DeepEthogram, a machine learning pipeline for supervised behavior classification from raw pixels
Figures
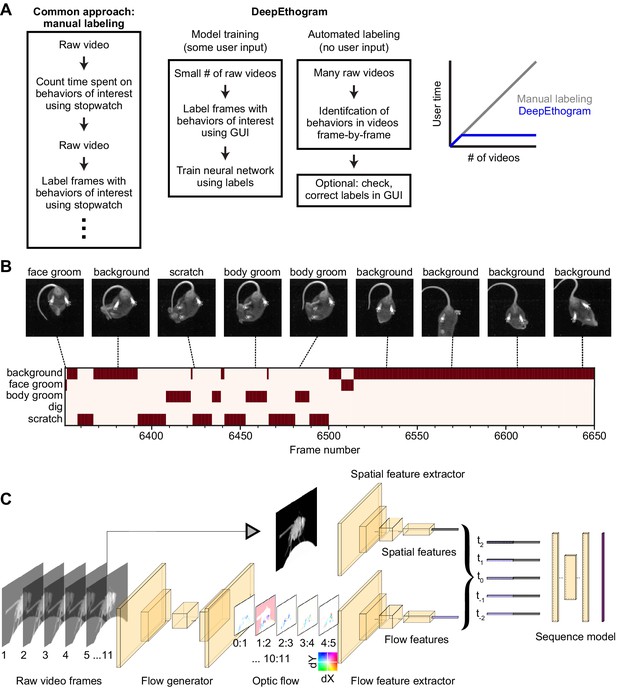
DeepEthogram overview.
(A) Workflows for supervised behavior labeling. Left: a common traditional approach based on manual labeling. Middle: workflow with DeepEthogram. Right: Schematic of expected scaling of user time for each workflow. (B) Ethogram schematic. Top: example images from Mouse-Ventral1 dataset. Bottom: ethogram with human labels. Dark colors indicate which behavior is present. Example shown is from Mouse-Ventral1 dataset. Images have been cropped, brightened, and converted to grayscale for clarity. (C) DeepEthogram-fast model schematic. Example images are from the Fly dataset. Left: a sequence of 11 frames is converted into 10 optic flows. Middle: the center frame and the stack of 10 optic flows are converted into 512-dimensional representations via deep convolutional neural networks (CNNs). Right: these features are converted into probabilities of each behavior via the sequence model.
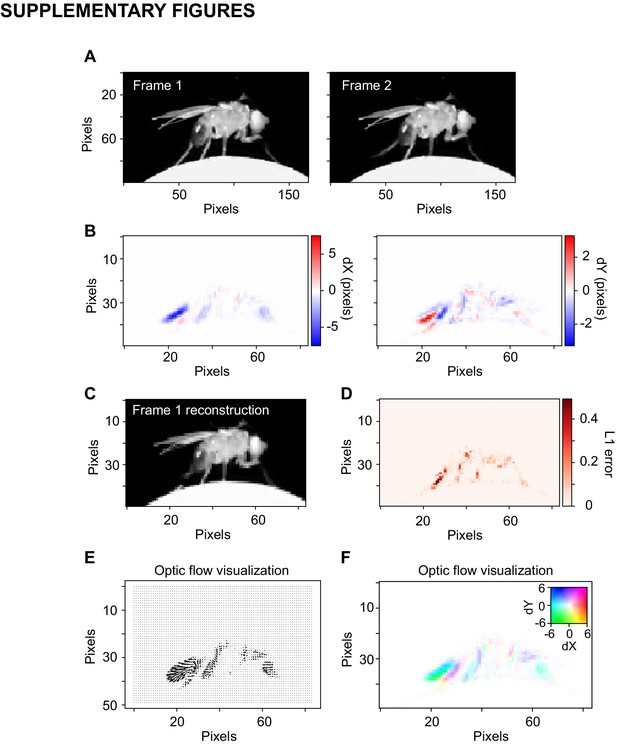
Optic flow.
(A) Example images from the Fly dataset on two consecutive frames. (B) Optic flow estimated with TinyMotionNet. Note that the image size is half the original due to the TinyMotionNet architecture. Displacements in the x dimension (left) and y dimension (right) between the frames in (A). (C) The reconstruction of frame 1 estimated by sampling frame 2 according to the optic flow calculation. The image was resized with bilinear interpolation before resampling. (D) Absolute error between frame 1 and the frame 1 reconstructed from optic flow. (E) Visualization of optic flow using arrow lengths to indicate the direction and magnitude flow. (F) Visualization of optic flow using coloring according the inset color scale. Left displacements are mapped to cyan, right displacements to red, and so on. Saturation indicates the magnitude of displacement.

Datasets and behaviors of interest.
(A) Left: raw example images from the Mouse-Ventral1 dataset for each of the behaviors of interest. Right: time spent on each behavior, based on human labels. Note that the times may add up to more than 100% across behaviors because multiple behaviors can occur on the same frame. Background is defined as when no other behaviors occur. (B–I) Similar to (A), except for the other datasets.
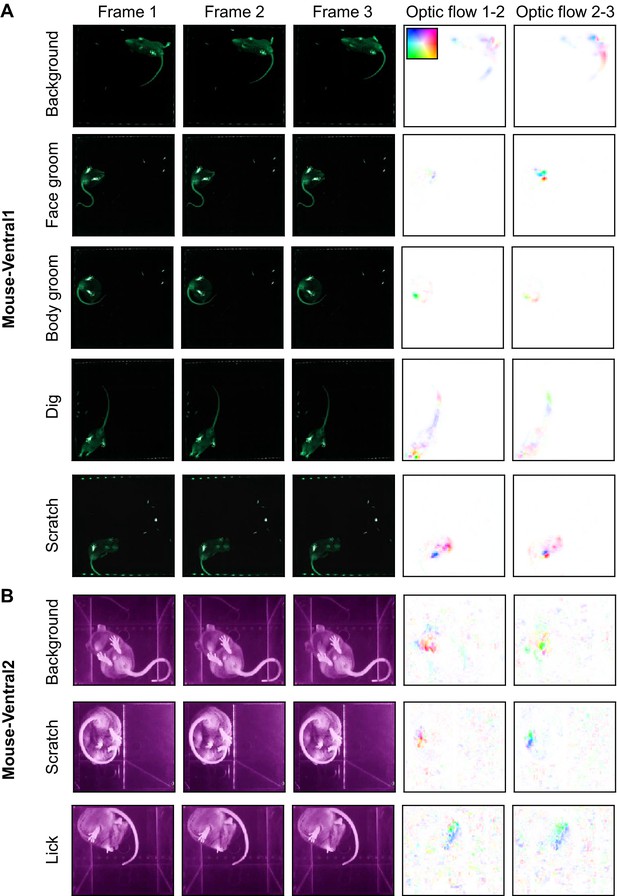
Example images from the datasets, part 1.
(A) Examples from the Mouse-Ventral1 dataset. Each row is three consecutive frames of the indicated behavior. Right columns: optic flow computed by TinyMotionNet and visualized as in Figure 1—figure supplement 1F. (B) Similar to (A), except for the Mouse-Ventral2 dataset.
