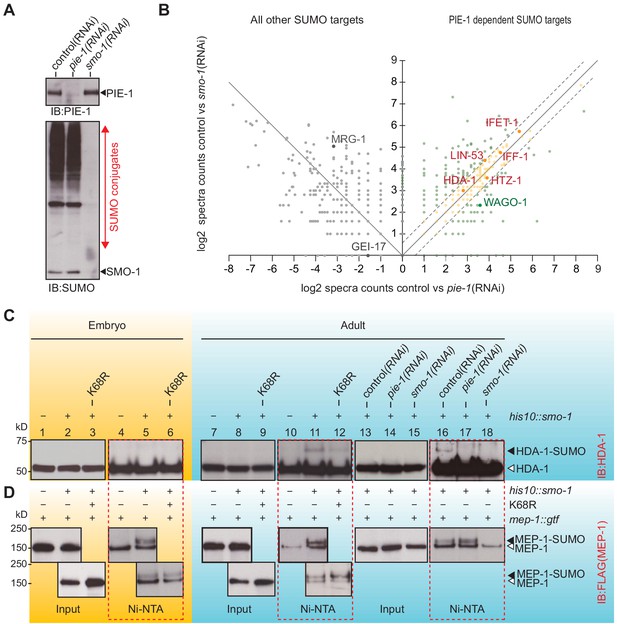
PIE-1 SUMOylation promotes HDA-1 SUMOylation in the adult germline.
(A) Western blot showing relative levels of SUMOylation in HIS10::SMO-1 worms treated with control (L4440), pie-1(RNAi), or smo-1(RNAi). (B) Scatter plot comparing the levels of SUMOylated proteins in pie-1(RNAi) worms (x axis) and smo-1(RNAi) worms (y axis). Eluates from affinity chromatography of control, pie-1(RNAi), and smo-1(RNAi) lysates were analyzed by mass spectrometry. The log of the difference between spectral counts in control and mutant was plotted for each protein. Positive values represent proteins whose spectral counts were reduced in pie-(RNAi) and smo-1(RNAi). Negative values on the x axis represent proteins whose spectral counts increased in pie-1(RNAi) compared to control. Dashed lines indicate the position of a 1.5-fold difference between the changes in smo-1(RNAi) and pie-1(RNAi) worms. A full list of PIE-1-dependent SUMO targets is provided in Supplementary file 4. (C and D) Western blot analyses of SUMOylated HDA-1 (C) or MEP-1 (D) enriched from embryo (yellow background) or adult (blue background) lysates of wild-type, pie-1, or smo-1 mutants. Ni-NTA pull-downs are outlined by dashed red boxes. Black triangles indicate SUMOylated proteins; white triangles indicate unmodified proteins.
-
Figure 3—source data 1
Comparison the levels of SUMOylated proteins in pie-1(RNAi) with in smo-1(RNAi).
- https://cdn.elifesciences.org/articles/63300/elife-63300-fig3-data1-v1.xlsx
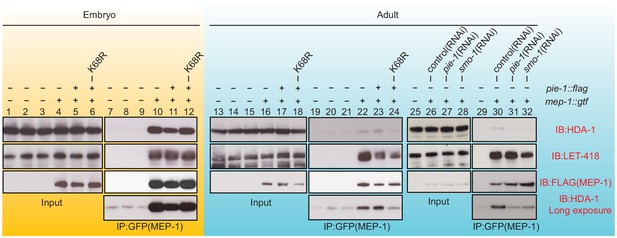
PIE-1 SUMOylation is required for the assembly of MEP-1/HDA-1 complex in the adult germline.
Western blot analyses of proteins that immunoprecipitate with MEP-1::GTF from embryo (yellow background) or adult lysates (blue background) of wild-type, pie-1, or smo-1 mutant worms. MEP-1::GTF was immunoprecipitated with GFP nanobody (see 'Materials and methods'). Blots were probed with HDA-1, LET-418, or anti-FLAG (MEP-1::GTF) antibodies. Longer exposure of the HDA-1 blots shows the reduced interaction of HDA-1 with MEP-1 in pie-1 and smo-1 mutants.
