RNF41 regulates the damage recognition receptor Clec9A and antigen cross-presentation in mouse dendritic cells
Figures
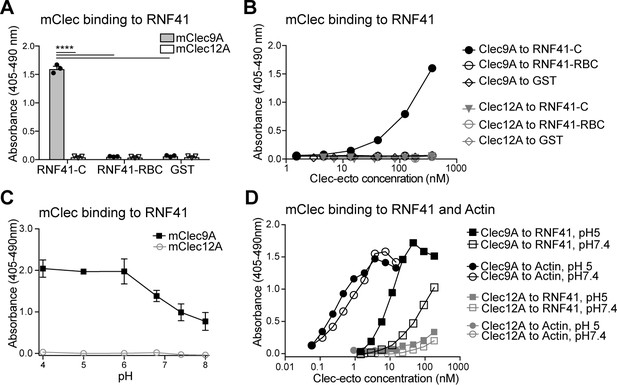
Clec9A binds to the E3 Ubiquitin ligase RNF41.
(A, B) Mouse Clec9A binds to the RNF41 C-terminal domain. ELISA plates were coated with GST-tagged RNF41-C-terminal domain (RNF41-C), RNF41-N-terminal domains (RNF41-RBC) and GST control. (A) Binding of FLAG-tagged mClec9A-ecto or the control FLAG-mClec12A-ecto (10 μg/ml) was detected with anti-FLAG HRP. Cumulative data of three experiments is shown, presented as mean ± SEM (unpaired t-test) ****p<0.0001. (B) Binding of FLAG-tagged mClec9A-ecto or the control FLAG-mClec12A-ecto (10 μg/ml to 0.04 μg/ml) was detected with anti-FLAG HRP. Cumulative data of three experiments is shown. (C, D) Mouse Clec9A binding to RNF41 is enhanced at low pH. ELISA plates were coated with GST-tagged RNF41-C, actin complexes or GST. Binding of biotinylated FLAG-tagged mClec9A-ecto or biotinylated control FLAG-mClec12A-ecto at different pH was detected with streptavidin-HRP. Data is presented as the absorbance 405–490 nm of (Clec binding to GST-RNF41 - Clec binding to GST). (C) Cumulative data of three experiments is shown, presented as mean ± SEM. mClec9A (2.5 μg/ml) binding to RNF41-C at pH 4, pH 5 and pH 6 was significantly greater than at pH 7.4 and at pH 8 (1-way ANOVA) *p<0.05. (D) Representative of three independent experiments.
-
Figure 1—source data 1
Clec9A binding to RNF41.
- https://cdn.elifesciences.org/articles/63452/elife-63452-fig1-data1-v1.xlsx
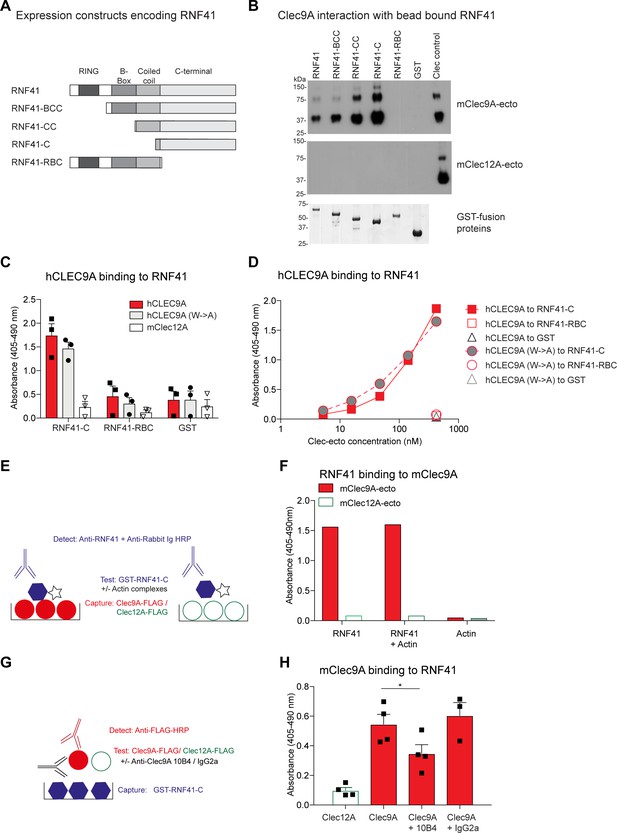
Clec9A binds to the E3 ubiquitin ligase RNF41.
(A) RNF41 constructs. GST-tagged RNF41 constructs were generated, which encoded either full-length GST-tagged RNF41, or truncated forms of RNF41. (B) mClec9A binds to the RNF41 C-terminal domain. FLAG-tagged mClec9A-ecto and control, FLAG-tagged mClec12A-ecto, were incubated with glutathione bead bound GST-RNF41 proteins, or a GST control. Bound proteins were eluted and detected using anti-FLAG-HRP. A sample of the purified Clec9A-ecto or Clec12A-ecto is shown in lane 7 (Clec control). The bottom panel shows Coomassie staining of GST-RNF41 fusion proteins eluted from glutathione beads. (C) Human CLEC9A binds to the C-terminal domain of RNF41. RNF41-C, RNF41-RBC and GST control were coated onto ELISA plates, and incubated with FLAG-tagged human wild-type CLEC9A (hCLEC9A-ecto; 10 μg/ml) or mutant CLEC9A (hCLEC9A W131A, W227A, represented as hCLEC9A-ecto (W->A); 10 μg/ml). Binding was detected using anti-FLAG-HRP. Cumulative data of three experiments is shown, presented as the mean ± standard error of the mean (SEM). (D) Human CLEC9A binding to RNF41. RNF41-C, RNF41-RBC and GST control were coated onto ELISA plates, and incubated with FLAG-tagged wild-type CLEC9A (hCLEC9A-ecto, 10 to 0.04 μg/ml) and mutant CLEC9A (hCLEC9A W131A, W227A, represented as hCLEC9A-ecto W->A, 10 to 0.04 μg/ml). Binding was detected using anti-FLAG-HRP. (E, F) RNF41 binding to Clec9A is not inhibited by Actin. mClec9A-ecto and the control, mClec12A-ecto (10 μg/ml) were coated onto ELISA plates as per schematic diagram (E), RNF41-C (10 μg/ml) and Actin complexes were either added individually, or mixed together then incubated onto ELISA plates. RNF41 binding to Clec9A was detected using rabbit anti-RNF41 and anti-rabbit Ig-HRP (F). Representative of two experiments. (G, H) Mouse Clec9A-ecto binding to RNF41 is partially inhibited using anti-Clec9A mAb 24/04-10B4. RNF41-C (10 μg/ml) was coated onto ELISA plates as per schematic diagram (G). FLAG-tagged mClec9A-ecto (3 μg/ml) was pre-incubated alone, or with anti-Clec9A mAb 24/04-10B4 or rat IgG2a isotype control GL117 (120 μg/ml). Clec9A-Ab complexes and controls (hCLEC12A-ecto) were incubated onto ELISA plates and Clec binding to RNF41 detected using anti-FLAG-HRP (H). Cumulative data of four experiments is shown, presented as the mean ± SEM. Analyzed by paired t-test. Clec9A binding to RNF41-C was significantly greater than Clec9A binding to RNF41-C in the presence of anti-Clec9A mAb 24/04-10B4, *p<0.05.
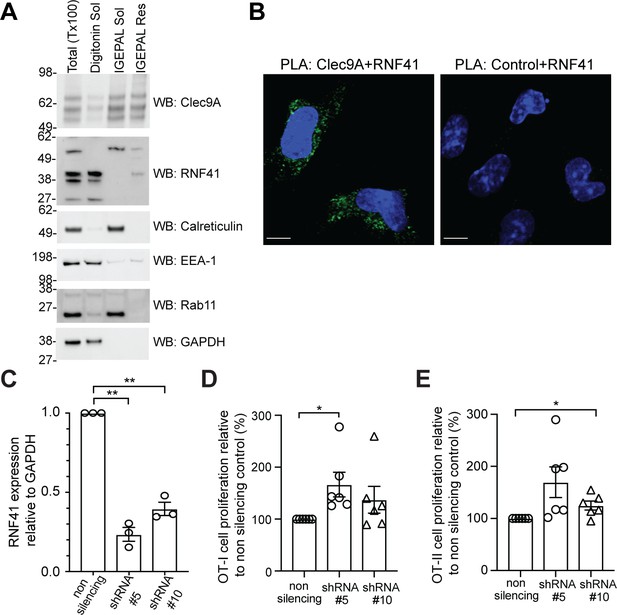
RNF41 regulates presentation of dead cell-associated Ag in DC.