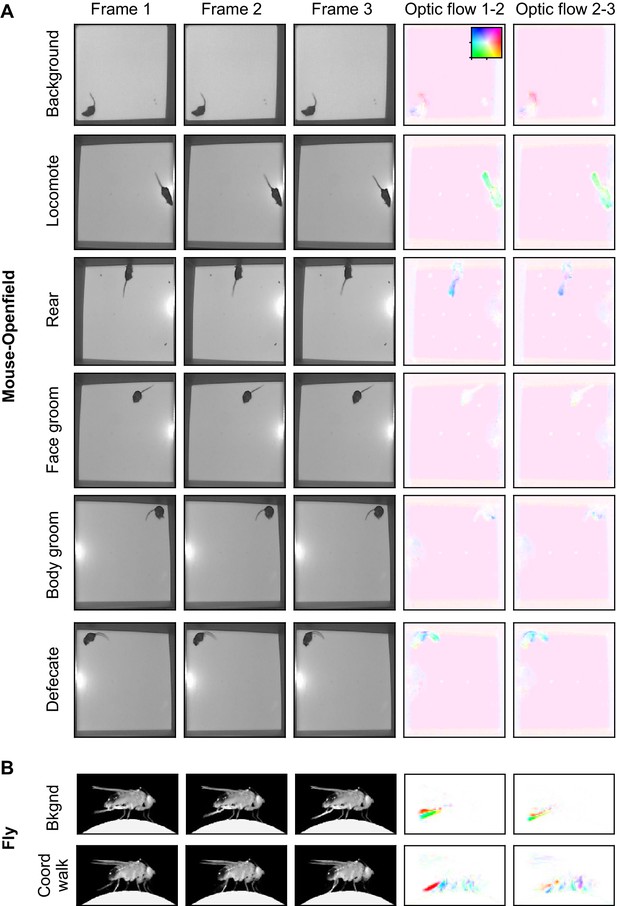
Example images from the datasets, part 2.
(A) Examples from the Mouse-Openfield dataset. Each row is three consecutive frames of the indicated behavior. Right columns: optic flow computed by TinyMotionNet and visualized as in Figure 1—figure supplement 1F. (B) Similar to (A), except for the Fly dataset.

Example images from the datasets, part 3.
Examples from the Mouse-Homecage dataset. Each row is three consecutive frames of the indicated behavior. Right columns: optic flow computed by TinyMotionNet and visualized as in Figure 1—figure supplement 1F.
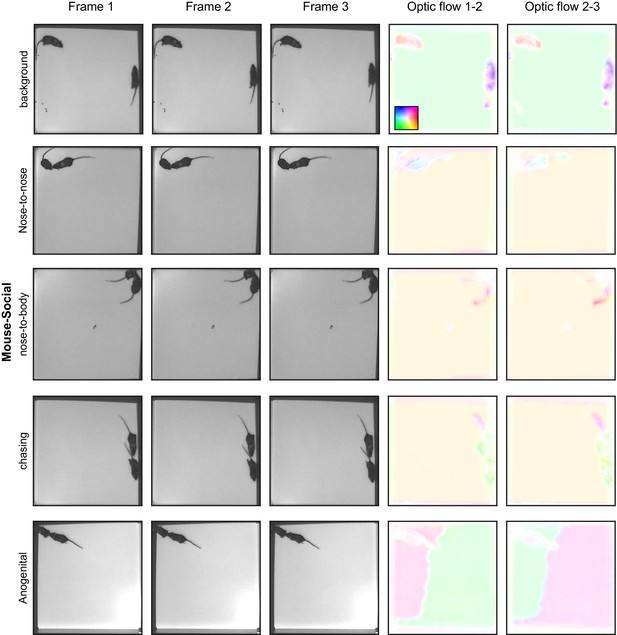
Example images from the datasets, part 4.
Examples from the Mouse-Social dataset. Each row is three consecutive frames of the indicated behavior. Right columns: optic flow computed by TinyMotionNet and visualized as in Figure 1—figure supplement 1F.

Example images from the datasets, part 5.
Examples from the Sturman-EPM dataset. Each row is three consecutive frames of the indicated behavior. Right columns: optic flow computed by TinyMotionNet and visualized as in Figure 1—figure supplement 1F. All data from Sturman et al., 2020.
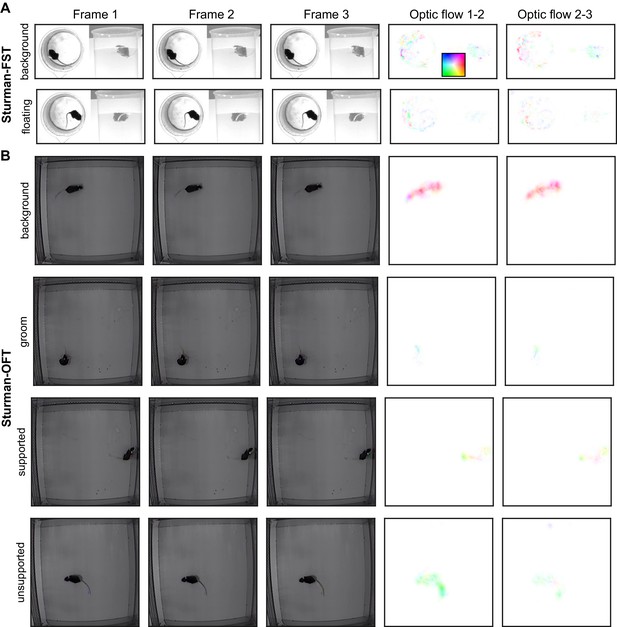
Example images from the datasets, part 6.
(A) Examples from the Sturman-FST dataset. Each row is three consecutive frames of the indicated behavior. Right columns: optic flow computed by TinyMotionNet and visualized as in Figure 1—figure supplement 1F. (B) Similar to (A), except for the Sturman-OFT dataset. All data from Sturman et al., 2020.
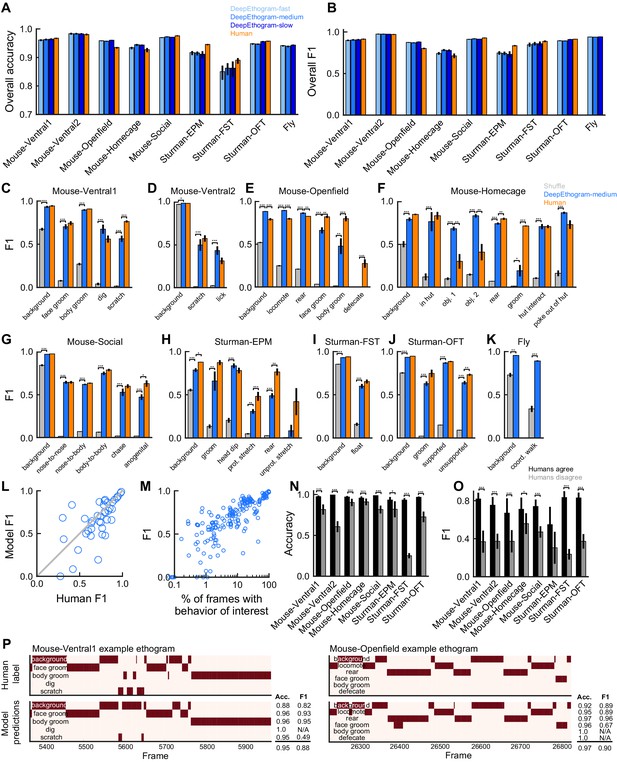
DeepEthogram performance.