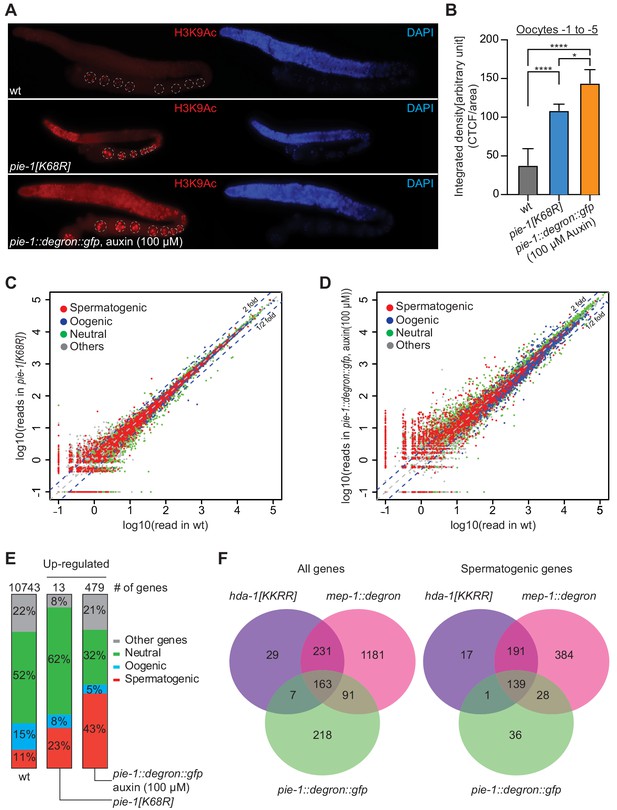
PIE-1 regulates histone H3K9Ac and spermatogenic genes in the adult germline.
(A) Immunofluorescence micrographs of H3K9Ac and DAPI staining in adult gonad of wild-type (wt), pie-1(ne4303[K68R]), and pie-1::degron::gfp animals (100 µM auxin exposure). Oocyte nuclei are indicated with white dashed circle. (B) Quantification of immunofluorescence intensity in oocytes (−1 to −5). H3K9Ac signal was measured by ImageJ. The mean of the correlated total cell fluorescence (CTCF)/area ± SEM is plotted on the y axis. Significance was measured using a Tukey’s test: ****p<0.0001; *p<0.05. (C and D) Scatter plots comparing mRNA-seq reads in (C) pie-1(ne4303[K68R]) or (D) pie-1::degron::gfp to those in wt. Blue dashed lines indicate twofold increased or decreased in the mutant. Genes were categorized as spermatogenic, oogenic, neutral, or other, as defined by Ortiz et al., 2014. A value of 0.1 was assigned to undetected genes, thus genes with an x value of ‘−1’ were not detected in wt. (E) Bar graph showing fractions of upregulated genes involved in spermatogenesis, oogenesis, neutral, or other categories. Genes expressed in wt gonads were used as a reference (10743 genes) (Kim et al., 2021). The number of upregulated genes in each mutant is labeled at the top. (F) Venn diagram showing overlap of genes upregulated in pie-1::degron::gfp, hda-1[KKRR], and mep-1::degron. The hda-1[KKRR] and mep-1::degron data are from Kim et al., 2021.
-
Figure 5—source data 1
HDAC immunostaining signal intensities.
- https://cdn.elifesciences.org/articles/63300/elife-63300-fig5-data1-v1.xlsx
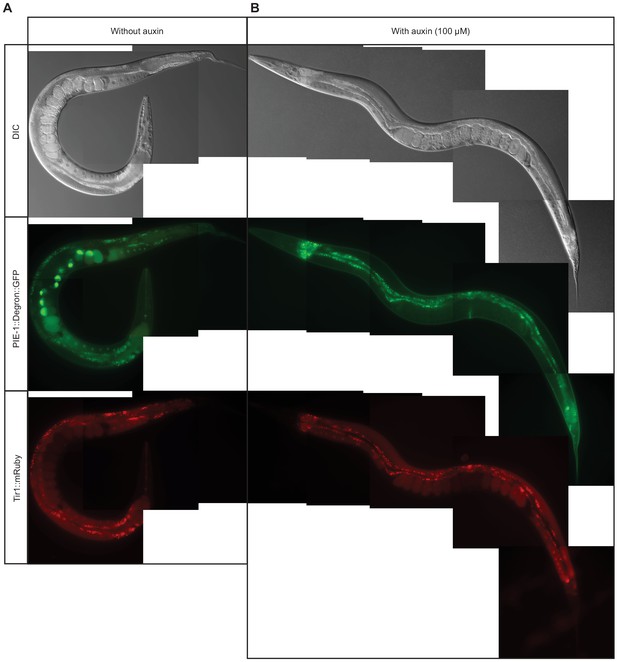
Auxin-induced depletion of PIE-1::DEGRON::GFP.
