Cross-compartment signal propagation in the mitotic exit network
Figures
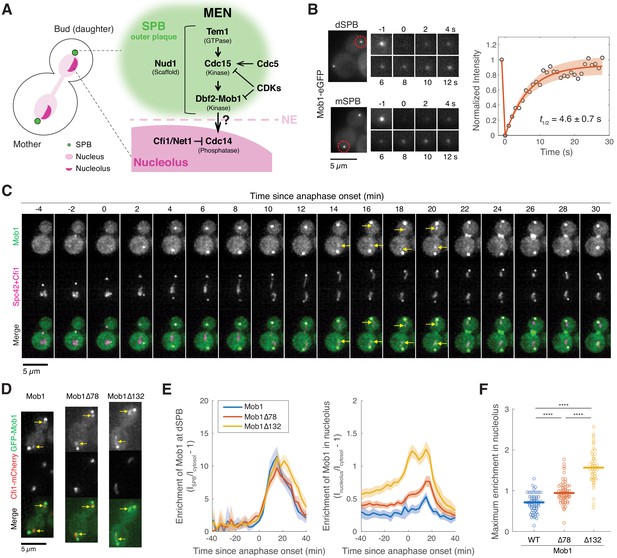
Dbf2-Mob1 transiently localizes to the nucleolus in late anaphase.
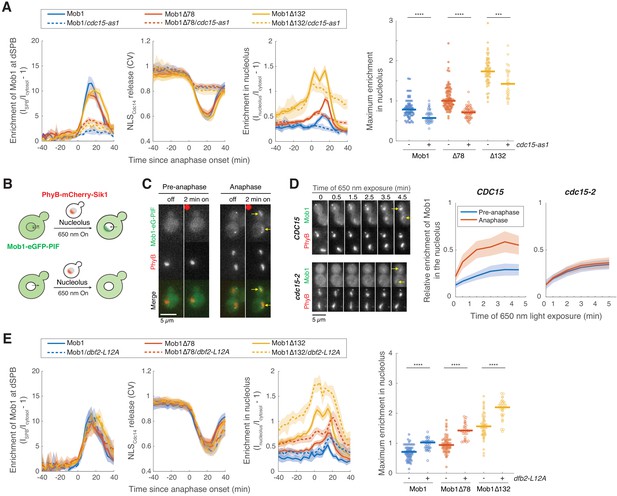
(A) Major components of the mitotic exit network (MEN) and their subcellular localization. (B) Fluorescence recovery after photobleaching (FRAP) analysis of Mob1-eGFP (A39695). Red circles indicate the area of photo-bleaching. Cells were grown and imaged at room temperature in SC medium + 2% glucose. Graph to the right represents average measurements of double normalized fluorescence intensities (n = 6 cells) after correcting for photo-bleaching during acquisition. Red curve is the average fit and shaded area represents standard deviation (SD) of the fits. Half recovery time ± SD is indicated. (C) Localization of Mob1 during the cell cycle. A40257 (with Mob1-eGFP, Cfi1-mCherry and Spc42-mCherry) cells were grown at room temperature in SC medium + 2% glucose and imaged every minute for 2 hr. Arrows highlight the nucleolar localization. (D) Nucleolar localization of full-length (A39931) and N-terminally truncated (A39933 and A39935) Mob1. Cells were grown at room temperature in SC medium + 2% glucose and imaged every 3 min for 4 hr. Arrows highlight the nucleolar localization. (E) Enrichment of Mob1 (A41211, n = 62 cells), Mob1Δ78 (A41212, n = 60 cells), and Mob1Δ132 (A41213, n = 48 cells) at the daughter spindle pole body (dSPB; left) and in the nucleolus (right) as a function of cell cycle progression. Cells were grown at 25°C in SC medium + 2% glucose and imaged every 3 min for 4 hr. Single cell traces were aligned based on anaphase onset, as defined as spindle length >3 μm (Figure 1—figure supplement 1F, measured based on SPB marker Spc42-mCherry), and averaged. Solid lines represent the average, and shaded areas represent 95% confidence intervals. (F) Maximum enrichment of full-length Mob1 (WT) and truncated Mob1 (Mob1Δ78 and Mob1Δ132) in the nucleolus in anaphase of cells in (E). Solid lines represent the median. ****p<0.0001 by two-sided Wilcoxon rank sum test.
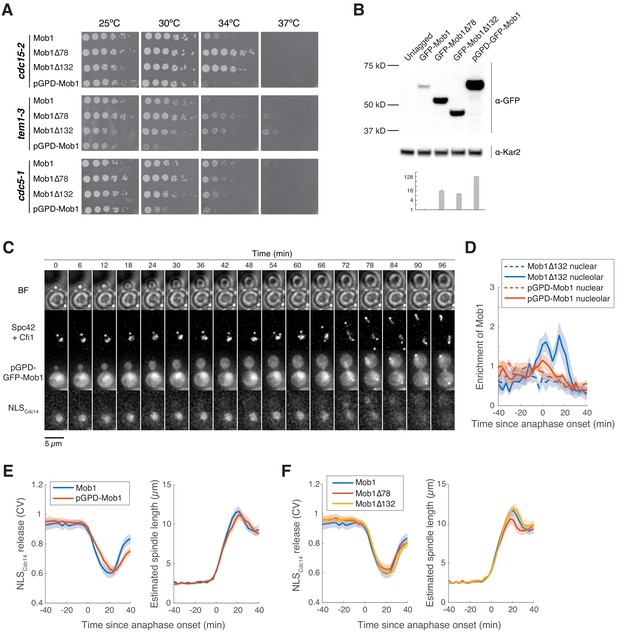
N-terminally truncated Mob1 is hyperactive.
(A) Fivefold serial dilutions of cdc15-2 (A41424, A41425, A41426, A41427), tem1-3 (A41429, A41430, A41431, A41428), and cdc5-1 (A41432, A41433, A41434, A41435) harboring the indicated MOB1 constructs in YEP + 2% glucose at the indicated temperatures. (B) Immunoblot (top) and quantification (bottom) of untagged (A2587), full-length (A41351), and truncated GFP-Mob1 (A41352, A41353) as well as full-length GFP-Mob1 expressed from the pGPD/TDH3 promoter (A41350). (C) Localization of GFP-Mob1 expressed under the control of pGPD promoter (A41595) during the cell cycle. Cells were grown and imaged as in Figure 1E. Increased nuclear but not nucleolar localization of Mob1 was observed. (D) Enrichment of Mob1Δ132 (A41213, n = 14 cells) and pGPD-Mob1 (A41595, n = 17 cells) in the nucleolus (solid lines) compared to the nucleus (dashed lines) as a function of cell cycle progression. Cells were grown and imaged as in Figure 1E. (E) Kinetics of mitotic exit network (MEN) activation as measured by the release of NLSCdc14 reporter from the nucleus (see Figure 1—figure supplement 2) and anaphase progression as indicated by spindle length for cells harboring Mob1 (A41213, n = 62 cells) or pGPD-Mob1 (A41595, n = 43 cells). pGPD-Mob1 slightly delays MEN activation and exit from mitosis. (F) Kinetics of MEN activation and anaphase progression for experiments shown in Figure 1E. Mob1Δ78 slightly accelerated mitotic exit. For graphs in (D–F), single cell traces were aligned based on anaphase onset and averaged. Solid lines represent the average, and shaded areas represent 95% confidence intervals.