All results are from the test sets only. (A) Overall accuracy for each model size and dataset. Error bars indicate mean ± SEM across five random splits of the data (three for Sturman-EPM). (B) Similar to (A), except for overall F1 score. (C) F1 score for DeepEthogram-medium for individual behaviors on the Mouse-Ventral1 dataset. Gray bars indicate shuffle (Materials and methods). *p≤0.05, **p≤0.01, ***p≤0.001, repeated measures ANOVA with a post-hoc Tukey’s honestly significant difference test. (D) Similar to (C), but for Mouse-Ventral2. Model and shuffle were compared with paired t-tests with Bonferroni correction. (E) Similar to (C), but for Mouse-Openfield. (F) Similar to (D), but for Mouse-Homecage. (G) Similar to (D), but for Mouse-Social. (H) Similar to (C), but for Sturman-EPM. (I) Similar to (C), but for Sturman-FST. (J) Similar to (C), but for Sturman-OFT. (K) Similar to (D), but for Fly dataset. (L) F1 score on individual behaviors (circles) for DeepEthogram-medium vs. human performance. Circles indicate the average performance across splits for behaviors in datasets with multiple human labels. Gray line: unity. Model vs. human performance: p=0.067, paired t-test. (M) Model F1 vs. the percent of frames in the training set with the given behavior. Each circle is one behavior for one split of the data. (N) Model accuracy on frames for which two human labelers agreed or disagreed. Paired t-tests with Bonferroni correction. (O) Similar to (N), but for F1. (P) Ethogram examples. Dark color indicates the behavior is present. Top: human labels. Bottom: DeepEthogram-medium predictions. The accuracy and F1 score for each behavior, and the overall accuracy and F1 scores are shown. Examples were chosen to be similar to the model’s average by behavior.
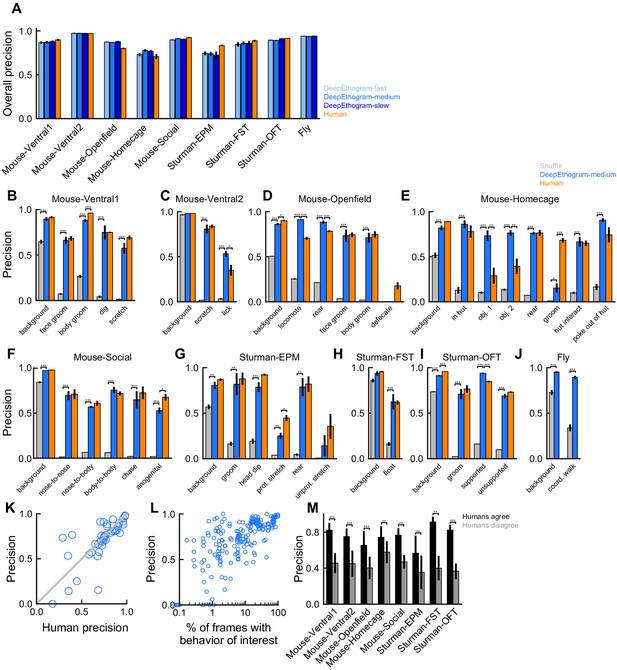
DeepEthogram performance, precision.
All results are from the test sets only. (A) Overall precision for each model size and dataset. Error bars indicate mean ± SEM across five random splits of the data (three for Sturman-EPM). (B) Precision for DeepEthogram-medium for individual behaviors on the Mouse-Ventral1 dataset. *p≤0.05, **p≤0.01, ***p≤0.001, repeated measures ANOVA with post-hoc Tukey’s honestly significant difference test. (C) Similar to (B), but for Mouse-Ventral2. Paired t-tests with Bonferroni correction. (D) Similar to (B), but for Mouse-Openfield. (E) Similar to (D), but for Mouse-Homecage. (F) Similar to (C), but for Mouse-Social. (G) Similar to (B), but for Sturman-EPM. (H) Similar to (B), but for Sturman-FST. (I) Similar to (B), but for Sturman-OFT. (J) Similar to (C), but for Fly dataset. (K) Precision on individual behaviors for DeepEthogram-medium vs. human performance. Circles are average performance across data splits for individual behaviors for all datasets with multiple human labels. Model performance vs. human performance: p=0.529, paired t-test. (L) Model precision vs. the percent of frames in the training set with the given behavior. Each point is for one behavior for one split of the data. (M) Model precision on frames for which two human labelers agreed or disagreed. Asterisks indicate p<0.05, paired t-test with Bonferroni correction.
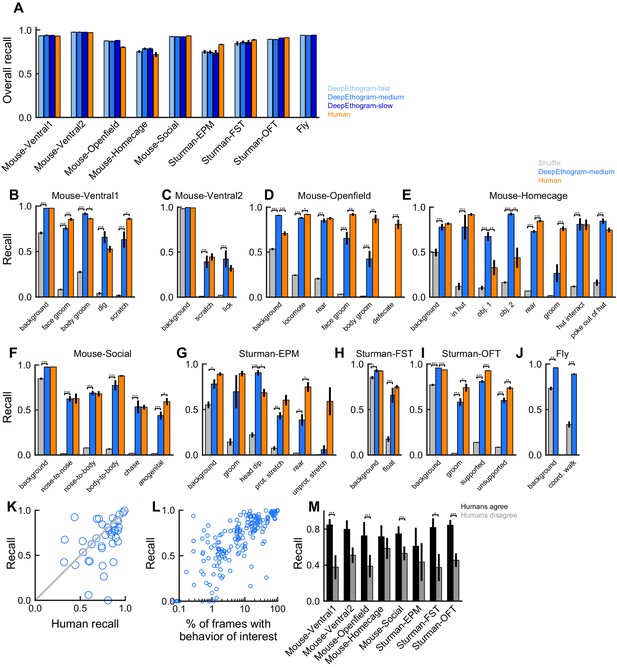
DeepEthogram performance, recall.
All results are from the test sets only. (A) Overall recall for each model size and dataset. Error bars indicate mean ± SEM across five random splits of the data (three for Sturman-EPM). (B) Recall for DeepEthogram-medium for individual behaviors on the Mouse-Ventral1 dataset. *p≤0.05, **p≤0.01, ***p≤0.001, repeated measures ANOVA with post-hoc Tukey’s honestly significant difference test. (C) Similar to (B), but for Mouse-Ventral2. Paired t-tests with Bonferroni correction. (D) Similar to (B), but for Mouse-Openfield. (E) Similar to (D), but for Mouse-Homecage. (F) Similar to (C), but for Mouse-Social. (G) Similar to (B), but for Sturman-EPM. (H) Similar to (B), but for Sturman-FST. (I) Similar to (B), but for Sturman-OFT. (J) Similar to (C), but for Fly dataset. (K) Recall on individual behaviors for DeepEthogram-medium vs. human performance. Shown is the average performance across splits for all datasets with multiple human labels. Circles are average performance across data splits for individual behaviors for all datasets with multiple human labels. Model performance vs. human performance: p<0.035, paired t-test. (L) Model precision vs. the percent of frames in the training set with the given behavior. Each point is for one behavior for one split of the data. (M) Model recall on frames for which two human labelers agreed or disagreed. Asterisks indicate p<0.05, paired t-test with Bonferroni correction.