(A) RNF41 is associated with membrane-associated cellular fractions. Cellular fractions were prepared from MutuDC 1940 by sequential detergent lysis and centrifugation conditions. In brief, DC were initially treated with digitonin and centrifugation, and digitonin soluble fractions harvested (containing GAPDH, EEA-1). Digitonin resistant fractions were then subjected to IGEPAL lysis and centrifugation, and IGEPAL soluble fractions (containing Calreticulin and Rab11) and IGEPAL resistant fractions harvested. Total lysates (Tx100) or cellular fractions (Digitonin soluble, Sol; IGEPAL soluble, Sol; IGEPAL resistant, Resistant) were analyzed by western blot (WB). Representative of five independent experiments. (B) Clec9A and RNF41 interact in DC. A proximity ligation assay (PLA) was performed using MutuDC 1940 with mouse polyclonal anti-Clec9A serum directed against the extracellular domain, or control unimmunized serum, and rabbit anti-RNF41 polyclonal Ab. PLA signal is shown in green, DAPI (nucleus) in blue. Images are representative of four independent experiments (Clec9A+RNF41: 214 cells, Control+RNF41: 168 cells). Scale bars represent 10 μm. (C–E) RNF41 knockdown enhances presentation of dead cell Ag. MutuDC 1940 were transduced with shRNA construct #5 or #10 to knockdown RNF41 expression, compared with a non-silencing control retrovirus. (C) Reduced RNF41 expression was confirmed by qPCR. Cumulative data from three independent experiments, demonstrating the mean ± SEM (paired t-test) **p<0.01. (D–E) Transduced MutuDC 1940 were cultured with dead CHO-K1 cells expressing OVA at a ratio of 1 DC: 2 dead CHO-K1 cells, followed by culture with (D) OT-I and (E) OT-II Transgenic T cells for 3 days. T cell proliferation relative to the non-silencing control. Cumulative data from six independent experiments, demonstrating the mean ± SEM (paired t-test) *p<0.05.
-
Figure 2—source data 1
RNF41 knockdown: RNF41 expression and Ag presentation.
- https://cdn.elifesciences.org/articles/63452/elife-63452-fig2-data1-v1.xlsx
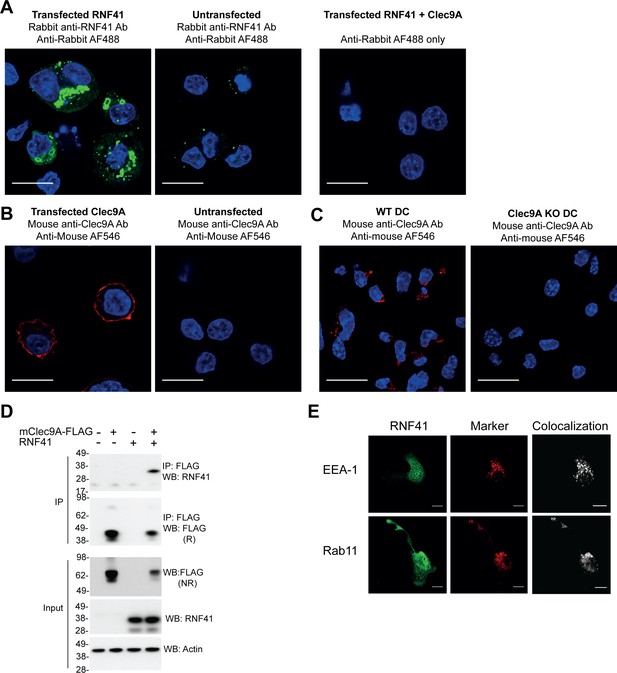
Detection of RNF41, Clec9A and their interaction.
(A, B) Detection of RNF41 and Clec9A expression in transfected cells. 293 F cells were transfected with full-length RNF41, FLAG-tagged full-length mClec9A (mClec9A-FLAG) or both. RNF41 was visualized by confocal microscopy using rabbit anti-RNF41 antibody; untransfected cells indicate staining of endogenous RNF41(untransfected cells: 17 cells, RNF41 transfected cells: 22 cells). Clec9A was visualized using polyclonal mouse anti-Clec9A Ab (untransfected cells: 11 cells, Clec9A transfected cells: 10 cells). Scale bars represent 10 μm. (C) Clec9A expression in Flt3L-DC. Flt3L-DC were generated from wild-type (WT DC) or Clec9a-/− mice (Clec9A KO DC). Cells were fixed and stained with polyclonal mouse anti-Clec9A Ab (WT DC: 59 cells, Clec9A KO DC: 37 cells). Scale bars represent 10 μm. (D) Co-immunoprecipitation of Clec9A and RNF41. 293 F cells were co-transfected with mClec9A-FLAG, RNF41 or vector alone (-). At 24 hr, immunoprecipitated (IP) Clec9A complexes and cellular lysates (input) were analyzed by WB under reducing (R) or non-reducing (NR) conditions. Representative of two independent experiments. (E) RNF41 colocalizes with endosomal vesicles in DC. RNF41, early endosomes (EEA-1) and recycling endosomes (Rab11) were visualized in Flt3L-DC by confocal microscopy (RNF41, indicated in green, EEA-1, and Rab11 in red). Colocalization images were generated using Image J; pixels that have a positive signal in both channels are shown in white. Images are representative of four independent experiments (EEA-1: 53 cells, Rab11: 69 cells). Scale bars represent 10 μm.
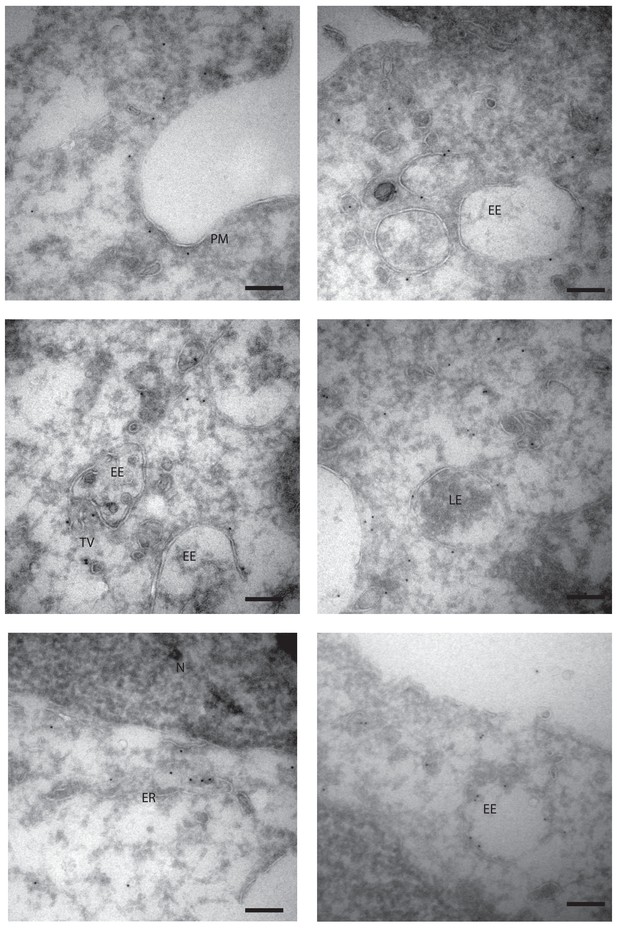
Subcellular localization of RNF41 in DC by immuno-electron microscopy.
MutuDC1940 were fixed for Tokuyasu immuno-electron microscopy and section labeled with anti-RNF41 antibody and 10 nm gold particles coupled to protein A. Localization to the plasma membrane (PM), early (EE) and late endosomes (LE), endosomal tubular-vesicular compartments (TV), as well as localization to the endoplasmic reticulum (ER) and cytoplasm is shown. Controls using Protein A Gold in the absence of primary antibody did not show any labeling. Scale bars represent 200 nm. Representative of 2 independent experiments.