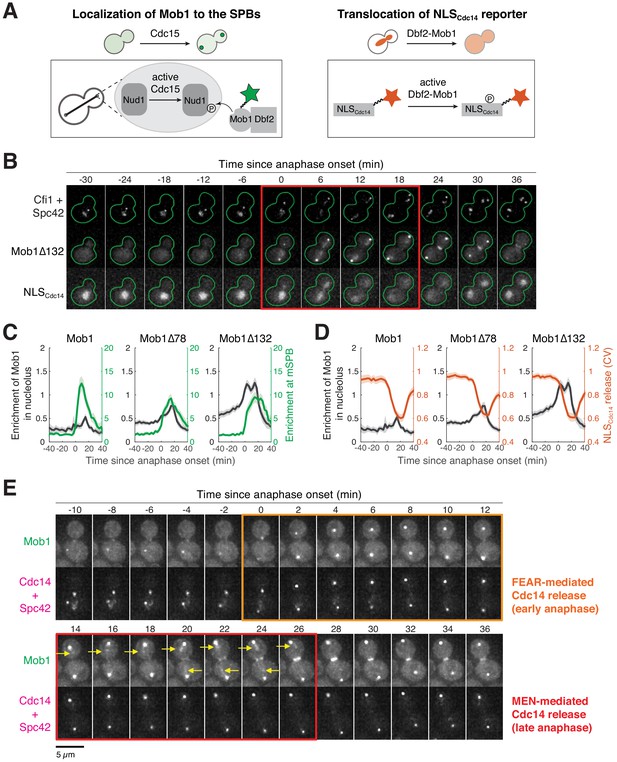
Mob1’s nucleolar localization correlates with mitotic exit network (MEN) activation and Cdc14 release from the nucleolus.
(A) Illustration of Mob1’s spindle pole body (SPB) localization and translocation of NLSCdc14 reporter in response to MEN activation. (B) Localization of Mob1Δ132 and the MEN activity reporter NLSCdc14 during the cell cycle. A41213 (GFP-MOB1Δ132, CFI1/NET1-mCherry, SPC42-mCherry, and NLSCdc14-ymiRFP670) cells were grown as in Figure 1E. Red square indicates anaphase. (C) Relative timing of nucleolar localization of Mob1 (black) and SPB localization of Mob1 (green) in cells harboring the indicated MOB1 alleles. (D) Relative timing of nucleolar localization of Mob1 (black) and release of the NLSCdc14 reporter from the nucleus (red) in cells harboring the indicated MOB1 alleles. (E) Localization of Mob1 and Cdc14 during the cell cycle. A40314 (MOB1-eGFP, SPC42-mCherry, and CDC14-mCherry) cells were grown at room temperature in SC medium + 2% glucose and imaged every 2 min for 4 hr. The colored squares indicate the frames where Cdc14 is released from the nucleolus mediated by either the Cdc fourteen early anaphase release (FEAR) network or the MEN. Yellow arrows highlight localization of Mob1 in the nucleolus.
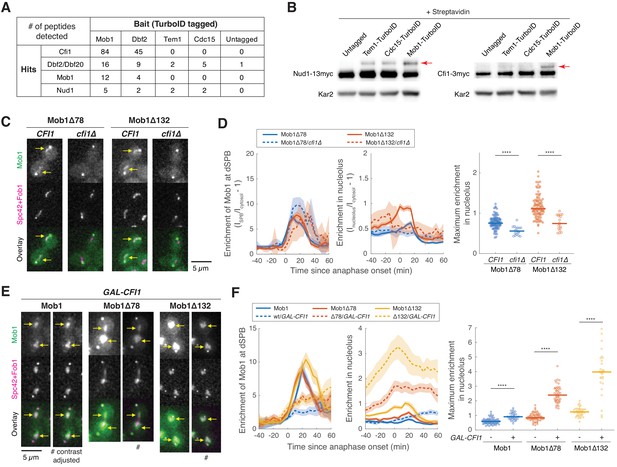
Dbf2-Mob1 localizes to the nucleolus through interacting with Cfi1/Net1.
(A) Results of proximity-based biotinylation with TurboID for mitotic exit network (MEN) proteins (A41367, A41370, A41368, A41369, and A2588). Cells were grown at room temperature in YEP + 2% glucose + 50 μM biotin. (B) Streptavidin gel-shift assays to probe the interactions of TurboID-labeled MEN proteins with the MEN scaffold Nud1 (left, A11869, A41381, A41382 and A41380) or Cfi1/Net1 (right, A1638, A41406, A41407 and A41372). Cells were grown at room temperature in YEP + 2% glucose and lysates were treated with streptavidin and immunoblotted as indicated. Red arrows highlight biotinylated proteins. (C and D) Representative images (C) and quantification (D) of Mob1Δ78 localization in wild-type CFI1/NET1 (A41344, n = 106 cells) or cfi1/net1Δ (A41347, n = 18 cells) cells and Mob1Δ132 localization in CFI1/NET1 (A41345, n = 95 cells) or cfi1/net1Δ (A41348, n = 18 cells) cells. Cells were grown at 25°C in SC medium + 2% glucose and imaged every 5 min for 4 hr. Arrows highlight nucleolar localization. (E and F) Representative images (E) and quantification (F) of Mob1 localization in wild-type (A41343, n = 110 cells) or GAL-CFI1/NET1 expressing cells (A41340, n = 71 cells), Mob1Δ78 localization in wild-type (A41344, n = 103 cells) or GAL-CFI1/NET1 expressing cells (A41341, n = 68 cells), and Mob1Δ132 localization in wild-type (A41345, n = 71 cells) or cells expressing GAL-CFI1/NET1 (A41342, n = 53 cells). # denotes that the image was linearly contrast adjusted to avoid over-saturation for Mob1Δ78 and Mob1Δ132. Cells were first grown at room temperature in SC medium + 2% raffinose. Cells were then mounted onto agarose pads made with SC medium + 1% raffinose + 1% galactose and imaged every 5 min for 5 hr at 25°C. Arrows highlight nucleolar localization. Solid lines represent the average of single cell traces aligned to anaphase onset while shaded areas represent 95% confidence intervals. For maximum enrichment, each dot represents a single cell. The solid lines represent the median. ****p<0.0001 by two-sided Wilcoxon rank sum test.