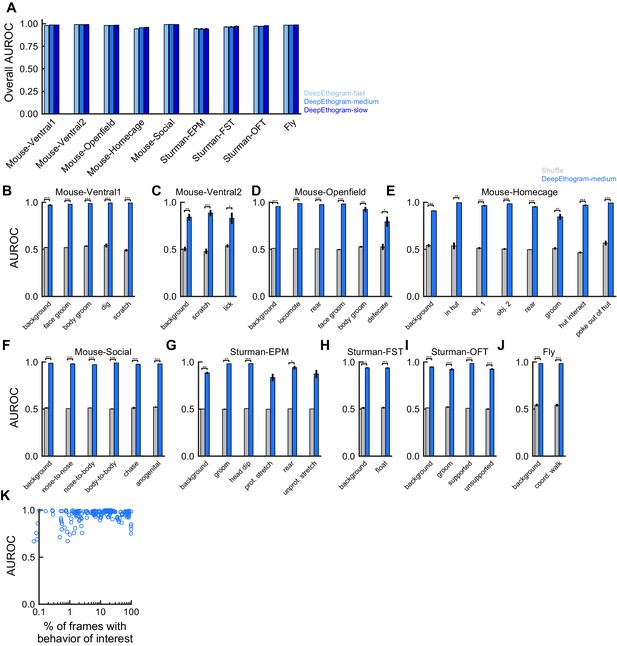
DeepEthogram performance, area under the receiver operating characteristic curve (AUROC).
All results are from the test sets only. (A) Overall recall for each model size and dataset. Error bars indicate mean ± SEM across five random splits of the data (three for Sturman-EPM). (B) AUROC for DeepEthogram-medium for individual behaviors on the Mouse-Ventral1 dataset. *p≤0.05, **p≤0.01, ***p≤0.001, paired t-test with Bonferroni correction. (C) Similar to (B), but for Mouse-Ventral2. (D) Similar to (B), but for Mouse-Openfield. (E) Similar to (B), but for Mouse-Homecage. (F) Similar to (B), but for Mouse-Social. (G) Similar to (B), but for Sturman-EPM. (H) Similar to (B), but for Sturman-FST. (I) Similar to (B), but for Sturman-OFT. (J) Similar to (B), but for Fly dataset. (K) Model AUROC vs. the percent of frames in the training set with the given behavior. Each point is for one behavior for one split of the data.
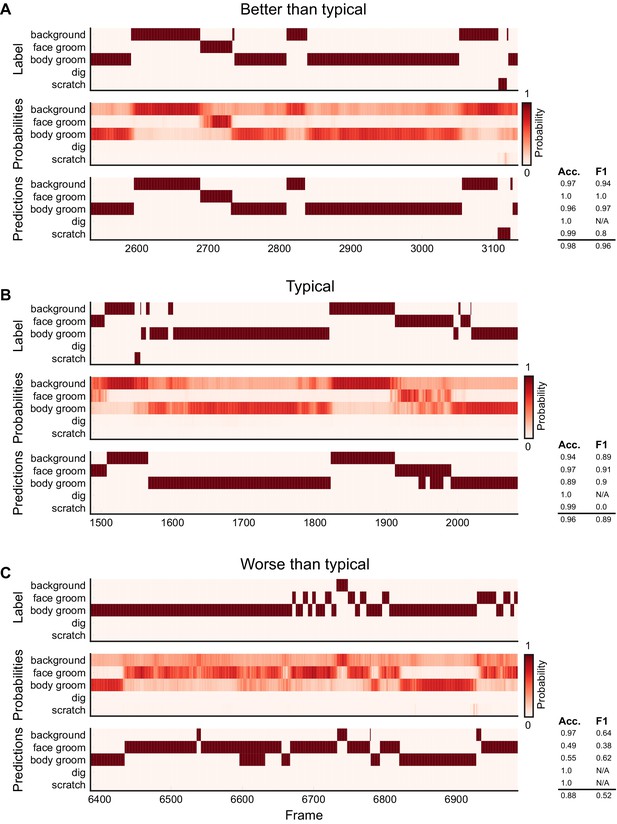
Ethogram examples for the Mouse-Ventral1 dataset.
(A) An example ethogram with above-average performance, showing the human labels, estimated probabilities for each behavior from DeepEthogram-medium, and the thresholded and postprocessed predictions, for data from the test set. The accuracy and F1 score for each behavior are shown, along with the overall accuracy and overall F1 score. (B, C) Similar to (A), except for approximately average performance and below-average performance.
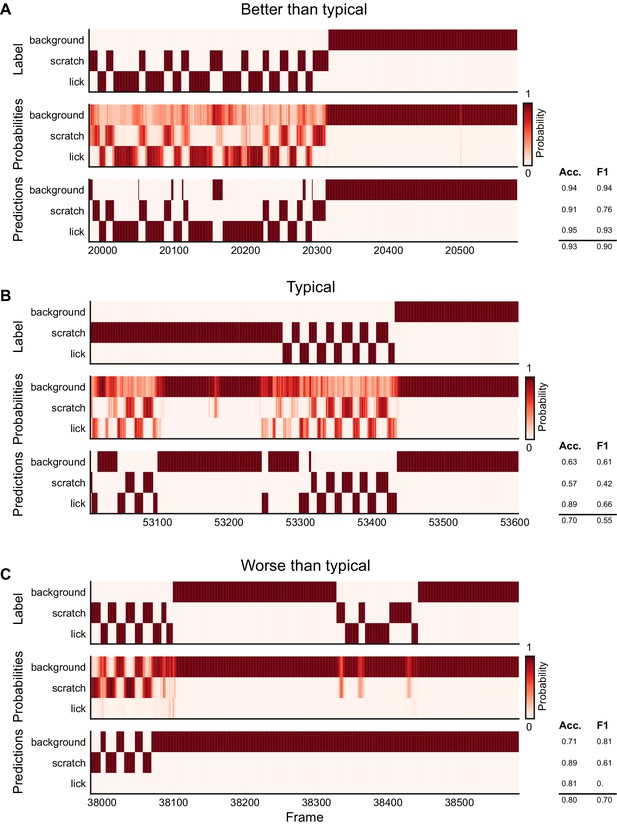
Ethogram examples for the Mouse-Ventral2 dataset.
(A) An example ethogram with above-average performance, showing the human labels, estimated probabilities for each behavior from DeepEthogram-medium, and the thresholded and postprocessed predictions, for data from the test set. The accuracy and F1 score for each behavior are shown, along with the overall accuracy and overall F1 score. (B, C) Similar to (A), except for approximately average performance and below-average performance.
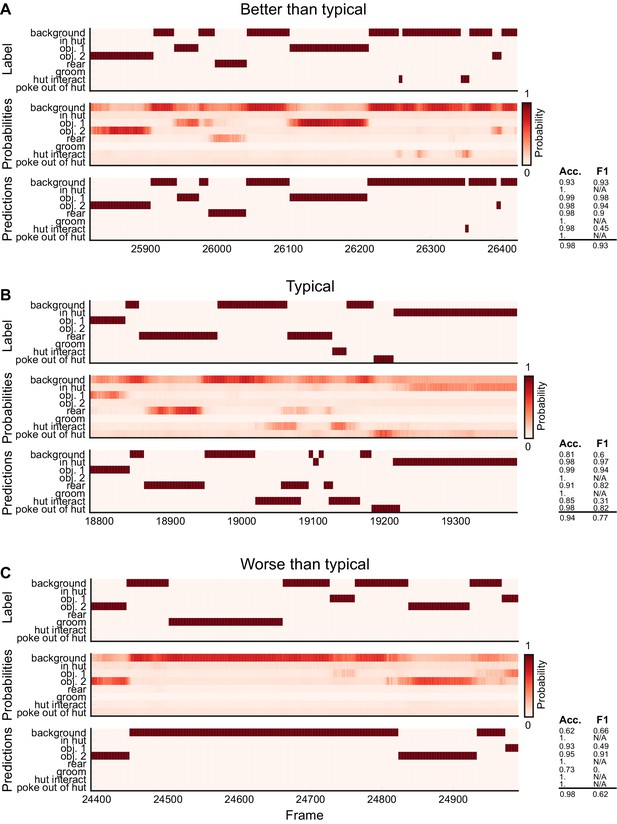
Ethogram examples for the Mouse-Openfield dataset.