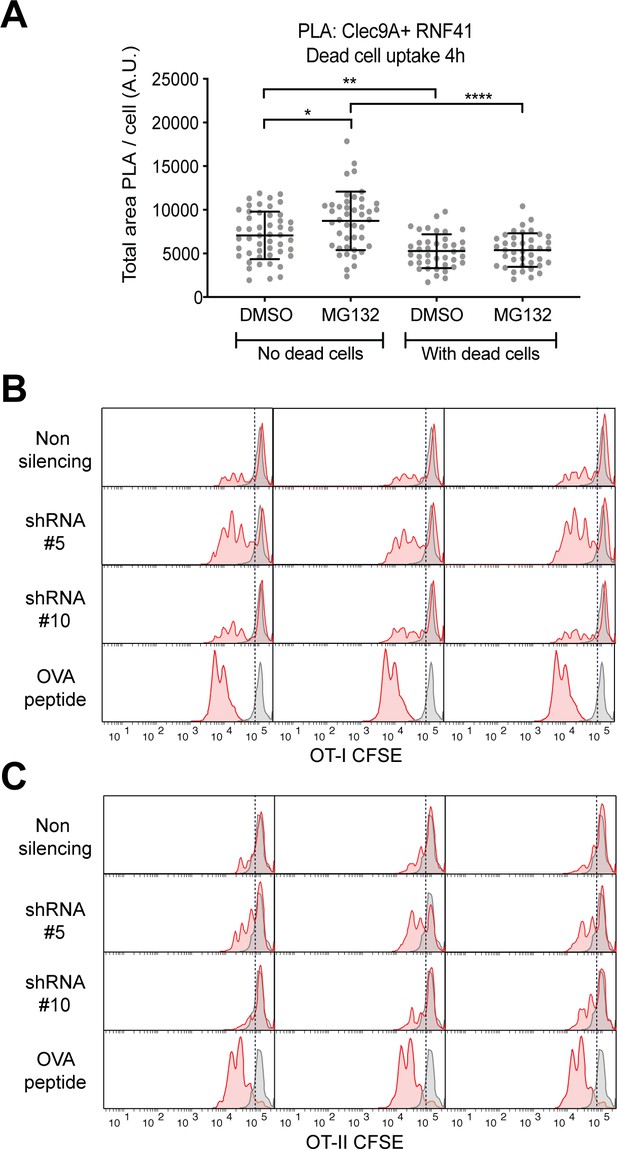
RNF41 regulates Clec9A and the presentation of dead cell-associated Ag in DC.
(A) Proximity Ligation Assay quantification of Clec9A and RNF41 interaction in DC. MutuDC 1940 were cultured for 4 hr +/− PKH26 labeled CHO-K1 dead cells, and the proteasomal inhibitor MG132 (5 μM) or the control DMSO. A PLA was performed for Clec9A and RNF41 (No dead cells; DMSO: 49 cells, MG132: 43 cells. With dead cells; DMSO: 40 cells, MG132: 40 cells). The total area of PLA per cell (Arbitrary Units, A.U.) was analyzed by one-way ANOVA with Tukey’s multiple comparisons. Bars represent mean ± SD.* p<0.05, **p<0.01, ****p<0.0001. (B, C) RNF41 knockdown enhances presentation of dead cell Ag. MutuDC1940 transduced with RNF41 shRNA construct #5, #10 or a non-silencing control, were cultured with dead CHO-K1 cells expressing OVA at a ratio of 1 DC: two dead CHO-K1 cells, followed by culture with (B) OT-I and (C) OT-II Transgenic T cells for 3 days. As a positive control, parental MutuDC1940 were incubated with OVA peptide (0.5 µg/ml). Proliferation of OT-I and OT-II cells is shown for three replicates from one representative experiment (red: DC cultured with dead CHO-OVA cells; gray: DC without dead CHO-OVA cells).
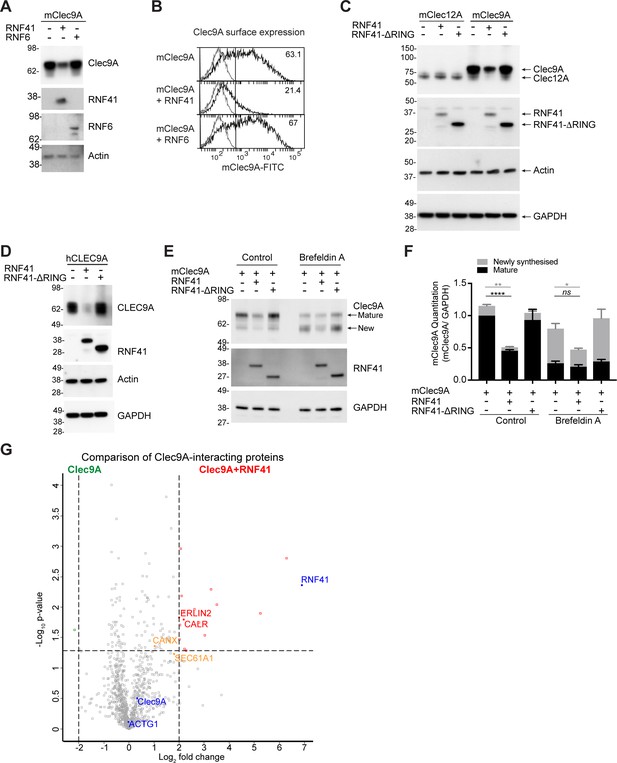
RNF41-mediated regulation of Clec9A receptor levels is associated with novel interactions.
(A, B) RNF41 co-expression reduces Clec9A expression. 293 F cells were co-transfected with FLAG-tagged full-length mClec9A (mClec9A-FLAG), RNF41 or RNF6-Myc. (A) Cellular lysates were analyzed by WB (24 hr). (B) Clec9A surface expression of transfected cells was analyzed by flow cytometry (Transfected Clec9A: solid line; untransfected: gray line; 48 hr). Representative of three independent experiments. Cells transfected with Clec9A and RNF41 had significantly reduced Clec9A surface levels compared with cells transfected with Clec9A; cumulative data from six experiments (paired t-test) **p<0.01. (C, D) RNF41 regulation of Clec9A is mediated by the RNF41 RING domain. 293 F cells were co-transfected with mClec9A-FLAG/mClec12A-FLAG/hCLEC9A-FLAG, and RNF41 or RNF41-ΔRING. Cellular lysates were analyzed by WB. Representative of (C) 8 and (D) three independent experiments. (E, F) RNF41 regulates newly synthesized and mature Clec9A. 293 F cells were co-transfected with mClec9A-FLAG and RNF41 or RNF41-ΔRING. At 14 hr post-transfection, cells were treated with Brefeldin A (5 μg/ml) or DMSO for a further 4 hr. (E) Cellular lysates were analyzed by WB. (F) Densitometric analysis of newly synthesized and mature Clec9A levels relative to GAPDH from four experiments is shown, demonstrating Clec9A levels as mean ± SEM (paired t-test). ns not significant, *p<0.05, **p<0.01, ****p<0.0001. (G) Identification of novel Clec9A-interaction partners that are enhanced by RNF41. 293 F cells were co-transfected with mClec9A-FLAG in the presence or absence of RNF41. At 22 hr post-transfection Clec9A-interacting proteins were immunoprecipitated (IP) and analyzed by LC-MS/MS. Volcano plot (X-axis: log2 fold-change; Y-axis: -Log10 p-value) comparing Clec9A-interacting proteins from cells transfected with mClec9A+RNF41 versus mClec9A alone. The dotted vertical lines indicate a 4-fold protein change. The dotted horizontal line indicates a p-value cut-off of 0.05. Selected proteins with >4 fold upregulation are annotated in red,>2 fold upregulation in orange,>4 fold downregulation in green. Clec9A and its interacting proteins, actin (ACTG1) and RNF41, are in blue.
-
Figure 3—source data 1
RNF41 regulation of Clec9A.
- https://cdn.elifesciences.org/articles/63452/elife-63452-fig3-data1-v1.xlsx
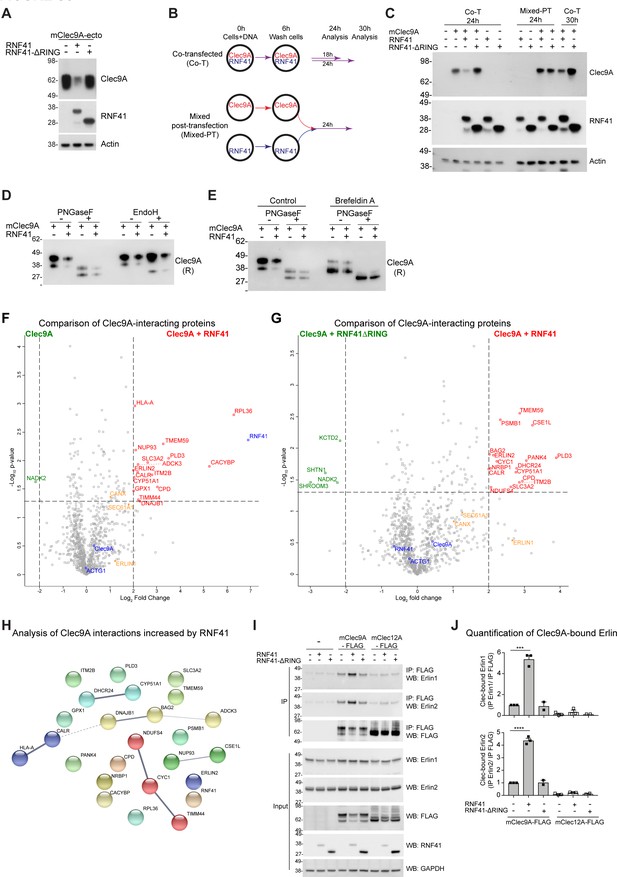
RNF41 mediated regulation of Clec9A is associated with novel interactions.