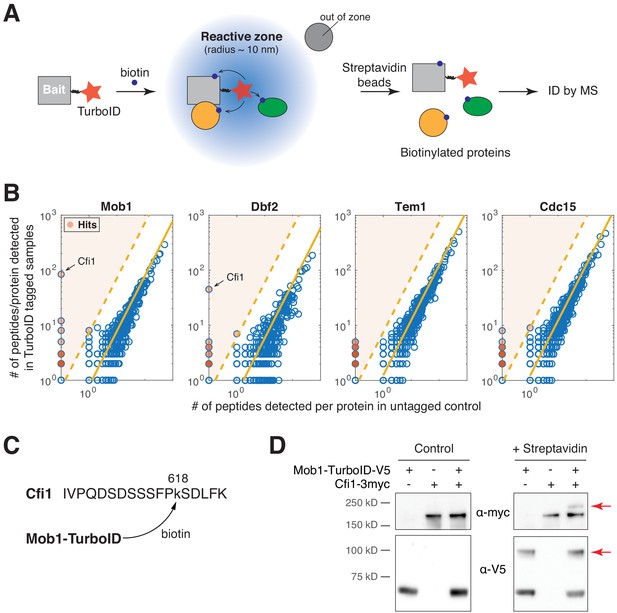
TurboID identified interaction between Dbf2-Mob1 and Cfi1/Net1.
(A) Overview of TurboID proximity-based biotinylation to identify interaction partners of a target protein. (B) Number of total peptides detected for MS-identified proteins in cells with TurboID tagged baits relative to untagged control cells (source data for Figure 2A). Shaded red dots represent proteins (hits) identified as interaction partners of the bait protein. Solid yellow lines denote the mean ratio of total peptide detected for all proteins. Dashed lines denote the threshold used for identifying hits (10 standard derivations above the mean). (C) Biotinylated peptide of Cfi1/Net1 detected in cells harboring Mob1-TurboID. (D) Streptavidin gel-shift assay to probe the interaction between Mob1 and Cfi1/Net1. Lysates of A41379, A1638, and A41372 cells with or without streptavidin treatment were immunoblotted as indicated. Red arrows highlight biotinylated proteins.
CDC15 regulates nuclear access of Dbf2-Mob1.
(A) Enrichment of Mob1 at the daughter spindle pole body (dSPB), in the nucleolus, and Dbf2-Mob1’s kinase activity were determined in cells going through anaphase in CDC15 (A41211, A41212, and A41213; n = 74, 94, and 55 cells respectively) or cdc15-as1 (A41214, A41215, and A41216; n = 37, 63, and 30 cells respectively) cells. Cells were grown at 25°C in SC medium + 2% glucose and 10 μM 1-NA-PP1 and imaged every 3 min for 4 hr. (B) Probing Dbf2-Mob1’s nuclear access by recruiting Mob1 to the nucleolus with the PhyB-PIF optogenetics system. By anchoring PhyB to the nucleolus, diffuse nuclear Dbf2-Mob1, if present, can be visualized by recruiting Mob1-eGFP-PIF to the nucleolus. (C) Recruiting Mob1 to the nucleolus at different cell cycle stages. A40260 cells were grown at 25°C in SC medium + 2% glucose, imaged after a 2 hr incubation with 31.25 μM PCB in the dark. Red dot denotes the frame where 650 nm light was applied to activate PhyB. Yellow arrows highlight the light-induced recruitment. (D) Recruitment of Mob1 to the nucleolus in CDC15 (A41360) or cdc15-2 (A41361) cells. Quantifications of Mob1’s enrichment in the nucleolus as a function of PhyB activation time in CDC15 (A41360, n = 27 and 16 cells for pre-anaphase and anaphase respectively) or cdc15-2 (A41361, n = 14 and 36 cells for pre-anaphase and anaphase respectively) cells. Cells were grown at room temperature in SC medium + 2% glucose, incubated with 12.5 μM PCB for 2 hr in the dark, and shifted to 34°C for 50 min before imaging. (E) Nucleolar enrichment of full-length and truncated Mob1 in wild-type DBF2 or dbf2-L12A cells (A41394, A41395, and A41396; n = 32, 28, and 31 cells respectively). Wild-type traces for comparison were the same as in Figure 1E. Cells were grown similarly as in Figure 1E. For graphs in (A) and (E), solid lines represent the average of single cell traces aligned to anaphase onset. Shaded areas represent 95% confidence intervals. For maximum enrichment, each dot represents a single cell. Solid lines represent the median. ****p<0.0001; ***p<0.001 by two-sided Wilcoxon rank sum test.
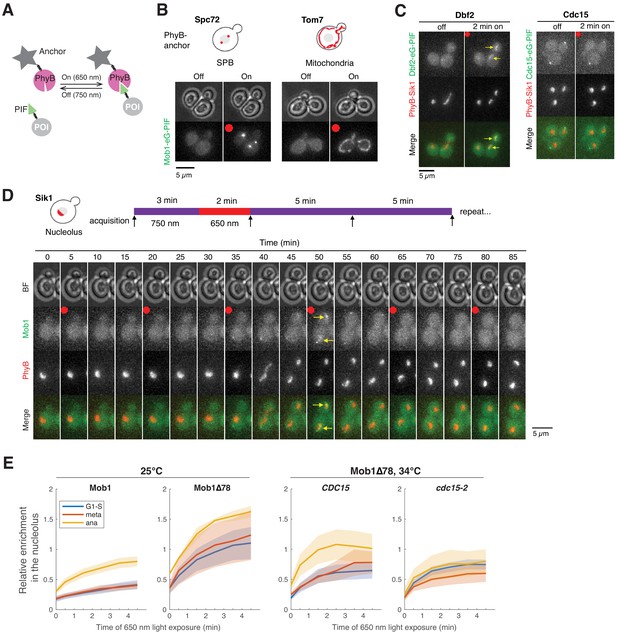
Examining nuclear access of Dbf2-Mob1 by optogenetics.