(A) An example ethogram with above-average performance, showing the human labels, estimated probabilities for each behavior from DeepEthogram-medium, and the thresholded and postprocessed predictions, for data from the test set. The accuracy and F1 score for each behavior are shown, along with the overall accuracy and overall F1 score. (B, C) Similar to (A), except for approximately average performance and below-average performance.
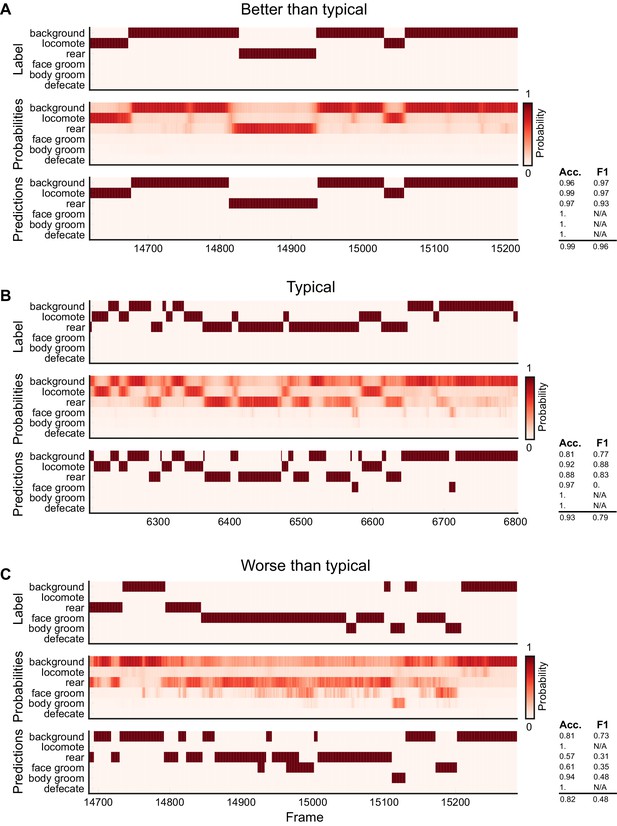
Ethogram examples for the Mouse-Homecage dataset.
(A) An example ethogram with above-average performance, showing the human labels, estimated probabilities for each behavior from DeepEthogram-medium, and the thresholded and postprocessed predictions, for data from the test set. The accuracy and F1 score for each behavior are shown, along with the overall accuracy and overall F1 score. (B, C) Similar to (A), except for approximately average performance and below-average performance.
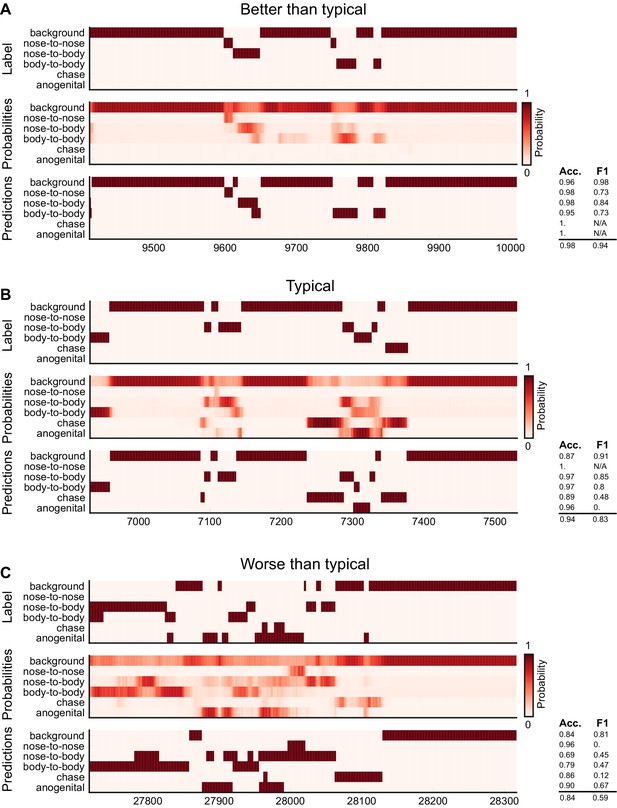
Ethogram examples for the Mouse-Social dataset.
(A) An example ethogram with above-average performance, showing the human labels, estimated probabilities for each behavior from DeepEthogram-medium, and the thresholded and postprocessed predictions, for data from the test set. The accuracy and F1 score for each behavior are shown, along with the overall accuracy and overall F1 score. (B, C) Similar to (A), except for approximately average performance and below-average performance.
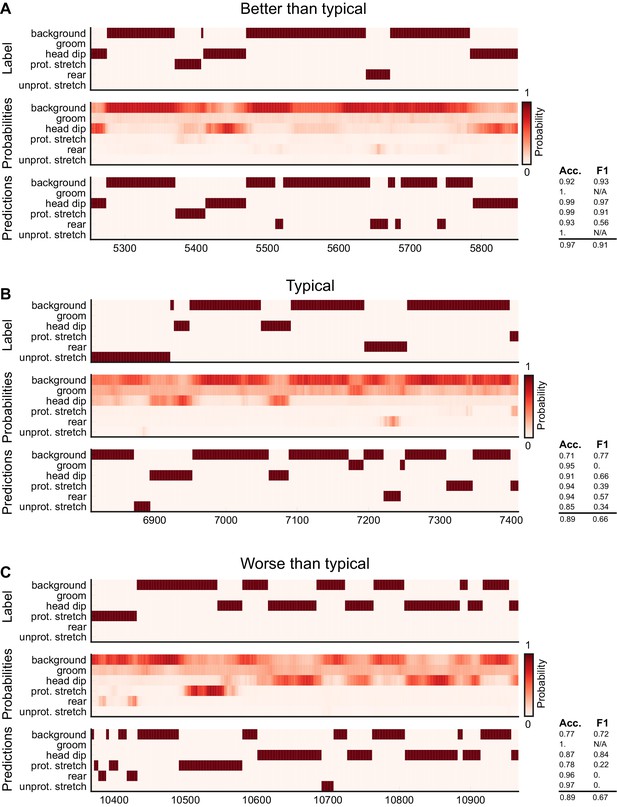
Ethogram examples for the Sturman-EPM dataset.
(A) An example ethogram with above-average performance, showing the human labels, estimated probabilities for each behavior from DeepEthogram-medium, and the thresholded and postprocessed predictions, for data from the test set. The accuracy and F1 score for each behavior are shown, along with the overall accuracy and overall F1 score. (B, C) Similar to (A), except for approximately average performance and below-average performance.