Differential interference contrast (DIC) (top) and epifluorescence images of live adult worms expressing PIE-1::DEGRON::GFP (middle) and TIR1::mRuby (bottom) in the absence (A) or presence (B) of 100 µM auxin. Intestinal autofluorescence is observed in the green and red channels.
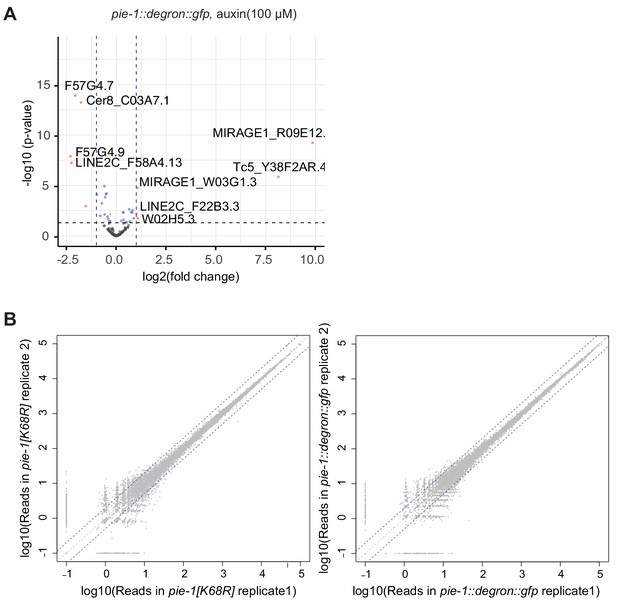
Transposons upregulated in pie-1::degron::gfp.
Volcano plot of transposon expression in pie-1::degron::gfp. The x axis shows the fold change in pie-1::degron::gfp vs. wild type, and vertical dashed lines indicate twofold change. The y axis is the adjusted p-value from DESeq2, and the horizontal dashed line indicates a p-value of 0.05. If available, transposon family names are shown with sequence names.
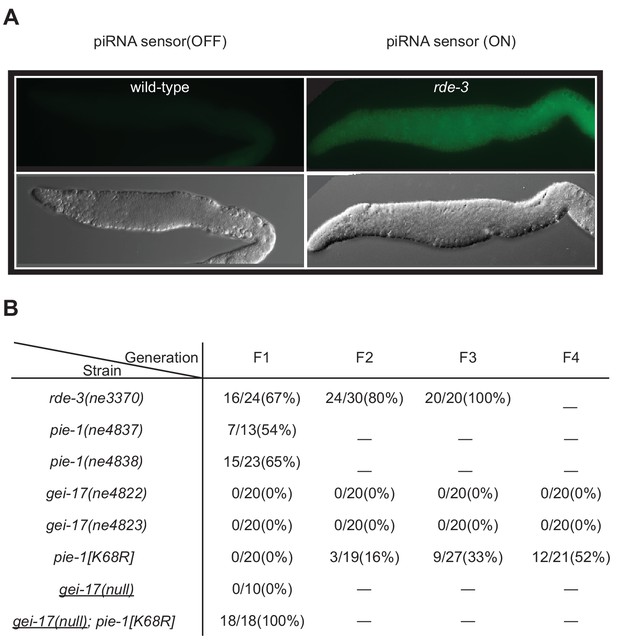
PIE-1 and GEI-17 function together to promote piRNA-mediated silencing.