(A) The PhyB-PIF based light-inducible organelle targeting system. When exposed to 650 nm light, anchored PhyB interacts with PIF and thus targets protein of interest (POI) to designated subcellular regions. This interaction is reversed with exposure to 750 nm light. (B) Recruiting Mob1 to spindle pole bodies (SPBs; A40346) or the mitochondrial surface (A40354) with the PhyB-PIF optogenetics system. Cells were grown at room temperature in SC medium + 2% glucose, incubated with 31.25 μM PCB for 2 hr in the dark prior to imaging. Red dots indicate application of red light (650 nm) for 5 min to activate PhyB. (C) Recruiting Dbf2 (A40262) or Cdc15 (A40258) to the nucleolus with PhyB-Sik1. Cells were grown similar to (B). Yellow arrows highlight the light-induced recruitment. (D) Probing Mob1’s nuclear access during the cell cycle with PhyB-Sik1. A40260 cells were grown similar to (B) and imaged every 5 min while the red light (650 nm) was applied for 2 min every 15 min. Red dots denote frames where 650 nm light was applied to activate PhyB prior to imaging as indicated in the cartoon above. Yellow arrows highlight the light-induced recruitment. (E) Quantifications of enrichment of Mob1 in the nucleolus as a function of PhyB activation time for full-length Mob1 (A41360; n = 22, 14, and 21 cells for G1-S, metaphase and anaphase respectively), Mob1Δ78 (A41366; n = 11, 6, and 2 cells for G1-S, metaphase and anaphase respectively) at 25°C and for Mob1Δ78 in CDC15 (A41366; n = 25, 4, and 9 cells for G1-S, metaphase and anaphase respectively) or cdc15-2 cells (A41365; n = 39, 9, and 10 cells for G1-S, metaphase and anaphase respectively) at 34°C. Cells were grown similar to (B).
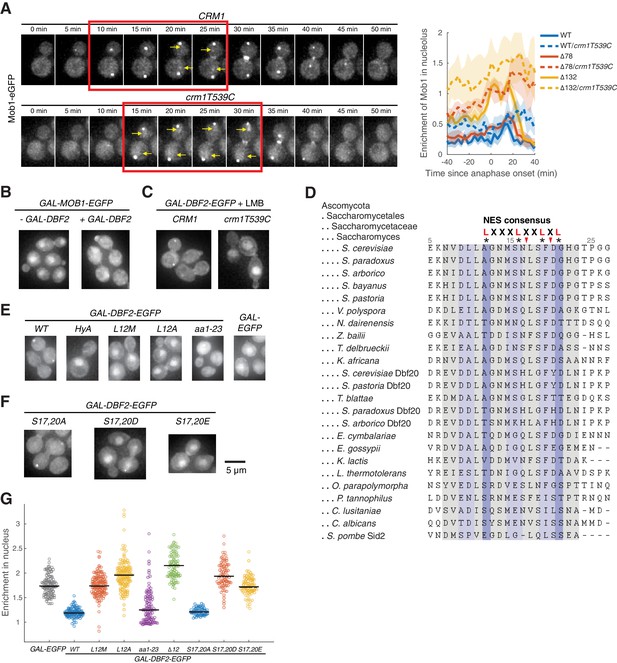
Identification of a functional nuclear export signal (NES) in Dbf2.
(A) Mob1’s cellular localization when nuclear export is inhibited. Left, cells (A39893, A41349) were grown at room temperature in SC medium + 2% glucose and leptomycin B (LMB). Red box indicates anaphase and yellow arrows highlight increased nuclear and nucleolar localization. Right, comparison of nucleolar enrichment of Mob1 in CRM1 (A41211, A41212, and A41213; n = 12, 18, and 11 cells respectively) and crm1T539C cells (A41373, A41374, and A41375; n = 13, 9, and 16 cells respectively). Cells were grown at 25°C in SC medium + 2% glucose with 100 ng/ml LMB and imaged every 3 min for 4 hr. (B) Localization of overexpressed Mob1-eGFP from the GAL1-10 promoter with (A41363) or without (A41364) co-overexpressing Dbf2 from the same promoter. Cells were grown in SC medium + 2% raffinose and mounted onto agarose pads made with SC medium + 1% raffinose and 1% galactose. (C) Localization of overexpressed Dbf2-eGFP with (A41383) or without (A41384) inhibiting Crm1 with LMB. Cells were grown in SC medium + 2% raffinose and mounted onto agarose pad made with SC medium + 1% raffinose, 1% galactose, and 100 ng/ml LMB. (D) Alignment of Dbf2 homologs in Saccharomycetes and S. pombe. (E–G) Localization of various Dbf2-eGFP mutants expressed from the GAL1-10 promoter (A41388, A41389, A41386, A41390, A41391, A41392, A41393, A41440, A41441, and A41442; n = 97, 98, 123, 135, 120, 88, 58, 76, and 79 cells respectively). Cells were grown as in (B) and analyzed after 5 hr of growth in galactose containing medium.
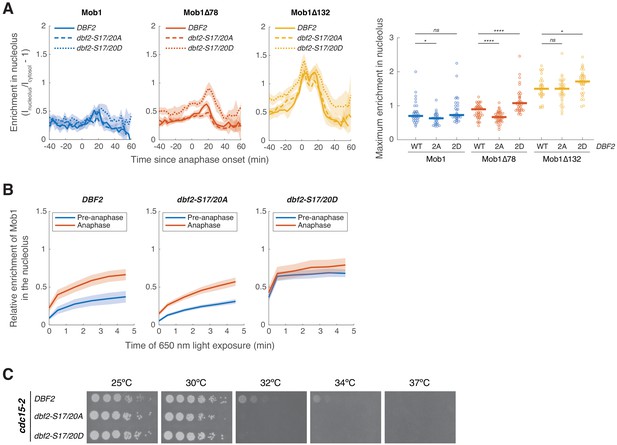
Phosphorylation of Dbf2’s nuclear export signal (NES) partially regulates Dbf2-Mob1’s nuclear access.
(A) Nucleolar enrichment of full-length and truncated Mob1 in wild-type DBF2, dbf2-S17/20A or dbf2-S17/20D cells (A41614, A41617, A41620, A41615, A41618, A41621, A41616, A41619, A41622; n = 32, 28, 33,39, 43, 37, 31, 35, and 37 cells respectively). Cells were grown and imaged as in Figure 1E. Solid lines represent the average; shaded areas represent 95% confidence intervals. For maximum enrichment, each dot represents a single cell. Solid lines represent the median. ****p<0.0001; *p<0.05 by two-sided Wilcoxon rank sum test. (B) Enrichment of Mob1 in the nucleolus as a function of PhyB activation time for cells expressing wild-type DBF2 (A41608; n = 42 and 24 cells for pre-anaphase and anaphase, respectively), dbf2-S17/20A (A41609; n = 55 and 41 cells for pre-anaphase and anaphase, respectively) or dbf2-S17/20D (A41610; n = 58 and 28 cells for pre-anaphase and anaphase, respectively). Cells were grown as in Figure 3—figure supplement 1. (C) Fivefold serial dilutions of cdc15-2 cells harboring wild-type DBF2 (A41624), dbf2-S17/20A (A41625), or dbf2-S17/20D (A41626) in YEP + 2% glucose at the indicated temperatures.