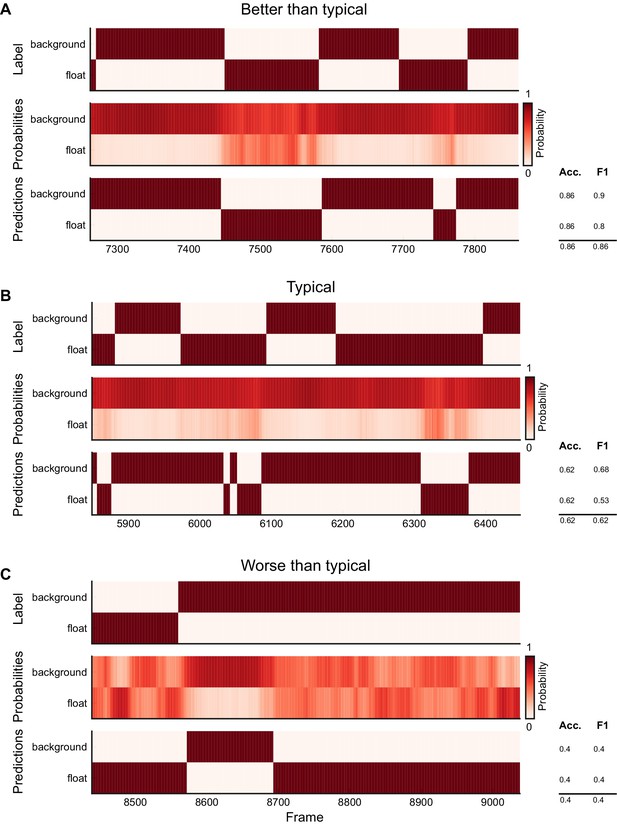
Ethogram examples for the Sturman-FST dataset.
(A) An example ethogram with above-average performance, showing the human labels, estimated probabilities for each behavior from DeepEthogram-medium, and the thresholded and postprocessed predictions, for data from the test set. The accuracy and F1 score for each behavior are shown, along with the overall accuracy and overall F1 score. (B, C) Similar to (A), except for approximately average performance and below-average performance.
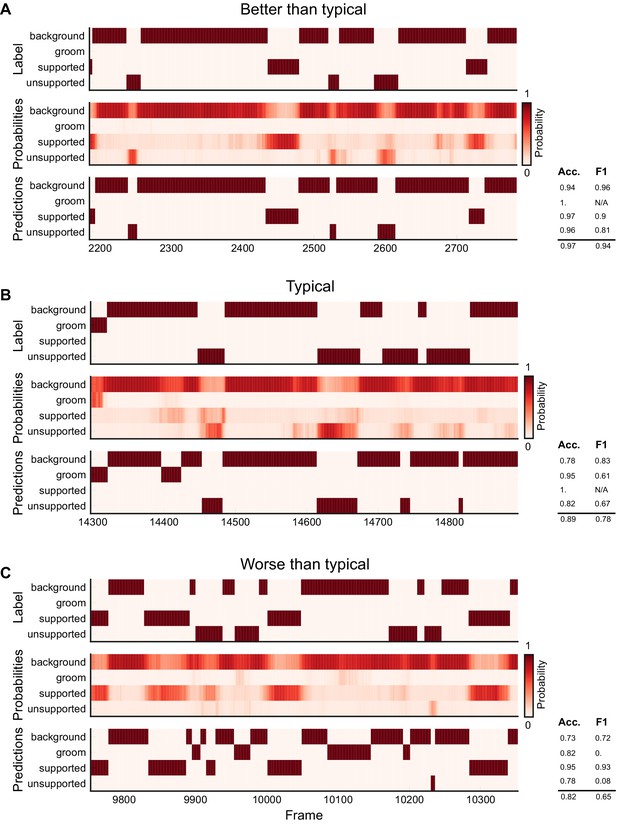
Ethogram examples for the Sturman-OFT dataset.
(A) An example ethogram with above-average performance, showing the human labels, estimated probabilities for each behavior from DeepEthogram-medium, and the thresholded and postprocessed predictions, for data from the test set. The accuracy and F1 score for each behavior are shown, along with the overall accuracy and overall F1 score. (B, C) Similar to (A), except for approximately average performance and below-average performance.
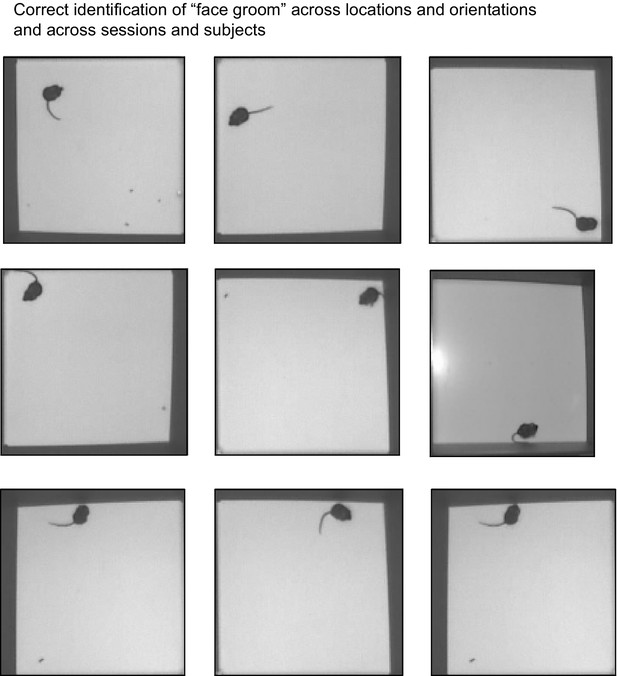
DeepEthogram exhibits position and heading invariance.
Nine randomly selected examples of the ‘face groom’ behavior from the Mouse-Openfield dataset. All examples were identified as ‘face groom’ by DeepEthogram-medium. The examples include different videos and different mice.
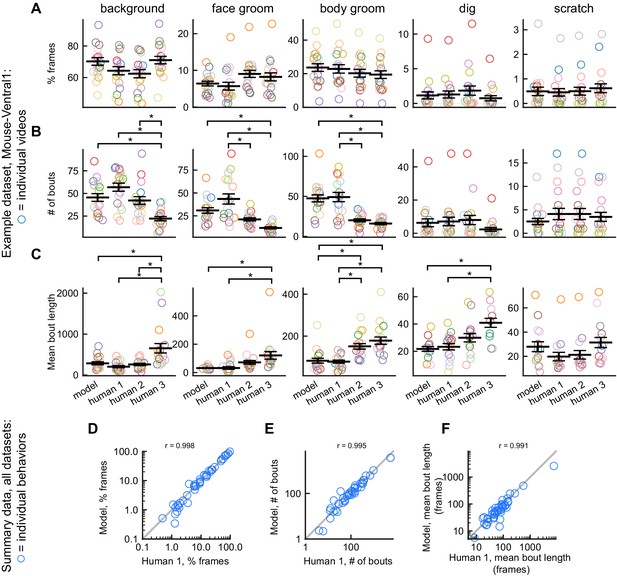
DeepEthogram performance on bout statistics.
All results from DeepEthogram-medium, test set only. (A–C) Comparison of model predictions and human labels on individual videos from the Mouse-Ventral1 dataset. Each point is one behavior from one video. Colors indicate video ID. Error bars: mean ± SEM (n = 18 videos). Asterisks indicate p<0.05, one-way ANOVA with Tukey’s multiple comparison test. No asterisk indicates p>0.05. (D–F) Comparison of model predictions and human labels on all behaviors for all datasets. Each circle is one behavior from one dataset, averaged across splits of the data. Gray line: unity.