(A) RNF41 regulation of Clec9A is mediated through the extracellular domain of Clec9A. 293 F cells were co-transfected with constructs encoding a soluble FLAG-tagged extracellular domain of mClec9A (mClec9A-ecto) and RNF41 or RNF41-ΔRING. Culture supernatants were analyzed for expression of mClec9A-ecto (anti-FLAG M2). Cellular lysates were analyzed for expression of RNF41 and Actin by western blot (WB). (B, C) RNF41 interacts with Clec9A within cells to regulate Clec9A levels. 293 F cells were co-transfected with FLAG-tagged full-length mClec9A (mClec9A-FLAG) and RNF41 or RNF41-ΔRING (Co-transfected; Co-T) and cultured for 24–30 hr. At the same time, 293 F cells were separately transfected with mClec9A-FLAG, RNF41 or RNF41-ΔRING, as per schematic. At 6 hr post-transfection, all transfected cells were washed to remove transfection reagents and plasmids, and cells that had been separately transfected with Clec9A (93% viable) or RNF41 (82% viable, 18% dead cells) mixed together for a further 24 hr (Mixed post-transfection; Mixed-PT). Cellular lysates were analyzed by WB (24 hr, 30 hr). Representative of two independent experiments. (D, E) RNF41 can regulate newly synthesized Clec9A. 293 F cells were co-transfected with mClec9A-FLAG, RNF41 or vector (-). (D) At 22 hr post-transfection, Clec9A complexes were immunoprecipitated (IP) using anti-FLAG beads, and treated with de-glycosylation enzymes, PNGaseF (500 U) or EndoH (500 U), and analyzed by WB using anti-FLAG under reducing conditions. Both monomeric forms of Clec9A were sensitive to PNGaseF, which removes complex and high mannose glycosylation, but only the smaller monomeric form of Clec9A was sensitive to EndoH that removes high mannose glycosylation. Representative of three independent experiments. (E) At 14 hr post-transfection, cells were treated with Brefeldin A (5 μg/ml) or DMSO for a further 4 hr. Clec9A complexes were IP using anti-FLAG beads, treated with PNGaseF (500 U) and analyzed by WB using anti-FLAG under reducing conditions. Representative of two independent experiments. (F, G) Identification of Clec9A-interaction partners that are enhanced by RNF41. Volcano plot (X-axis: log2 fold-change; Y-axis: -Log10 p-value) comparing Clec9A-interacting proteins from cells transfected with (F) mClec9A + RNF41 versus mClec9A alone and (G) mClec9A + RNF41 versus mClec9A + RNF41 ΔRING. The dotted vertical lines indicate a 4-fold protein change. The dotted horizontal line indicates a p-value cut-off of 0.05. Selected proteins with >4 fold upregulation are annotated in red,>2 fold upregulation in orange,>4 fold downregulation in green. Clec9A and its interacting proteins, actin and RNF41, are in blue. (H) Clec9A-RNF41 interactome. The protein-protein interaction network between 25 proteins that showed over 4-fold increase in association with Clec9A in the presence of RNF41 compared with either vector alone or RNF41 ΔRING, with a p-value cutoff of 0.05, was predicted using STRING analysis (https://string-db.org). Protein nodes were MCL clustered with an inflation parameter of 3. The strength of data support for each interaction is represented by the line thickness. (I, J) RNF41 mediates increased Clec9A-Erlin interaction. IP Clec9A complexes from cells transfected with mClec9A/mClec12A, RNF41/RNF41-ΔRING and Ub-Myc were analyzed for co-IP of Erlin1 and Erlin2 by WB. Quantification of Clec9A-bound Erlin1/2 was performed by densitometric analysis of IP Erlin relative to IP Clec9A from three experiments, demonstrated as mean ± SEM (unpaired t-test) ***p<0.001, ****p<0.0001.
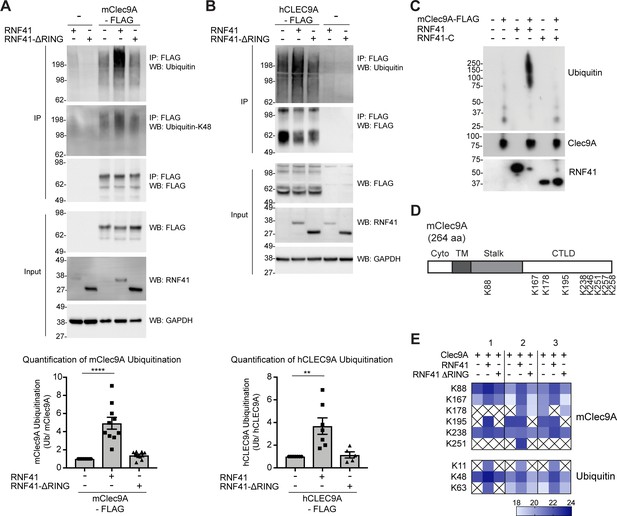
RNF41 interacts with Clec9A to mediate Clec9A ubiquitination.
(A, B) RNF41 mediates mClec9A ubiquitination in vivo. 293 F cells were co-transfected with mClec9A-FLAG or hCLEC9A-FLAG, Myc-tagged ubiquitin (Ub-Myc), and RNF41 or RNF41-ΔRING. At 22 hr, IP Clec9A complexes were analyzed for ubiquitination by WB using anti-Ubiquitin Ab, representative of (A) 10 experiments, (B) seven experiments, and anti-Ubiquitin K-48 Ab, representative of (A) two experiments. Densitometric analysis of ubiquitinated Clec9A relative to total Clec9A from IP of Clec9A complexes from (A) 10 experiments and (B) seven experiments is shown, as mean ± SEM (ANOVA) **p<0.01, ****p<0.0001. (C) RNF41 mediates Clec9A ubiquitination in vitro. mClec9A-FLAG was immunoprecipitated from transfected 293 F cells, then incubated with GST-RNF41 or GST-RNF41-C in the presence of E1, E2 (UbcH5a) and biotinylated ubiquitin. Ubiquitination of Clec9A complexes was analyzed by WB for ubiquitin (Streptavidin-HRP), total Clec9A (anti-FLAG-M2) and total RNF41. Representative of two independent experiments. (D, E) Proteomic analysis of RNF41 mediated ubiquitination of Clec9A complexes. 293 F cells were co-transfected with mClec9A-FLAG, Ub-Myc and RNF41 or RNF41-ΔRING. At 22 hr, IP Clec9A complexes (pH 6.0) were analyzed by LC-MS/MS. (D) Nine major Clec9A ubiquitination sites were identified, from at least four independent experiments, and the position of these sites is indicated. (E) Heat map showing quantification results of 6 and 3 ubiquitinated peptides derived from Clec9A and Ubiquitin, respectively, across three independent experiments as indicated by 1,2,3 above the heat map. The intensities of the ubiquitinated peptides are shown as log2. An X indicates that the ubiquitinated peptide was not confidently detected or quantified.
-
Figure 4—source data 1
RNF41 mediates ubiquitination of mouse and human Clec9A.
- https://cdn.elifesciences.org/articles/63452/elife-63452-fig4-data1-v1.xlsx
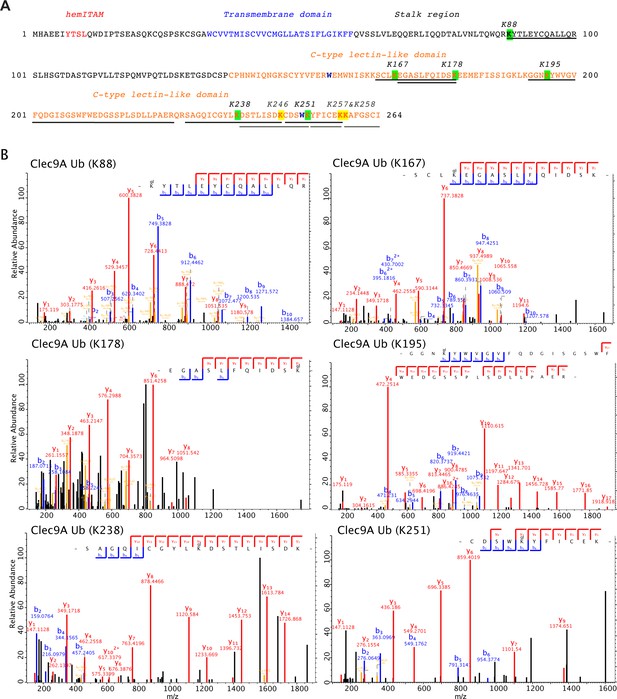
Identification of ubiquitination sites for Clec9A.