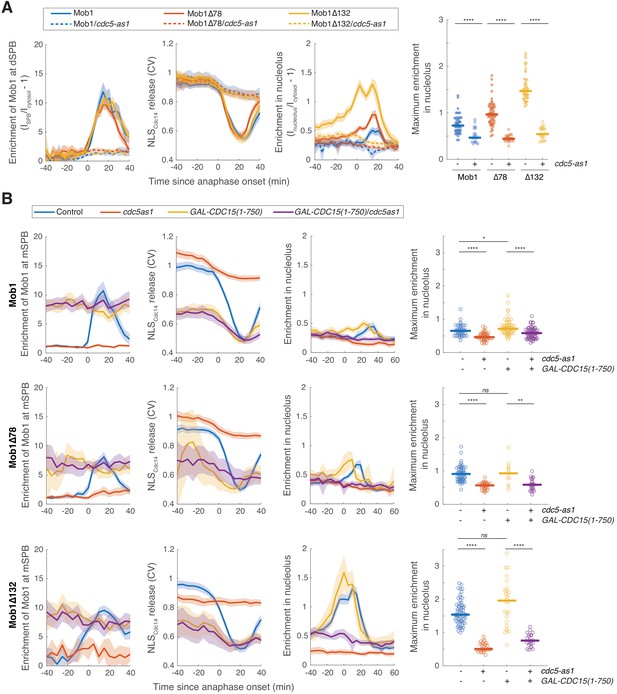
Nucleolar localization of Dbf2-Mob1 is regulated by CDC5 independently of CDC15.
(A) Enrichment of Mob1 at the daughter spindle pole body (dSPB), in the nucleolus, and Dbf2-Mob1’s kinase activity in cells wild type for CDC5 (A41211, A41212, and A41213; n = 49, 60, and 47 cells respectively) or harboring a cdc5-as1 allele (A41334, A41335, and A41336; n = 23, 30, and 28 cells respectively). Cells were grown at 25°C in SC medium + 2% glucose and 5 μM CMK and imaged every 3 min for 4 hr. (B) Cells harboring GAL-CDC15(1-750) and cdc5-as1 either containing eGFP-MOB1 (A41211, A41334, A41376, and A41337; n = 44, 41, 58, and 61 cells respectively), or eGFP-MOB1Δ78 (A41212, A41335, A41377, and A41338; n = 54, 30, 12, and 22 cells respectively), or eGFP-MOB1Δ132 (A41213, A41336, A41378, and A41339; n = 62, 28, 26, and 22 cells respectively) were analyzed to determine Mob1 localization. Localization to the mother SPB (mSPB) instead of dSPB was quantified here because cells expressing GAL-CDC15(1-750) often exit from mitosis in the mother (without movement of a SPB into the bud). For cells exited with two SPBs in the mother cell, maximum intensities of the two SPBs were used. Cells were grown at 25°C in SC medium + 1% raffinose, 1% galactose, and 5 μM CMK and imaged every 5 min for 5 hr. Solid lines represent the average of single cell traces aligned to anaphase onset. Shaded areas represent 95% confidence intervals. For maximum enrichment, each dot represents a single cell. Solid lines represent the median. ****p<0.0001; **p<0.01; *p<0.05 by two-sided Wilcoxon rank sum test.
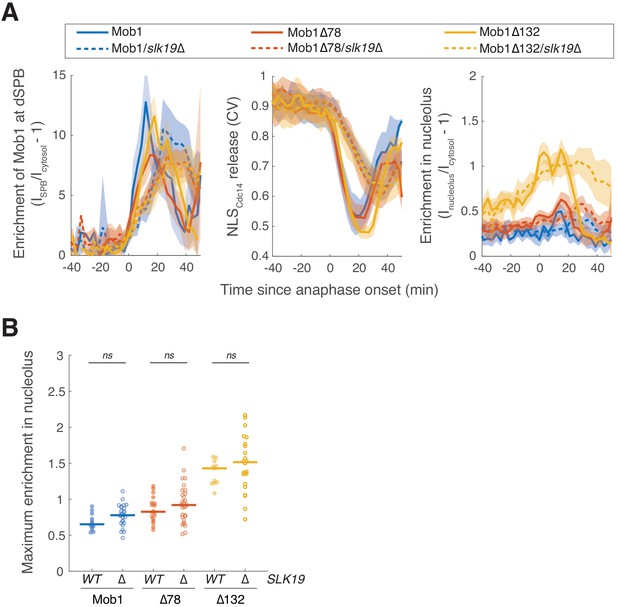
Nucleolar localization of Dbf2-Mob1 does not depend on the Cdc fourteen early anaphase release (FEAR) network.
(A) Enrichment of Mob1 at the daughter spindle pole body (dSPB), in the nucleolus, and Dbf2-Mob1’s kinase activity in wild-type (A41211, A41212 and A41213; n = 12, 18, 11) or slk19Δ (A41357, A41358, A41359; n = 26, 29, 22 cells) cells. Cells were grown at 25°C in SC medium + 2% glucose and imaged every 3 min for 4 hr. Solid lines represent the average of single cell traces aligned to anaphase onset. Shaded areas represent 95% confidence intervals. (B) Maximum enrichment of Mob1 in the nucleolus. Each dot represents a single cell. The solid lines represent the median. ns, not significant (p>0.05) by two-sided Wilcoxon rank sum test.