(A) Epifluorescence images (upper panels) of piRNA sensor expression in dissected gonads from wild-type and rde-3(ne3370) worms. The lower panels show differential interference contrast images of the gonads in the upper panels. (B) Synergistic effects of desilencing piRNA sensors in pie-1[K68R]; gei-17 double mutants. The desilenced piRNA sensor (gfp::csr-1) was scored in the indicated alleles. gei-17(null) alleles were generated by CRISPR editing.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat# F1804; RRID:AB_262044 | IB(1:1000) |
| Antibody | Rabbit polyclonal anti-MRG-1 | Novus Biologicals | Cat# 49130002; RRID:AB_10011724 | IB(1:1000) |
| Antibody | Rabbit polyclonal anti-HDA-1 | Novus Biologicals | Cat# 38660002; RRID:AB_10708816 | IB(1:2500) |
| Antibody | Rabbit polyclonal anti-LET-418 | Novus Biologicals | Cat# 48960002; RRID:AB_10708820 | IB(1:1000) |
| Antibody | Rat monoclonal anti-tubulin | Bio-Rad | Cat# MCA77G; RRID:AB_325003 | IB(1:2000) |
| Antibody | Mouse monoclonal anti-histone H3, acetyl K9 | Abcam | Cat# ab12179; RRID:AB_298910 | IF(1:100) |
| Antibody | Rabbit polyclonal anti-PRG-1 | Batista et al., 2008 | N/A | IB(1:1000) |
| Antibody | Mouse monoclonal anti-SMO-1 | Pelisch et al., 2017 | Gift from Hay Lab | IB(1:500) Freshly purified from hybridoma cell culture |
| Antibody | Mouse monoclonal anti-PIE-1(P4G5) | Mello et al., 1996 | N/A | IB(1:100) |
| Antibody | Goat anti-mouse IgG (HRP-conjugated) | Thermo Fisher Scientific | Cat# 62–6520; RRID:AB_2533947 | IB(1:2500) |
| Antibody | Mouse anti-rabbit IgG light (HRP-conjugated) | Abcam | Cat# ab99697; RRID:AB_10673897 | IB(1:3000) |
| Antibody | Anti-rat IgG (HRP-conjugated) | Jackson ImmunoResearch Labs | Cat# 712-035-150; RRID:AB_2340638 | IB(1:5000) |
| Antibody | Goat anti-mouse IgG (H + L) Alexa Fluor 594 | Thermo Fisher Scientific | (Cat# A-11005; RRID:AB_2534073) | IF(1:1000) |
| Strain, strain background | C. elegans strains | This study | Supplementary file 5 | |
| Strain, strain background | E. coli: strain OP50 | Caenorhabditis Genetics Center | WormBase: OP50 | |
| Strain, strain background | E. coli: strain HT115 | Caenorhabditis Genetics Center | WormBase: HT115 | |
| Strain, strain background | E. coli: Ahringer collection | Laboratory of C. Mello | N/A | |
| Peptide, recombinant protein | Ex Taq DNA polymerase | Takara | Cat# RR001C | |
| Peptide, recombinant protein | iProof high fidelity DNA polymerase | Bio-Rad | Cat#1725302 | |
| Peptide, recombinant protein | BsaI | New England Biolabs | Cat# R3535S | |
| Peptide, recombinant protein | NheI | New England Biolabs | Cat# R3131S | |
| Peptide, recombinant protein | HaeIII (screen for G56R) | New England Biolabs | Cat# R0108S | |
| Peptide, recombinant protein | Alt-R S.p. Cas9 Nuclease V3 | Integrated DNA Technologies (IDT) | Cat# 1081058 | CRISPR reagent |
| Peptide, recombinant protein | GFP-binding protein beads | Homemade | N/A | |
| Chemical compound, drug | Isopropyl-β-D-thiogalactoside | Sigma-Aldrich | Cat# 11411446001 | |
| Chemical compound, drug | Ampicillin | Sigma-Aldrich | Cat# A9518 | |
| Chemical compound, drug | Tetracycline | Sigma-Aldrich | Cat# 87128 | |
| Chemical compound, drug | Indole-3-acetic acid | Alfa Aesar | Cat# A10556 | |