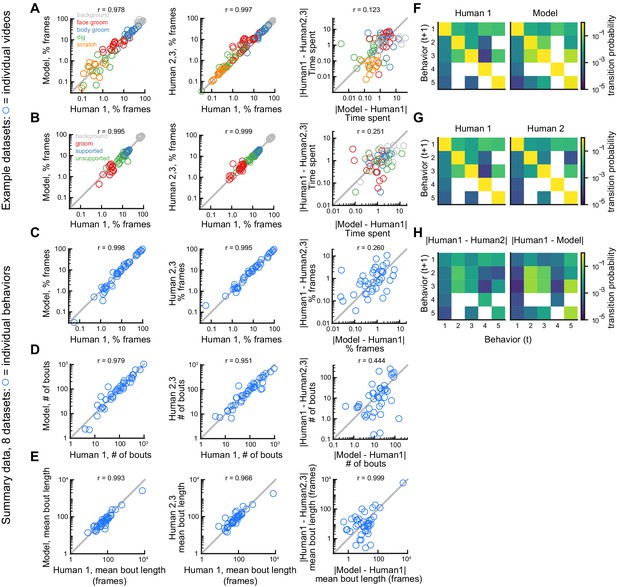
Comparison of model performance to human performance on bout statistics.
All model data are from DeepEthogram-medium, test set data. r values indicate Pearson’s correlation coefficient. (A) Performance on Mouse-Ventral1 dataset for time spent. Each circle is one behavior from one video. Left: Human 1 vs. model. Middle: Human 1 vs. Humans 2 and 3. Both Humans 2 and 3 are shown on the y-axis. Right: absolute error between Human 1 and model vs. absolute error between Human 1 and each of Humans 2 and 3. Model difference vs. human difference: p<0.001, paired t-test. (B) Similar to (A), but for Sturman-OFT dataset. Right: model difference vs. human difference: p<0.001, paired t-test. (C–E) Performance on all datasets with multiple human labelers (Mouse-Ventral1, Mouse-Openfield, Sturman-OFT, Sturman-EPM, Sturman-FST). Each point is one behavior from one dataset, averaged across data splits. Performance for Humans 2 and 3 were averaged. Similar to Figure 4D–F, but only for datasets with multiple labelers. Left: Human 1 vs. model. Middle: Human 1 vs. Humans 2 and 3. Right: absolute error between Human 1 and model vs. absolute error between Human 1 and each of Humans 2 and 3. p>0.05, paired t-test with Bonferroni correction, in (C–E) right panels. (F–H) Example transition matrices for Mouse-Ventral1 dataset. For humans and models, transition matrices were computed for each data split and averaged across splits.
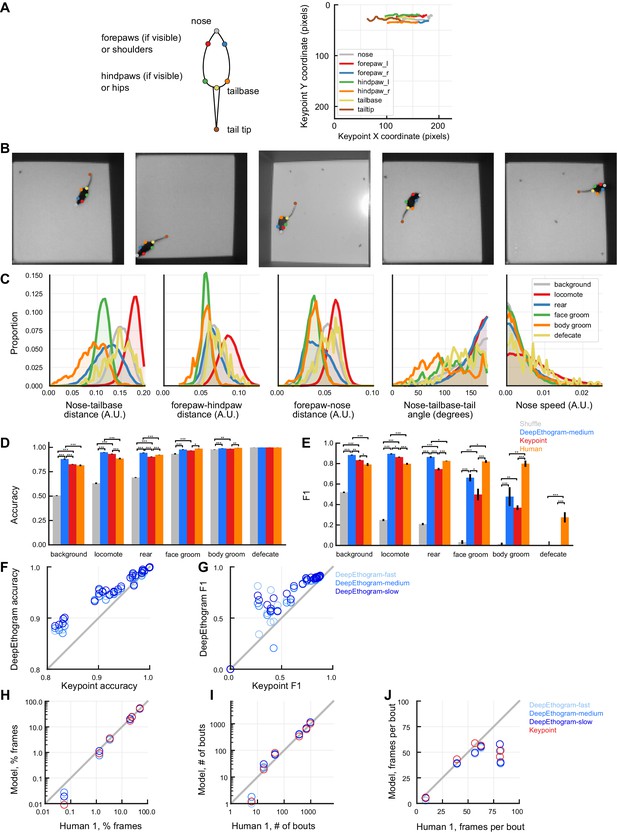
Performance of keypoint-based behavior classification on the Mouse-Openfield dataset.
(A) Left: keypoints identified, labeled, and predicted using DeepLabCut. Right: example keypoint sequence predicted by DeepLabCut from a held-out video. (B) Example images from held-out videos showing good DeepLabCut performance. (C) Histograms of behavioral features derived from keypoints for each behavior. (D) Accuracy on the test set. Error bars: mean ± SEM, n = 5 data splits. *p≤0.05, **p≤0.01, ***p≤0.001, repeated measures ANOVA with post-hoc Tukey’s honestly significant difference test. Human vs. shuffle results not shown for clarity. (E) Similar to (A), but for F1. (F) Accuracy of keypoint-based behavioral classification vs. DeepEthogram. Each point is one behavior from one model type (colored as in D) and one data split. (G) similar to (F), but for F1. (H) Human vs. model time spent exhibiting each behavior. Each point is one behavior from one model type, averaged across data splits. (I) similar to (H), but for average bout number. (J) similar to (H), but for average frames per bout.
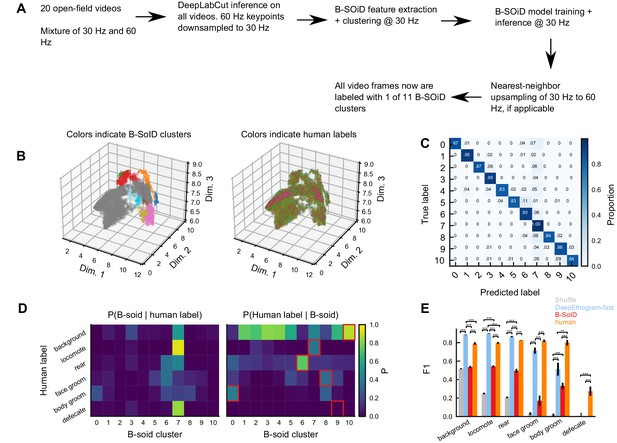
Comparison with unsupervised methods.
(A) B-SoID pipeline. (B) B-SoID behavioral space. Shown are a random sample of points that B-SoID labeled confidently (57% of total data). Left: colors are B-SoID cluster assignments. Right: colors (0–5) indicate human labeled behaviors. Note the overall lack of clustering of human-identified colors. (C) B-SoID classifier confusion matrix. X-axis: label predicted by the B-SoID classifier (Random forest). Note the good performance; the classifier successfully recaptures the HDBScan clustering, indicating the B-SoID is performing as expected. (D) Comparison between B-SoID cluster assignments and human labels. Left: each element is the proportion of B-soid clusters that correspond to the given human label. Rows sum to one. Right: each element is the proportion of human labels corresponding to the given B-SoID cluster. Columns sum to 1. Red outlines indicate the B-SoID cluster with maximum correspondence to the human label. Note the overall lack of a consistent structure between human-identified behaviors and B-SoID clusters. One exception: 74% of cluster six corresponds to the ‘rearing’ behavior. (E) Performance comparison between the unsupervised pipeline and DeepEthogram-fast. *p≤0.05, **p≤0.01, ***p≤0.001, repeated measures ANOVA with post-hoc Tukey’s honestly significant difference test. Human vs. shuffle results not shown for clarity.