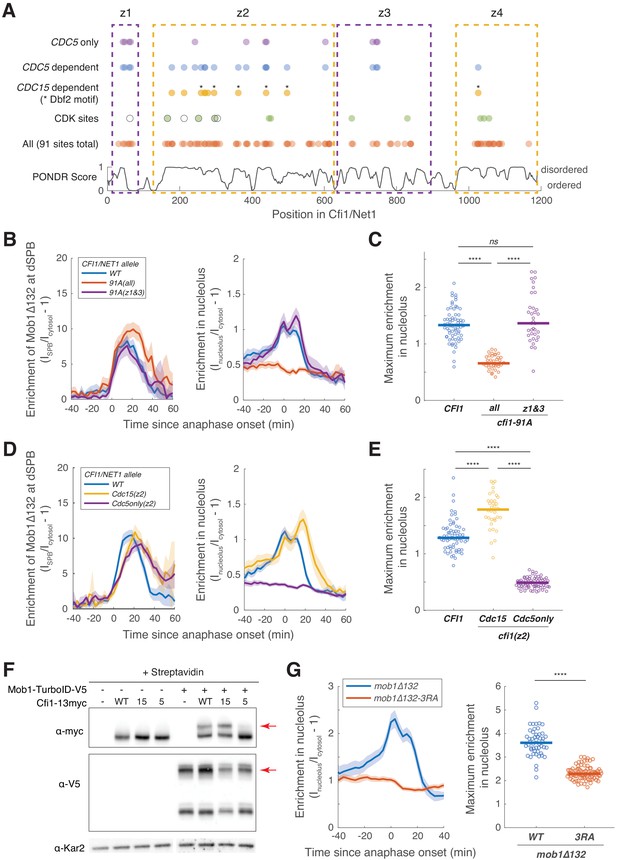
Cdc5 promotes Dbf2-Mob1’s nucleolar localization by phosphorylating Cfi1/Net1.
(A) Distribution of all, CDK sites, CDC15- and CDC5-dependent phosphorylation sites (Supplementary file 4) and disordered regions in Cfi1/Net1. For CDK sites, open circles represent sites identified and mutated in Azzam et al., 2004 (cfi1/net1-6Cdk) and filled circles represent sites identified in Holt et al., 2009. Dashed boxes denote the four zones. (B–E) Localization of Mob1Δ132 in CFI1/NET1 (A41411, n = 67 cells for B and C and 66 cells for D and E), cfi1-91A mutants (A41412 and A41413, n = 36 and 35 cells), cfi1-Cdc15(z2) (A41593, n = 34 cells) or cfi1-Cdc5only(z2) (A41594, n = 69 cells). Cells were grown at 25°C in SC medium + 2% glucose and imaged every 3 min for 4 hr. (F) Streptavidin gel-shift assays to probe the interactions between TurboID-tagged Mob1 and different CFI1/NET1 alleles (from left to right: A2587, A41596, A41597, A41598, A41379, A41611, A41612, A41613). -, not tagged; WT, wild-type Cfi1-13myc; 15, Cfi1-Cdc15(z2)−13myc; 5, Cfi1-Cdc5only(z2)−13myc. Cells were grown at room temperature in YEP + 2% glucose and lysates were treated with streptavidin and immunoblotted as indicated. Red arrows highlight biotinylated proteins. (G) Enrichment of Mob1Δ132 (A41664, n = 50 cells) or Mob1Δ132-3RA (A41665, n = 85 cells) in the nucleolus. Cells were grown at 25°C in SC medium + 2% glucose and imaged every 3 min for 4 hr. For graphs in (B–E and G), solid lines represent the average of single cell traces aligned to anaphase onset. Shaded areas represent 95% confidence intervals. For maximum enrichment, each dot represents a single cell. Solid lines represent the median. ****p<0.0001 by two-sided Wilcoxon rank sum test.

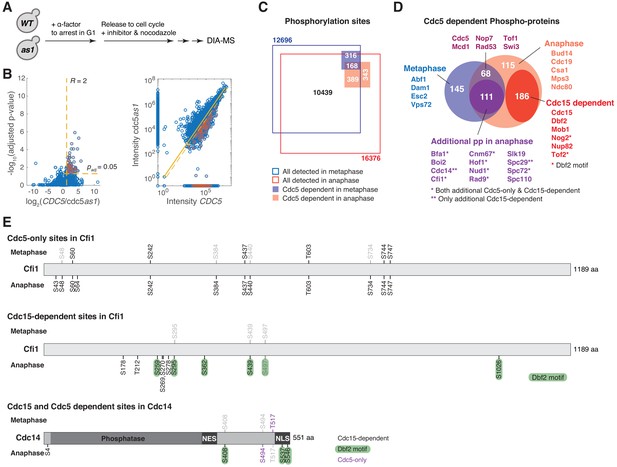
Phosphoproteomics identifies CDC15 (mitotic exit network [MEN]) and CDC5 dependent phosphorylation in anaphase.
(A) Overview of the sample preparation for phosphoproteomics analysis to map CDC15 (MEN) and CDC5 dependent phosphorylation in anaphase. Paired wild-type (WT, A2587) and cdc15-as1 (A10991) or paired wild-type and cdc5-as1 (A40903) cultures were synchronized in G1 with α-factor (5 μg/ml). After 150 min cells were released into fresh medium with the corresponding inhibitors in the absence of pheromone at room temperature. After ~100 min cells were harvested to extract proteins and processed for data-independent acquisition mass spectrometry (DIA-MS) analysis with seven technical replicates for each sample. Immuno-fluorescence (IF) using an anti-tubulin antibody was performed on the collected cells to determine the percentage of cells with anaphase spindle (~70% for wild-type cells and ~95% for cdc5-as1 and cdc15-as1 cells). (B) Volcano plots of −log10 transformed FDR adjusted P-value versus log2(fold change or ratio) of intensities measured for peptides identified in anaphase cells with WT or analog-sensitive (as1) alleles of CDC15 or CDC5. Yellow dashed lines indicate the cutoff (R > 2 and padj<0.05) used to identify peptides whose phosphorylation depends on the corresponding kinase as marked with red shaded dots. (C) Correlations of peptide intensity in WT and as1 samples for Cdc15 and Cdc5 inhibition in anaphase. Data points on the axis represent peptides that were only detected in one sample but not the other. Red shaded dots denote hits for CDC15 or CDC5-dependent phosphopeptides identified based on the cutoff described in (B) and for peptides that were detected in at least five out of seven replicates in WT samples but were missing in as1 samples (thus no fold change could be calculated). (D and E) Summary of phosphorylation sites (D) and phospho-proteins (E) determined as CDC15- or CDC5-dependent. Potential Dbf2-Mob1 targets were identified as CDC15-dependent and to fit the Dbf2 phosphorylation consensus motif RXXS*, where * denotes the site of phosphorylation. Cdc5-only sites are sites that are CDC5-dependent but not CDC15-dependent. (F) CDC15- and CDC5-dependent sites in Mob1 and Dbf2. Light gray sites represent sites that were detected but were not determined as either CDC15- or CDC5-dependent.