(A) Domain annotated amino acid sequence of mouse Clec9A showing the positions of the identified ubiquitination sites in the Clec9A extracellular domains: the stalk region and the C-type lectin-like domain. Quantified ubiquitinated lysine residues are highlighted in green and the corresponding peptide sequences are underlined in black. Three additional ubiquitinated lysines, which have only been identified when using Byonic as a search engine, are highlighted in yellow and the corresponding peptide sequences are underlined in gray. (B) Mass spectrometric analysis of the ubiquitination sites in Clec9A. Fully annotated ms2 spectra are shown for the six ubiquitinated peptide sequences that were identified and quantified by MaxQuant (Supplementary file 1).
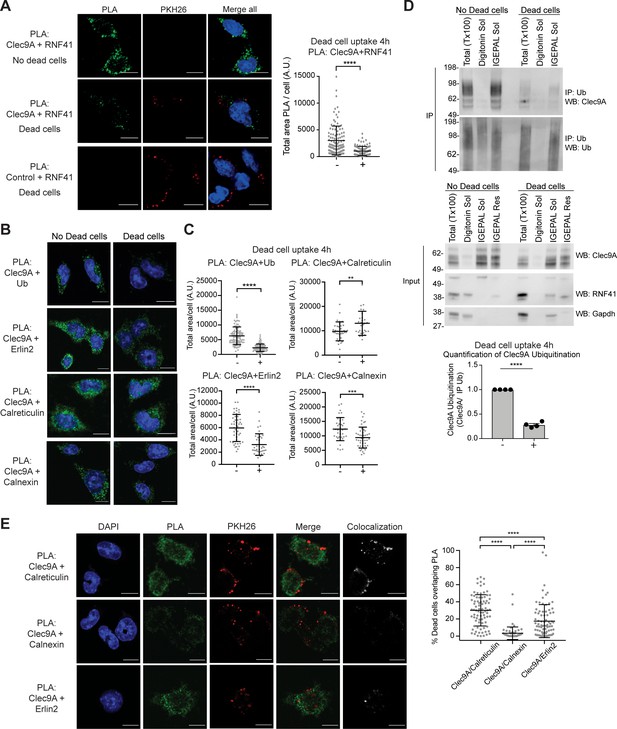
Dead cell uptake diverts Clec9A away from a degradative pathway.
(A) Clec9A and RNF41 interaction in DC is reduced at early stages following dead cell uptake. MutuDC 1940 were cultured in the absence (-) and presence (+) of PKH26-labeled CHO-K1 dead cells for 4 hr, and a PLA performed for Clec9A and RNF41. Representative images from three independent experiments. PLA signal is in green, dead cells (PKH26) in red, DAPI in blue. Scale bars represent 10 μm. The total area of Clec9A+RNF41 PLA per cell (Arbitrary Units, A.U.) was analyzed by Mann–Whitney test. Cumulative data from three independent experiments (4 h-dead cells: 112 cells; 4 h+dead cells: 96 cells). Bars represent mean ± SD. ****p<0.0001. (B–C) Clec9A-Ubiquitination and Clec9A interactions in DC are differentially regulated in the presence of dead cells. MutuDC 1940 were cultured +/− CHO-K1 dead cells for 4 hr, and a PLA performed for Clec9A with Ubiquitin, Erlin2, Calreticulin or Calnexin. (B) Representative images from two independent experiments (Clec9A+Erlin2 PLA: no dead cells; 85, +dead cells: 83 cells. Clec9A+Calreticulin PLA; no dead cells: 83 cells, +dead cells: 81 cells. Clec9A+Calnexin PLA; no dead cells: 84 cells, +dead cells: 73 cells; Clec9A+Ubiquitin PLA: no dead cells;101, + dead cells; 106 cells). PLA is in green, DAPI in blue. Scale bars represent 10 μm. (C) The total area of PLA per cell (A.U.) was analyzed by Mann–Whitney test (Clec9A+Ub PLA; no dead cells: 101 cells; +dead cells: 106 cells. Clec9A+Erlin2 PLA; no dead cells: 46 cells, +dead cells: 36 cells. Clec9A+Calreticulin PLA; no dead cells: 36 cells, +dead cells: 29 cells.) and unpaired t-test (Clec9A+Calnexin; no dead cells: 37 cells, +dead cells: 46 cells). Data is cumulative from two experiments for Clec9A+Ub; representative of three experiments for Clec9A+Erlin2; representative of two experiments for Clec9A+Calreticulin and Clec9A+Calnexin, Bars represent mean ± SD. **p<0.01, ***p<0.001, ****p<0.0001. (D) Clec9A ubiquitination in DC is reduced following dead cell uptake. MutuDC 1940 were cultured +/− CHO-K1 dead cells for 4 hr and cellular fractionation performed. Ubiquitinated proteins were IP using TUBE beads and ubiquitinated Clec9A detected by WB using anti-Clec9A. Representative of four independent experiments. Densitometric analysis of ubiquitinated Clec9A relative to ubiquitinated proteins from IP of ubiquitinated proteins from four experiments is shown, as mean ± SEM (unpaired t-test) ****p<0.0001. (E) Differential association of Clec9A-Calreticulin, Clec9A-Calnexin and Clec9A-Erlin2 interactions with dead cells. MutuDC 1940 were cultured for 4 hr with PKH26-labeled CHO-K1 dead cells, and a PLA performed for Clec9A with Calreticulin, Calnexin and Erlin2. PLA is in green, PKH26-labeled dead cells in red, DAPI in blue. Colocalization images were generated using Image J; pixels that have a positive signal in both PLA and PKH26 are shown in white. Scale bars represent 10 μm. Images are representative of two experiments (PLA Clec9A+Calreticulin: 81 cells; PLA Clec9A+Calnexin: 73 cells; PLA Clec9A+Erlin2: 83 cells). Colocalization of dead cells with PLA was assessed by Manders’ coefficient, bars represent mean ± SD (Kruskal-Wallis test with Dunn’s multiple comparisons). Cumulative data from two independent experiments ****p<0.0001.
-
Figure 5—source data 1
Clec9A interactions are regulated in the presence of dead cells.
- https://cdn.elifesciences.org/articles/63452/elife-63452-fig5-data1-v1.xlsx
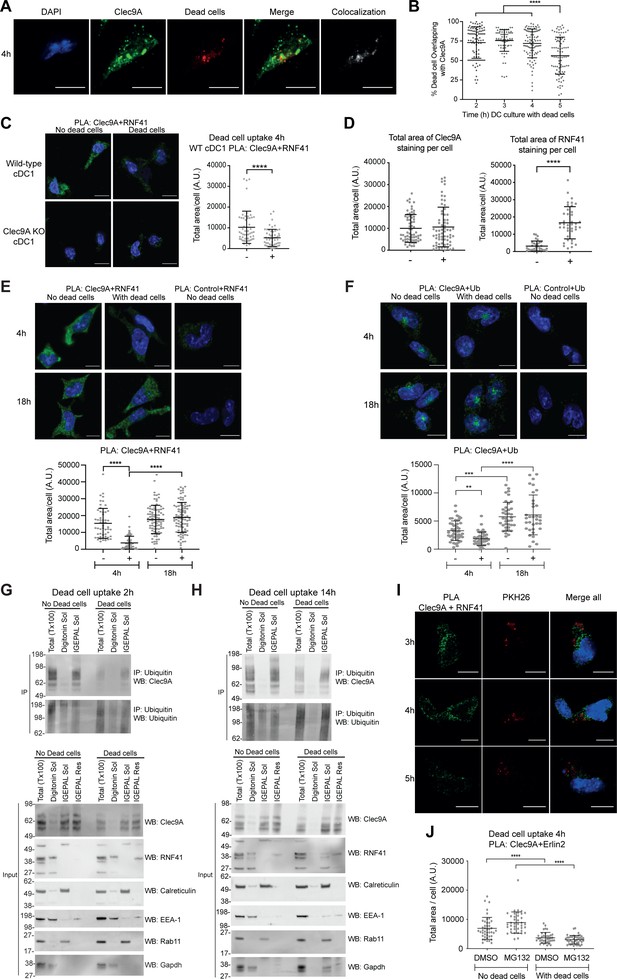
Dead cell processing alters the Clec9A regulatory pathway.