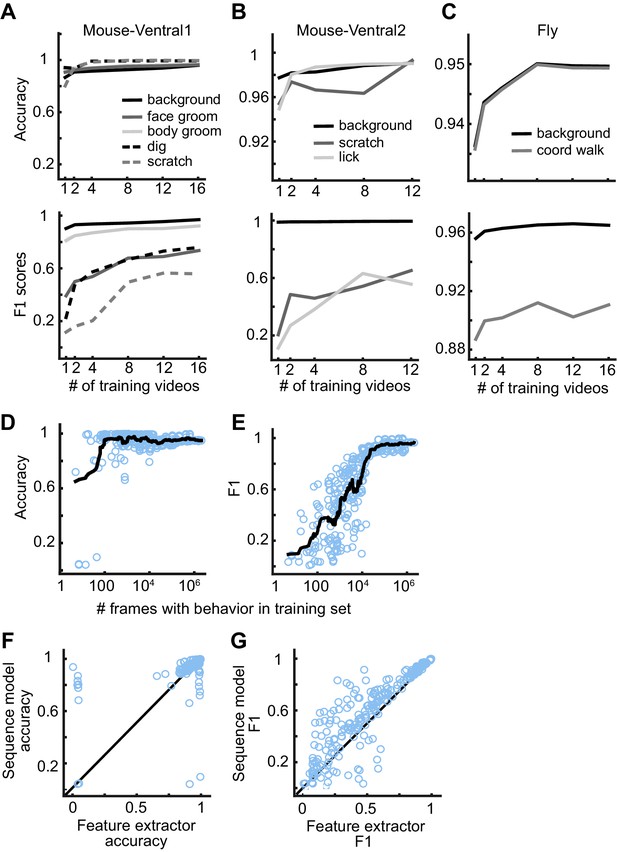
DeepEthogram performance as a function of training set size.
(A) Accuracy (top) and F1 score (bottom) for DeepEthogram-fast as a function of the number of videos in the training set for Mouse-Ventral1, shown for each behavior separately. The mean is shown across five random selections of training videos. (B, C) Similar to (A), except for the Mouse-Ventral2 dataset and Fly dataset. (D) Accuracy of DeepEthogram-fast as a function of the number of frames with the behavior of interest in the training set. Each point is one behavior for one random split of the data, across datasets. The black line shows the running average. For reference, 104 frames is ~5 min of behavior at 30 frames per second.(E) Similar to (D), except for F1 score. (F) Accuracy for the predictions of DeepEthogram-fast using the feature extractors only or using the sequence model. Each point is one behavior from one split of the data, across datasets, for the splits used in (D, E). (G) Similar to (F), except for F1 score.
Videos
DeepEthogram example from the Mouse-Ventral1 dataset.
Video is from the test set. Top: raw image. Title indicates frame number in video. Tick legends indicate pixels. Middle: human labels. Black box indicates the current frame. Bottom: DeepEthogram predictions from a trained DeepEthogram-medium model.
DeepEthogram example from the Mouse-Ventral2 dataset.
Video is from the test set. Top: raw image. Title indicates frame number in video. Tick legends indicate pixels. Middle: human labels. Black box indicates the current frame. Bottom: DeepEthogram predictions from a trained DeepEthogram-medium model.
DeepEthogram example from the Mouse-Openfield dataset.
Video is from the test set. Top: raw image. Title indicates frame number in video. Tick legends indicate pixels. Middle: human labels. Black box indicates the current frame. Bottom: DeepEthogram predictions from a trained DeepEthogram-medium model.
DeepEthogram example from the Mouse-Homecage dataset.
Video is from the test set. Top: raw image. Title indicates frame number in video. Tick legends indicate pixels. Middle: human labels. Black box indicates the current frame. Bottom: DeepEthogram predictions from a trained DeepEthogram-medium model.
DeepEthogram example from the Mouse-Social dataset.
Video is from the test set. Top: raw image. Title indicates frame number in video. Tick legends indicate pixels. Middle: human labels. Black box indicates the current frame. Bottom: DeepEthogram predictions from a trained DeepEthogram-medium model.
DeepEthogram example from the Sturman-EPM dataset.
Video is from the test set. Top: raw image. Title indicates frame number in video. Tick legends indicate pixels. Middle: human labels. Black box indicates the current frame. Bottom: DeepEthogram predictions from a trained DeepEthogram-medium model.
DeepEthogram example from the Sturman-FST dataset.
Video is from the test set. Top: raw image. Title indicates frame number in video. Tick legends indicate pixels. Middle: human labels. Black box indicates the current frame. Bottom: DeepEthogram predictions from a trained DeepEthogram-medium model.
DeepEthogram example from the Sturman-OFT dataset.
Video is from the test set. Top: raw image. Title indicates frame number in video. Tick legends indicate pixels. Middle: human labels. Black box indicates the current frame. Bottom: DeepEthogram predictions from a trained DeepEthogram-medium model.
DeepEthogram example from the Flies dataset.
Video is from the test set. Top: raw image. Title indicates frame number in video. Tick legends indicate pixels. Middle: human labels. Black box indicates the current frame. Bottom: DeepEthogram predictions from a trained DeepEthogram-medium model.
Tables
Inference speed.
| Dataset | Resolution | Inference time (FPS) | |||||
|---|---|---|---|---|---|---|---|
| Titan RTX | Geforce 1080 Ti | ||||||
| DEG_f | DEG_m | DEG_s | DEG_f | DEG_m | DEG_s | ||
| Mouse-Ventral1 | 256 × 256 | 235 | 128 | 34 | 152 | 76 | 13 |
| Mouse-Ventral2 | 256 × 256 | 249 | 132 | 34 | 157 | 79 | 13 |
| Mouse-Openfield | 256 × 256 | 211 | 117 | 33 | 141 | 80 | 13 |
| Mouse-Homecage | 352 × 224 | 204 | 102 | 28 | 132 | 70 | 11 |
| Mouse-Social | 224 × 224 | 324 | 155 | 44 | 204 | 106 | 17 |
| Sturman-EPM | 256 × 256 | 240 | 123 | 34 | 157 | 83 | 13 |
| Sturman-FST | 224 × 448 | 157 | 75 | 21 | 106 | 51 | 9 |
| Sturman-OFT | 256 × 256 | 250 | 125 | 34 | 159 | 84 | 13 |
| Flies | 128 × 192 | 623 | 294 | 89 | 378 | 189 | 33 |
Model summary.
| Model name | Flow generator (parameters) | Feature extractor (parameters) | Sequence model (parameters) | # frames input to flow generator | # frames input to RGB feature extractor | Total parameters |
|---|---|---|---|---|---|---|
| DeepEthogram-fast | TinyMotionNet (1.9M) | ResNet18 × 2 (22.4M) | TGM (250K) | 11 | 1 | ~24.5M |
| DeepEthogram-medium | MotionNet (45.8M) | ResNet50 × 2 (49.2M) | TGM (250K) | 11 | 1 | ~ 95.2M |
| DeepEthogram-slow | TinyMotionNet3D (0.4M) | ResNet3D-34 × 2 (127M) | TGM (250K) | 11 | 11 | ~ 127.6M |