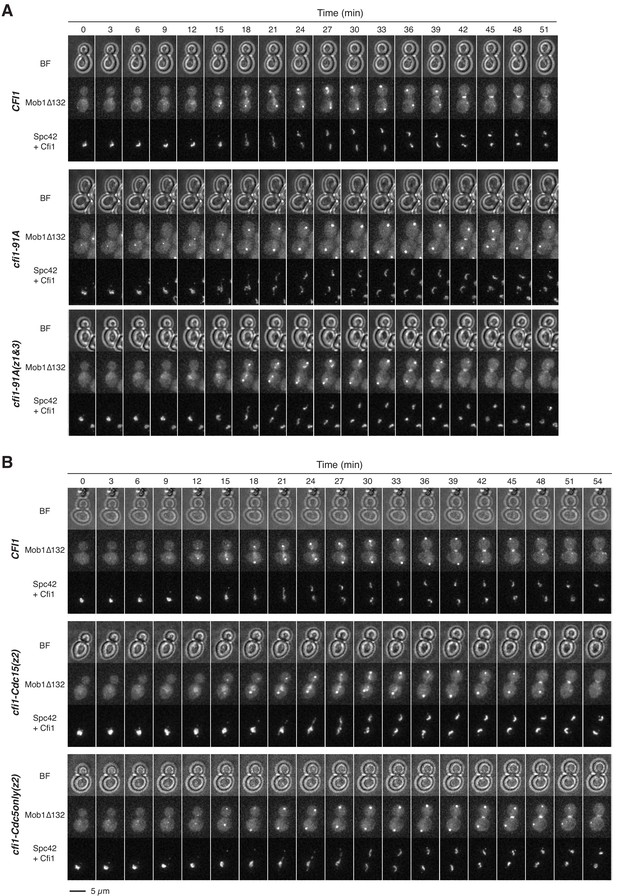
Phosphorylation of Cfi1/Net1 modulates Dbf2-Mob1’s nucleolar localization.
(A and B) Representative images of Mob1Δ132 localization in CFI1/NET1 (A41411), cfi1-91A (A41412 and A41413), cfi1-Cdc15(z2) (A41593), or cfi1-Cdc5only(z2) (A41594) cells.
Identification of CDC5-dependent phosphorylation sites in metaphase.
(A) Overview of the sample preparation for phosphoproteomic analysis to map CDC5-dependent phosphorylation sites in metaphase. Same procedures were followed as in Figure 5—figure supplement 1A except that following release of cells from the G1 arrest, cells were resuspended in medium containing the inhibitor CMK (5 μM) and nocodazole (15 μg/ml). (B) Volcano plot and correlation of peptide intensities with and without inhibition of Cdc5 (cdc5-as1 or WT) in metaphase. Yellow dashed lines indicate the cutoff (R > 2 and padj<0.05, or detected in at least four out of six replicates in WT sample but missing in cdc5as1 sample) used to identify peptides whose phosphorylation depends on CDC5 as marked with red shaded dots. (C and D) Summary and comparison of phosphorylation sites (C) and phospho-proteins (D) detected in metaphase and anaphase. (E) CDC5-only and CDC15-dependent sites in Cfi1/Net1 and Cdc14 mapped in metaphase and anaphase. Light gray sites in metaphase/anaphase represent phosphorylation sites that were detected but were not determined as CDC5-dependent.
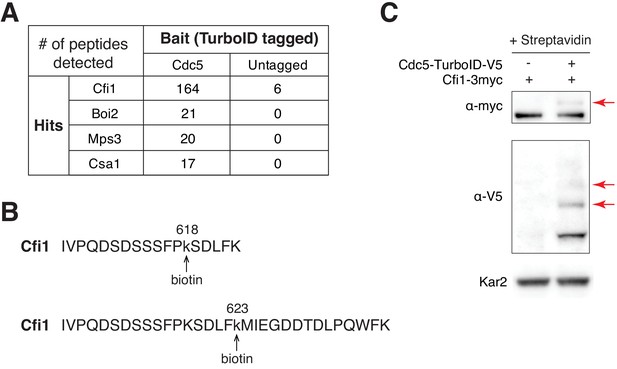
Cdc5 interacts with Cfi1/Net1 in vivo.
(A) Results of TurboID proximity-based biotinylation for Cdc5 (A41385 and A2588). (B) Biotinylated peptides of Cfi1/Net1 detected in cells with Cdc5-TurboID. (C) Streptavidin gel-shift assay to probe the interaction between Cdc5-TurboID and Cfi1/Net1 (A1638 and A41418). Cells were grown at room temperature in YPED. Lysates were treated with streptavidin and immunoblotted as indicated. Red arrows highlight biotinylated proteins.
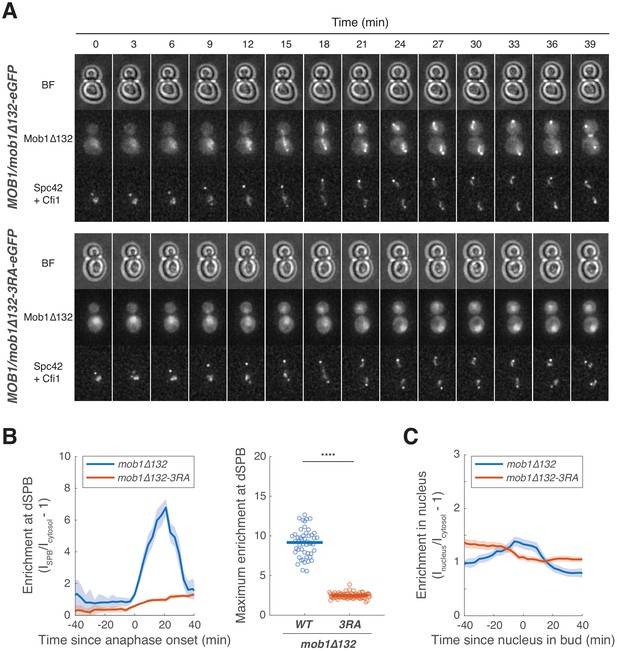
Dbf2-Mob1 localizes to the nucleolus via Mob1’s phosphoserine-threonine binding domain.
(A) Representative images of Mob1Δ132 (A41664) and Mob1Δ132-3RA (A41665) localization. (B) Enrichment of Mob1Δ132 (A41664, n = 50 cells) or Mob1Δ132-3RA (A41665, n = 85 cells) at the daughter spindle pole body (dSPB). Cells were grown like in Figure 5G. Solid lines represent the average of single cell traces aligned to anaphase onset. Shaded areas represent 95% confidence intervals. For maximum enrichment, each dot represents a single cell. Solid lines represent the median. ****p<0.0001 by two-sided Wilcoxon rank sum test. (C) Enrichment of Mob1Δ132 (A41666, n = 54 cells) or Mob1Δ132-3RA (A41667, n = 77 cells) in the nucleus. Cells were grown at 25°C in SC medium + 2% glucose and imaged every 3 min for 4 hr. Solid lines represent the average of single cell traces aligned to anaphase onset. Shaded areas represent 95% confidence intervals.