| Chemical compound, drug | Tetramisole hydrochloride | Sigma-Aldrich | Cat# L9756-5G | |
| Chemical compound, drug | Paraformaldehyde 16% solution | Electron Microscopy Science | Cat# Nm15710 | |
| Chemical compound, drug | PBS | Life Technologies | Cat# AM9615 | |
| Chemical compound, drug | Tween20 | Fisher BioReagents | Cat# BP337-500 | |
| Chemical compound, drug | Bovine serum albumin | Life Technologies | Cat# AM2618 | |
| Chemical compound, drug | 1M HEPES, pH 7.4 | TEKnova | Cat# H1030 | |
| Chemical compound, drug | Sodium citrate dihydrate | Thermo Fisher Scientific | Cat# BP337500 | |
| Chemical compound, drug | Triton X-100 | Sigma-Aldrich | Cat# T8787-250ml | |
| Chemical compound, drug | Complete EDTA-freeprotease inhibitor cocktail | Roche | Cat# 11836170001 | |
| Chemical compound, drug | NP-40 | EMD Millipore | Cat# 492018 | |
| Chemical compound, drug | Tris (Base) | Avantor | Cat# 4099–06 | |
| Chemical compound, drug | Boric acid | AMRESCO | Cat# M139 | |
| Chemical compound, drug | Ethylenediaminetetraacetic acid disodium salt dihydrate | Sigma-Aldrich | Cat# E1644 | |
| Chemical compound, drug | Sodium dodecyl sulfate | Sigma-Aldrich | Cat# L3771-100G | |
| Chemical compound, drug | Sodium chloride | Genesee Scientific | Cat# 18–214 | |
| Chemical compound, drug | Magnesium chloride | Sigma-Aldrich | Cat# M8266 | |
| Chemical compound, drug | DL-dithiothreitol | Sigma-Aldrich | Cat# D0632-10G | |
| Chemical compound, drug | Potassium acetate | Fisher BioReagents | Cat# BP364-500 | |
| Chemical compound, drug | Ammonium acetate | Sigma-Aldrich | Cat# A7262 | |
| Chemical compound, drug | Deoxy-bigCHAP | Alfa Aesar | Cat# J64578-MD | |
| Chemical compound, drug | Potassium chloride | Sigma-Aldrich | Cat# P9541 | |
| Chemical compound, drug | Guanidine-HCl | Sigma-Aldrich | Cat# G3272 | |
| Chemical compound, drug | Imidazole | Sigma-Aldrich | Cat# 792527 | |
| Chemical compound, drug | β-Mercaptoethanol | Sigma-Aldrich | Cat# M6250 | |
| Chemical compound, drug | Sodium phosphate, dibasic | Sigma-Aldrich | Cat# S7907 | |
| Chemical compound, drug | Sodium phosphate, monobasic | Sigma-Aldrich | Cat# S0751 | |
| Chemical compound, drug | Urea | Thermo Fisher Scientific | Cat# Ac327380010 | |
| Chemical compound, drug | Trichloroacetic acid | Sigma-Aldrich | Cat# T0699 | |
| Chemical compound, drug | 1-Bromo-3-chloropropane | Sigma-Aldrich | Cat# B9673 | |
| Chemical compound, drug | TE buffer, pH 8.0 | Thermo Fisher Scientific | Cat# AM9858 | |
| Chemical compound, drug | Tris(2-carboxyethyl)phosphine hydrochloride | Sigma-Aldrich | Cat# C4706 | |
| Chemical compound, drug | Trypsin | New England Biolabs | Cat# P8101S | |
| Chemical compound, drug | TRI reagent | Sigma-Aldrich | Cat# T9424 | |
| Chemical compound, drug | Iodoacetamide | Sigma-Aldrich | Cat# I1149 | |
| Commercial assay, kit | Ni-NTA resin | Qiagen | Cat# 30210 | |
| Commercial assay, kit | SlowFade Diamond antifade Mountant with DAPI | Life Technologies | Cat# S36964 | |
| Commercial assay, kit | Quick start Bradford 1× dye reagent | Bio-Rad | Cat# 5000205 | |
| Commercial assay, kit | GlycoBlueCoprecipitant | Thermo Fisher Scientific | Cat# AM9515 | |