(A, B) Association of Clec9A with dead cells taken up by DC. MutuDC 1940 were cultured with PKH26 labeled CHO-K1 dead cells for 2–5 hr, then fixed and stained for Clec9A. (A) Images show Clec9A expression (green), dead cells (red), DAPI (blue) at 4 hr. Colocalization images, generated using Image J, show pixels that have a positive signal in both Clec9A and dead cell channels in white, whereas single positive and negative pixels are shown in black. Images are representative of four independent experiments (101 cells). Scale bars represent 10 μm. (B) Colocalization of dead cells with Clec9A was assessed by Manders’ coefficient to determine the percentage of internalized dead cell material that overlapped with Clec9A. Analysis was performed using one-way ANOVA with Tukey’s multiple comparisons. Cumulative data from four independent experiments (2 hr: 91 cells, 3 hr: 74 cells, 4 hr: 101 cells, 5 hr: 85 cells). Bars represent mean ± SD. ****p<0.0001. (C) Clec9A-RNF41 interaction is reduced at early stages following dead cell uptake in primary cDC1. Spleen cDC1 were enriched from wild-type (WT) or Clec9a-/− mice (Clec9A KO), then cultured in the presence (+) or absence (-) of dead CHO-K1 cells for 4 hr. A PLA was performed for Clec9A and RNF41. Representative images from two independent experiments. PLA signal is in green, DAPI (nucleus) in blue. Scale bars represent 10 μm. (WT DC no dead cells: 53 cells; WT DC + dead cells: 56 cells; Clec9A KO DC no dead cells: 44 cells; Clec9A KO DC + dead cells: 23 cells). The total area of Clec9A + RNF41 PLA per cell (Arbitrary Units, A.U.) was analyzed by Mann–Whitney test. Cumulative data from two independent experiments. Bars represent mean ± SD. ****p<0.0001. (D) Clec9A levels and RNF41 levels are not reduced in the presence of dead cells. MutuDC 1940 were cultured with PKH26-labeled CHO-K1 dead cells for 4 hr, fixed and stained for either Clec9A (Top panel; No dead cells: 67 cells; Dead cells: 68 cells) or RNF41 (Bottom panel; No dead cells: 32 cells; Dead cells: 39 cells). The total area of Clec9A or RNF41 staining per cell (A.U.), was analyzed by Mann–Whitney test. Cumulative data from two independent experiments for Clec9A expression, one experiment for RNF41 expression. Bars represent mean ± SD. ****p<0.0001. (E) Clec9A and RNF41 interaction in DC is reduced at early stages following dead cell uptake. MutuDC 1940 were cultured with unlabeled dead CHO-K1 cells for 4 or 18 hr and a PLA performed for Clec9A and RNF41, with PLA signal indicated in green. Representative images from two independent experiments are shown for 4 hr and 18 hr. Scale bars represent 10 μm. The total area of PLA per cell (A.U.) was analyzed by Kruskal-Wallis test with Dunn’s multiple comparisons. Cumulative data from two independent experiments. (4 h-dead cells: 54 cells; 4 h+dead cells: 57 cells; 18 h-dead cells: 89 cells; 18 h+dead cells: 90 cells). Bars represent mean ± SD. ****p<0.0001. (F) Clec9A-Ubiquitination in DC, visualized by PLA, following dead cell uptake. MutuDC 1940 were cultured +/− CHO-K1 dead cells for 4 or 18 hr, and a PLA performed for Clec9A and Ubiquitin, with Clec9A-Ub PLA signal indicated in green, DAPI in blue. 4 hr images are from one experiment (4 h-dead cells: 47 cells; 4 h+dead cells: 39 cells), 18 hr images are representative of two independent experiments (18 h-dead cells: 82 cells; 18 h+dead cells: 73 cells). Scale bars represent 10 μm. The total area of PLA per cell (A.U.) was analyzed by Kruskal-Wallis test with Dunn’s multiple comparisons (4 h-dead cells: 47 cells; 4 h+dead cells: 39 cells; 18h-dead cells: 39 cells; 18h+dead cells: 34 cells). Bars represent mean ± SD. **p<0.01, ***p<0.001, ****p<0.0001. (G, H) Clec9A ubiquitination Clec9A in DC is reduced at early stages following dead cell uptake. MutuDC 1940 were cultured +/− CHO-K1 dead cells for (G) 2 hr or (H) 14 hr, and cellular fractionation performed. Ubiquitinated proteins were IP using TUBE beads and ubiquitinated Clec9A detected by WB using anti-Clec9A Ab. Cellular fractions (Input) were analyzed by WB. Representative of three independent experiments for 2 hr, and two experiments for14h culture. (I) Clec9A and RNF41 interaction does not appear to colocalize with dead cell uptake in DC. MutuDC 1940 were cultured with PKH26 labeled dead CHO-K1 cells (red) for 3, 4 and 5 hr, and a PLA performed for Clec9A and RNF41, with PLA signal indicated in green. Representative images from two independent experiments for 4 hr, and 1 experiment at 3 and 5 hr. (I: 3 hr: 32 cells, 4 hr: 84 cells, 5 hr: 42 cells). Scale bars represent 10 μm. (J) Clec9A and Erlin2 interaction in DC in the absence and presence of dead cells +/− MG132. MutuDC 1940 cells were cultured for 4 hr +/− PKH26 labeled CHO-K1 dead cells, and the proteasomal inhibitor MG132 (5 μM) or the control DMSO. A PLA was performed for Clec9A and Erlin2 (No dead cells; DMSO: 44 cells, MG132: 39 cells. Dead cells; DMSO: 38 cells, MG132: 42 cells). The total area of PLA per cell (A.U.) was analyzed by Kruskal-Wallis test with Dunn’s multiple comparisons. Bars represent mean ± SD. ****p<0.0001.
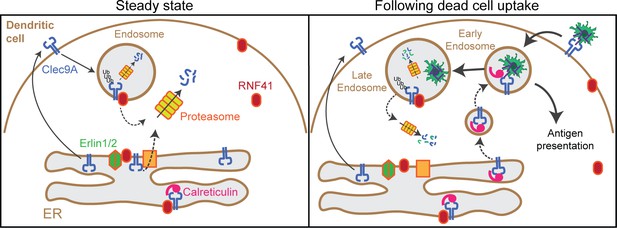
Model of RNF41 as a negative regulator of Clec9A and cross-presentation.
We propose that at steady state, Clec9A interacts with RNF41, Erlin1/2 and Calreticulin. RNF41 mediates Clec9A ubiquitination at the ER and endosomes, targeting it for degradation to control Clec9A receptor levels. At early stages following dead cell uptake by DC, Clec9A is diverted away from Erlin1/2 and RNF41-mediated degradation, and is recruited with Calreticulin to dead cell-containing endosomes, to facilitate cross-presentation of dead cell Ag. At later stages following dead cell uptake, RNF41 interacts with Clec9A at endosomes to regulate Clec9A following presentation of dead cell Ag.
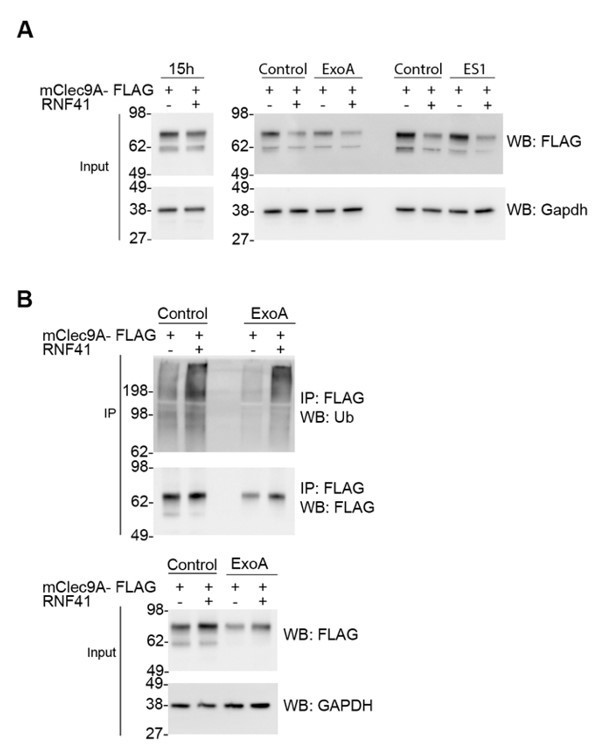
RNF41-mediated regulation is not dependent on Sec61 or VCP/p97.