| Commercial assay, kit | NuPage LDS sample buffer (4×) | Thermo Fisher Scientific | Cat# NP0008 | |
| Commercial assay, kit | pCR-Blunt II-TOPO cloning kit | Thermo Fisher Scientific | Cat# K280020 | |
| Commercial assay, kit | Pierce Silver Stain Kit | Thermo Fisher Scientific | Cat# 24612 | |
| Commercial assay, kit | Lumi-Light Plus western blotting substrate | Sigma-Aldrich | Cat# 12015196001 | |
| Commercial assay, kit | Hyperfilm ECL | Thermo Fisher Scientific | Cat# 45001507 | |
| Commercial assay, kit | KAPA RNA HyperPrep with RiboErase (KK8560) | Roche | Cat# 08098131702 | |
| Commercial assay, kit | KAPA single-indexed adapter kit (KK8700) | Roche | Cat# 08005699001 | |
| Commercial assay, kit | Illumina NextSeq 500/550 v2.5 kit (150 cycles) | Illumina | Cat# 20024907 | |
| Recombinant DNA reagent | Peft3::cas9 vector (backbone: blunt II topo vector in this study) | Friedland et al., 2013 | N/A | Backbone is changed to blunt II topo vector in this study |
| Recombinant DNA reagent | pRF4: injection marker, rol-6(su1006) | Mello et al., 1991 | N/A | Backbone is changed to blunt II topo vector in this study |
| Recombinant DNA reagent | sgRNA plasmid | This study | See Materials and methods; Supplementary file 6 | |
| Sequence-based reagent | gRNA and ss oligo donor sequences | This study | Supplementary file 6 | |
| Sequence-based reagent | Alt-R CRISPR-Cas9 tracrRNA | Integrated DNA Technologies (IDT) | Cat# 1072534 | CRISPR reagent |
| Software, algorithm | GraphPad Prism version 8.2.1 | GraphPad Software | http://www.graphpad.com | |
| Software, algorithm | ImageJ | Rueden et al., 2017 | https://imagej.net | |
| Software, algorithm | Strata 15.1 | Strata Statistical Software | http://www.strata.com | |
| Software, algorithm | Salmon | Patro et al., 2017 | Version 1.1.0 | |
| Software, algorithm | DESeq2 | Love et al., 2014 | Version 1.26.0 | |
| Software, algorithm | Prolucid | Xu et al., 2006 | N/A | |
| Software, algorithm | DTASelect 2 | Tabb et al., 2002 | N/A |
Additional files
-
Supplementary file 1
List of PIE-1 interactors identified in the yeast two-hybrid screen.
- https://cdn.elifesciences.org/articles/63300/elife-63300-supp1-v1.docx
-
Supplementary file 2
List of SUMO-conjugated worm proteins identified by affinity chromatography and mass spectrometry.
- https://cdn.elifesciences.org/articles/63300/elife-63300-supp2-v1.xlsx
-
Supplementary file 3
List of C. elegans SUMO targets also identified by Kaminsky et al., 2009 or by Drabikowski et al., 2018.
- https://cdn.elifesciences.org/articles/63300/elife-63300-supp3-v1.xlsx
-
Supplementary file 4
List of PIE-1-dependent SUMO targets.
- https://cdn.elifesciences.org/articles/63300/elife-63300-supp4-v1.xlsx
-
Supplementary file 5
Strains and alleles used in this study.
- https://cdn.elifesciences.org/articles/63300/elife-63300-supp5-v1.docx
-
Supplementary file 6
sgRNA sequences for CRISPR.
- https://cdn.elifesciences.org/articles/63300/elife-63300-supp6-v1.docx
-
Supplementary file 7
RNA-seq data from dissected gonads of wild type, pie-1(ne4303[K68R]), and pie-1::degron::gfp.
- https://cdn.elifesciences.org/articles/63300/elife-63300-supp7-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63300/elife-63300-transrepform-v1.docx