(A) RNF41 mediated regulation of Clec9A levels is not dependent on Sec61 or VCP/p97. 293F cells were co-transfected with constructs encoding FLAG-tagged full-length mClec9A in the absence and presence of RNF41. At 15 hours, an aliquot of cells was harvested (left panel; prior to RNF41-mediated effects on Clec9A levels), and remaining cells were cultured for a further 6 h in the presence or absence of 20 μg/ml of ExoA or 10 μM ES1. Cellular lysates (input) were analyzed by WB. (B) RNF41 mediated ubiquitination of mClec9A is not dependent on Sec61. 293F cells were co-transfected with mClec9A-FLAG, Ub-Myc, and RNF41. At 15h, cells were cultured for 6 hours in the presence or absence of 12.5 μg/ml of ExoA. IP Clec9A complexes were analyzed for ubiquitination by WB using anti-Ubiquitin Ab. Cellular lysates (input) were analyzed by WB (representative of two experiments).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | JM109 | Promega | Cat# L2001 | Competent cells |
| Strain, strain background (Escherichia coli) | BL21(DE3)pLysS | Promega | Cat# L1195 | Competent cells for protein expression |
| Strain, strain background (Mus musculus) | OT-I/Ly5.1 transgenic mice | Own colony (Lahoud et al., 2011 doi: 10.4049/jimmunol.1101176) | ||
| Strain, strain background (Mus musculus) | OT-II/Ly5.1 transgenic mice | Own colony (Lahoud et al., 2011 doi: 10.4049/jimmunol.1101176) | ||
| Strain, strain background (Mus musculus) | Clec9A-/− mice | Own colony (Caminschi et al., 2012 doi: 10.1016/j.molimm.2011.11.008) | ||
| Cell line (Homo sapiens) | Freestyle 293F cells | Thermo Fisher | Cat# R79007 | |
| Cell line (Mus musculus) | MutuDC1940 | Fuertes Marraco et al., 2012 doi: 10.3389/fimmu.2012.00331 | Gift from Prof. Acha-Orbea | |
| Cell line (Cricetulus griseus) | CHO-K1/ CHO-K1 cells expressing OVA-FLAG | Caminschi et al., 2007 doi: 10.1084/jem.20071351 | ||
| Transfected construct (Homo sapiens) | FLAG-tagged full length human CLEC9A (hCLEC9A-FLAG) | Caminschi et al., 2008 doi: 10.1182/blood-2008-05-155176 | Construct to transiently transfect Freestyle 293F | |
| Transfected construct (Mus musculus) | FLAG-tagged full length mouse Clec9A (mClec9A-FLAG) | Caminschi et al., 2008 doi: 10.1182/blood-2008-05-155176 | Construct to transiently transfect Freestyle 293F | |
| Transfected construct (Homo sapiens) | FLAG-tagged recombinant human CLEC9A ecto-domains (hCLEC9A-ecto) | Zhang et al., 2012 doi: 10.1016/j.immuni.2012.03.009 | Construct to transiently transfect Freestyle 293F | |
| Ttransfected construct (Mus musculus) | FLAG-tagged recombinant mouse Clec9A ecto-domains (mClec9A-ecto) | Zhang et al., 2012 doi: 10.1016/j.immuni.2012.03.009 | Construct to transiently transfect Freestyle 293F | |
| Ttransfected construct (Homo sapiens) | RNF41 full-length mammalian expression construct in pcDNA3.1+ RNF41 region: 1-MGYD…VEEI-317 | This paper, synthesized by GeneArt, Life Technologies | Construct to transiently transfect Freestyle 293F. Materials & Methods: Generation of Recombinant Proteins | |
| Transfected construct (Homo sapiens) | RNF41- ΔRING mammalian expression construct in pcDNA3.1+ RNF41 region: 72-MRNM…VEEI-317 | This paper, synthesized by GeneArt, Life Technologies | Construct to transiently transfect Freestyle 293F. Materials & methods: Generation of Recombinant Proteins | |
| Antibody | anti-FLAG (clone M2, Mouse monoclonal, HRP-conjugate) | Sigma | Cat# A8592 | WB (1:6000) |
| Antibody | anti-FLAG, FITC (rat monoclonal) | WEHI Antibody Facility | clone 9H1 | FACS (1:200) |
| Antibody | anti-Myc (mouse monoclonal) | WEHI Antibody Facility | clone 9E10 | WB (1:3000) |
| Antibody | anti-RNF41 (rabbit polyclonal) | Sigma | Cat# HPA016812 | WB (1:3000) |
| Antibody | anti-RNF41 (rabbit polyclonal) | Bethyl Laboratories | Cat# A300-048A | WB (1:3000) |
| Antibody | anti-RNF41 (rabbit polyclonal) | Bethyl Laboratories | Cat# A300-049A | IF (1:50) PLA (1:100) |
| Antibody | anti-mouse Clec9A (rat monoclonal) | Caminschi et al., 2008 doi: 10.1182/blood-2008-05-155176 | clone 24/04-10B4 | WB (2μg/ml) |
| Antibody | anti-mouse Clec9A (mouse polyclonal) | This paper | Used for confocal microscopy and PLA | IF (1:50) PLA (1:100) |
| Antibody | anti-Actin, HRP (goat polyclonal) | Santa Cruz | Cat# sc-1616HRP clone I-19 | WB (1:5000) |
| Antibody | anti-GAPDH, HRP (mouse monoclonal) | Thermo Fisher | Cat# MA5-15738-HRP Clone GA1R | WB (1:15000) |
| Antibody | anti-Ubiquitin, HRP (mouse monoclonal) | Santa Cruz | Cat# sc-8017 Clone P4D1 | WB (1:3000) |
| Antibody | anti-Ubiquitin (rabbit polyclonal) | Abcam | Cat#ab7780 | PLA (1:250) |
| Antibody | anti-K48 specific Ubiquitin (rabbit monoclonal) | Merck Millipore | Cat# 05-1307 Clone Apu2 | WB (1:3000) |
| Antibody | anti-Erlin1 (rabbit polyclonal) | Proteintech | Cat# 17311-1-AP | WB (1:3000) |
| Antibody | anti-Erlin2 (rabbit polyclonal) | Sigma | Cat # HPA002025 | WB (1:3000) IF (1:100) PLA (1:100) |
| Antibody | anti-Calreticulin (rabbit polyclonal) | Abcam | Cat#Ab2907 | WB (1:4000) IF (1:100) PLA (1:100) |
| Antibody | Anti-calnexin (rabbit polyclonal) | Abcam | Cat#ab22595 | PLA (1:250) |
| Antibody | anti-EEA-1 (mouse monoclonal) | Santa Cruz | Cat#sc-365652 Clone E-8 | WB (1:1500) IF (1:100) |
| Antibody | anti-Rab11 (mouse monoclonal) | BD Biosciences | Cat#610656 Clone 47 | WB (1:1000) IF (1:100) |
| Antibody | anti-mouse CD45.1, APC (mouse monoclonal) | BD Pharmingen | Cat# 558701 Clone A20 | FACS (1:200) |
| Antibody | anti-mouse CD45.2, Pacific Blue (mouse monoclonal) | BioLegend | Cat# 109820 Clone 104 | FACS (1:200) |
| Antibody | anti-mouse MHC II (hamster monoclonal) | WEHI | Clone N22 | IF (10μg/ml) |
| Antibody | anti-mouse CD8, PE (rat monoclonal) | WEHI | Clone YTS169.4 | FACS (1:200) |
| Antibody | Anti-mouse CD4 (rat monoclonal) | WEHI | Clone GK1.5 | FACS (1:400) |
| Antibody | anti-Rat Ig, HRP (goat polyclonal) | Southern Biotech | Cat# 3010-05 | WB (1:5000) |
| Antibody | anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (donkey polyclonal) | Life Technologies | Cat#A-21206 | IF (1:200) |
| Antibody | anti-Rabbit IgG, HRP (goat polyclonal) | Bio-rad | Cat# 1706515 | WB (1:5000 - 1:10000) |
| Antibody | anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 (goat polyclonal) | Life Technologies | Cat#A-11030 | IF (1:200) |
| Antibody | anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (goat polyclonal) | Life Technologies | Cat#A-21235 | IF (1:200) |
| Antibody | anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, (Goat polyclonal, Alexa Fluor 488- conjugate) | Life Technologies | Cat#A-11029 | IF (1:200) |
| Recombinant DNA reagent | pLMP-Cherry (plasmid) | Majewski et al., 2008 doi: 10.1371/journal.pbio.0060093 | Knockdown plasmid for shRNA | |
| Recombinant DNA reagent | Vsv-g (plasmid) | Majewski et al., 2008 doi: 10.1371/journal.pbio.0060093 | Envelope plasmid for shRNA knockdown | |
| Recombinant DNA reagent | pMD1-gag-pol (plasmid) | Majewski et al., 2008 doi: 10.1371/journal.pbio.0060093 | Packaging plasmid for shRNA knockdown | |
| Recombinant DNA reagent | RNF41-RBCC bacterial expression construct in pGex2T RNF41 region: 1-MGYD…VEEI-317 | This paper | For recombinant protein production in bacteria. Materials & methods: Generation of Recombinant Proteins | |
| Recombinant DNA reagent | RNF41-BCC bacterial expression construct in pGex2T RNF41 region: 72-MRNM…VEEI-317 | This paper | For recombinant protein production in bacteria. Materials & methods: Generation of Recombinant Proteins | |
| Recombinant DNA reagent | RNF41-CC bacterial expression construct in pGex2T RNF41 region: 135-IKHL…VEEI-317 | This paper | For recombinant protein production in bacteria. Materials & methods: Generation of Recombinant Proteins | |
| Recombinant DNA reagent | RNF41-C bacterial expression construct in pGex2T RNF41 region: 169-DIQL…VEEI-317 | This paper | For recombinant protein production in bacteria. Materials & methods: Generation of Recombinant Proteins | |
| Recombinant DNA reagent | RNF41-RBC bacterial expression construct in pGex2T RNF41 region: 1-MGYD…RAIR-181 | This paper | For recombinant protein production in bacteria. Materials & methods: Generation of Recombinant Proteins | |
| Sequence-based reagent | RNF41 shRNA#5_F | Integrated DNA Technologies | Oligonucleotides for generation for shRNA | TCGAGAAGGTATATTGCTGTTGACAGTGAGCGCTCCTTTGGTGTTGTTTGTTTATAGTGAAGCCACAGATGTATAAACAAACAACACCAAAGGAATGCCTACTGCCTCGG |
| Sequence-based reagent | RNF41 shRNA#5_R | Integrated DNA Technologies | Oligonucleotides for generation for shRNA | AATTCCGAGGCAGTAGGCATTCCTTTGGTGTTGTTTGTTTATACATCTGTGGCTTCACTATAAACAAACAACACCAAAGGAGCGCTCACTGTCAACAGCAATATACCTTC |
| Sequence-based reagent | RNF41 shRNA#10_F | Integrated DNA Technologies | Oligonucleotides for generation for shRNA | TCGAGAAGGTATATTGCTGTTGACAGTGAGCGACTGGAGATGCCCAAAGATGAATAGTGAAGCCACAGATGTATTCATCTTTGGGCATCTCCAGGTGCCTACTGCCTCGG |
| Sequence-based reagent | RNF41 shRNA#10_R | Integrated DNA Technologies | Oligonucleotides for generation for shRNA | AATTCCGAGGCAGTAGGCACCTGGAGATGCCCAAAGATGAATACATCTGTGGCTTCACTATTCATCTTTGGGCATCTCCAGTCGCTCACTGTCAACAGCAATATACCTTC |
| Sequence-based reagent | RNF41-q14_F | Geneworks | qPCR primers | GTGAGCACAACCCGAAGC |
| Sequence-based reagent | RNF41-q15_R | Geneworks | qPCR primers | GTTCATCTTTGGGCATCTCC |
| Peptide, recombinant protein | MHC class I (257-264) restricted OVA peptide | Mimotopes | Positive control for Ag presentation to CD8+ T cells | |
| Peptide, recombinant protein | MHC class II (323-339) restricted OVA peptide | Mimotopes | Positive control for Ag presentation to CD4+ T cells | |
| Peptide, recombinant protein | Mouse IgGκ binding protein, HRP | Santa Cruz | Cat#sc-516102 | WB (1:5000) |
| Peptide, recombinant protein | Streptavidin-HRP | GE Healthcare | Cat# RPN1231 | ELISA (1:15 000) |
| Peptide, recombinant protein | Streptavidin-PE | BD Biosciences | Cat# 554061 | FACS (1:200) |
| Commercial assay or kit | West Pico Super Signal ECL substrate | Thermo Fisher | Cat# 34080 | |
| Commercial assay or kit | Luminata Forte Western HRP substrate | Merck Millipore | Cat# WBLUF0500 | |
| Commercial assay or kit | Pierce BCA Protein Assay kit | Thermo Fisher | Cat# 23225 | |
| Commercial assay or kit | Ubiquitinylation kit | Enzo Life | Cat# BML-UW9920 | |
| Commercial assay or kit | BirA | Avidity | Cat# bulk BirA | |
| Commercial assay or kit | ProtoArray Human Protein MicroArray Version 4.1 PPI Kit for biotinylated proteins | Thermo Fisher | Cat# PAH05241011 | |
| Commercial assay or kit | PKH26 labeling | Sigma-Aldrich | Cat#MINI26-1KT | |
| Commercial assay or kit | Duolink In Situ PLA Probe Anti-Rabbit PLUS | Sigma-Aldrich | Cat#DUO92002 | |
| Commercial assay or kit | Duolink In Situ PLA Probe Anti-Mouse MINUS | Sigma-Aldrich | Cat#DUO92004 | |
| Commercial assay or kit | Duolink In Situ Detection Reagents Orange | Sigma-Aldrich | Cat#DUO92007 | |
| Commercial assay or kit | Duolink In Situ Detection Reagents FarRed | Sigma-Aldrich | Cat#DUO92013 | |
| Commercial assay or kit | Duolink In Situ Detection Reagents Green | Sigma-Aldrich | Cat#DUO92014 | |
| Chemical compound, drug | MG132 in Solution | Calbiochem | Cat# 474791 | |
| Chemical compound, drug | PR619 | Life Sensors | Cat# SI9619 | |
| Chemical compound, drug | Brefeldin A | Sigma-Aldrich | Cat#B5936-200UL | |
| Software, algorithm | Prospector v5 (Thermo Fisher) | https://www.thermofisher.com/au/en/home/life-science/protein-biology/protein-assays-analysis/protein-microarrays/technical-resources/data-analysis.html | ||
| Software, algorithm | Image Lab v5.2.1 (Bio-rad) | http://www.bio-rad.com/en-au/product/image-lab-software?ID=KRE6P5E8Z#fragment-6 | Image Lab Software, RRID:SCR_014210 | |
| Software, algorithm | FIJI/ImageJ version 2.0.0 | https://fiji.sc | ImageJ, RRID:SCR_003070 | |
| Software, algorithm | FlowJo 10.5.0 (Treestar Inc) | https://www.flowjo.com/solutions/flowjo/downloads | FlowJo, RRID:SCR_008520 | |
| Software, algorithm | Rotor-gene Q series software (Qiagen) | https://www.qiagen.com/au/resources/resourcedetail?id=9d8bda8e-1fd7-4519-a1ff-b60bba526b57&lang=en | Rotor-Gene Q Series Software, RRID:SCR_015740 | |
| Software, algorithm | Weasel v3.0.2 | http://www.frankbattye.com.au/Weasel/ | ||
| Software, algorithm | GraphPad Prism 7.0b | https://www.graphpad.com/scientific-software/prism/ | GraphPad Prism, RRID:SCR_002798 | |
| Software, algorithm | Byonic (Protein Metrics) | https://www.proteinmetrics.com/products/byonic/ | PMI-Byonic, RRID:SCR_016735 | |
| Software, algorithm | MaxQuant v1.6.0.16 | https://www.biochem.mpg.de/5111795/maxquant | MaxQuant, RRID:SCR_014485 | |
| Software, algorithm | Perseus v1.6.1.3 | https://www.biochem.mpg.de/5111810/perseus | Perseus, RRID:SCR_015753 | |
| Other | Bond-Elut Omix-mini Bed 96 C18 | Agilent Technologies | Cat# A57003MBK | |
| Other | Anti-FLAG M2 affinity gel | Sigma-Aldrich | Cat# A2220 | |
| Other | TUBE-agarose | Life Sensors | Cat# UM401 | |
| Other | Glutathione Sepharose 4B | GE Healthcare | Cat# 17-0756-05 | |
| Other | Protein G Sepharose | GE Healthcare | Cat# 17061801 | |
| Other | PNGase F | New England Biolabs | Cat# P0704S | |
| Other | Endo H | New England Biolabs | Cat# P0702S | |
| Other | 293fectin transfection reagent | Thermo Fisher | Cat# 12347500 | |
| Other | Freestyle MAX transfection reagent | Thermo Fisher | Cat# 16447100 |
Additional files
-
Supplementary file 1
Identification of human proteins that bind specifically to mouse Clec9A.
Table 2: Glycan modification of Clec9A dimers. Table 3: Proteins that showed an enhanced association with Clec9A in the presence of RNF41. Table 4: Quantification of Clec9A and Ubiquitin ubiquitinated peptides.
- https://cdn.elifesciences.org/articles/63452/elife-63452-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63452/elife-63452-transrepform-v1.pdf