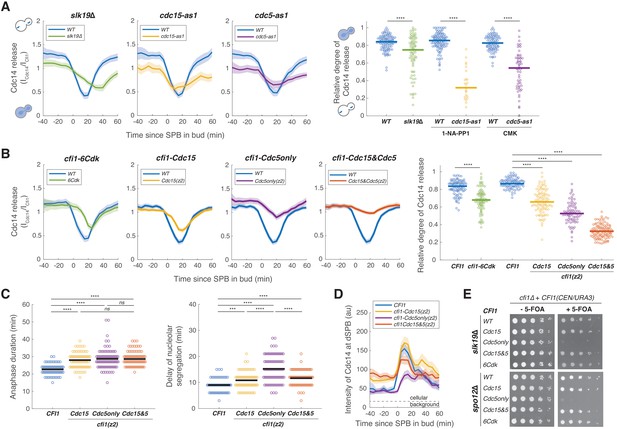
Mitotic exit network (MEN) and Cdc5 promote release of Cdc14 from the nucleolus by phosphorylating Cfi1/Net1.
(A) Cdc14 nucleolar release kinetics in wild-type (A41387, n = 134, 123, and 96 cells for each condition), slk19Δ (A41410, n = 86 cells), cdc15-as1 (A41408, n = 38 cells), or cdc5-as1 mutant (A41409, n = 61 cells). Cells were grown at 25°C in SC medium + 2% glucose with corresponding inhibitors and imaged every 5 min for 5 hr. Release of Cdc14 from the nucleolus was quantified as the ratio of fluorescence intensity of Cdc14-eGFP to Cfi1/Net1-mScarlet-I in the nucleolus (ICdc14/ICfi1). Relative degree of Cdc14 release from the nucleolus was calculated with the normalized minimal Cdc14 level in the nucleolus as 1 - (ICdc14(tmin)/ICfi1(tmin))/ (ICdc14(t-20)/ICfi1(t-20)), where tmin represents the frame with minimal Cdc14 level in the nucleolus and t-20 represents 20 min before movement of the spindle pole body (SPB) into bud. (B) Cdc14 nucleolar release kinetics in cells harboring wild-type CFI1/NET1 (A41387, n = 102 and 114 cells) or CFI1/NET1 phospho-mutants for CDK sites (A41420, n = 95 cells), Cdc15 sites (A41587, n = 104 cells), Cdc5 sites (A41588, n = 86 cells), and Cdc15&Cdc5 sites (A41589, n = 131 cells). Cells were grown at 25°C in SC medium + 2% glucose and imaged every 5 min for 5 hr. (C) Distribution of anaphase duration and relative delay of nucleolar segregation for different CFI1/NET1 phospho-mutants (A41436, A41590, A41591, and A41592; n = 76, 85, 99, and 92 cells respectively) measured using the SPB marker Spc42-eGFP and the nucleolar marker Cfi1/Net1-mScarlet-I (see Figure 6—figure supplement 3 for details). Cells were grown at 25°C in SC medium + 2% glucose and imaged every 3 min for 4 hr. (D) Intensities of Cdc14-eGFP at dSPBs in different CFI1/NET1 phospho-mutant cells (A41387, A41587, A41588, and A41589; n = 80, 82, 77, and 89 cells respectively). Cells were grown and imaged as in (B). (E) Genetic interactions between different CFI1/NET1 phospho-mutants and slk19Δ (A41645, A41646, A41647, A41648, A41649) or spo12Δ (A41650, A41651, A41652, A41653, A41654) analyzed by plasmid shuffling (see Materials and methods for details). Fivefold serial dilutions were spotted onto plates with or without 5’-fluoroorotic acid (5-FOA) and incubated at 25°C for 2–3 days. The presence of 5-FOA selects cells that are viable after losing the CFI1(URA3/CEN) plasmid. For all graphs, single cell traces were aligned to the frame where the dSPB entered the bud and averaged. Solid lines represent the average. Shaded areas represent 95% confidence intervals. For distributions, each dot represents a single cell. Solid lines represent the median for (A and B) and the mean for (C). ****p<0.0001; ***p<0.001; **p<0.01; *p<0.05 by two-sided Wilcoxon rank sum test.
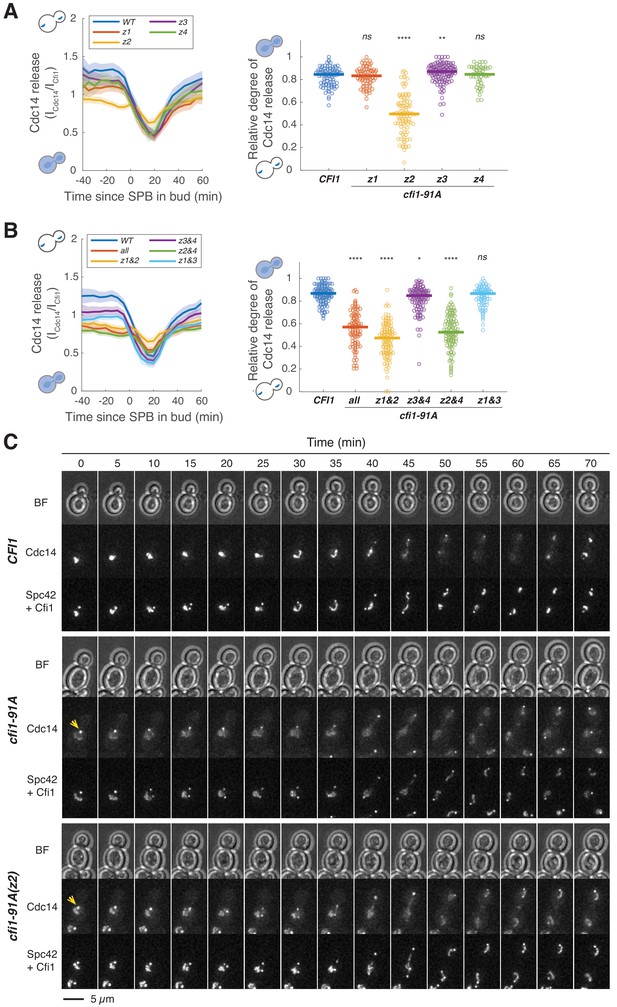
Phosphorylation in zone 2 of Cfi1/Net1 regulates Cdc14 release from the nucleolus.
(A and B) Kinetics of Cdc14 release from the nucleolus in cells harboring wild-type CFI1/NET1 (A41387, n = 103 cells for A, n = 130 cells for B) or different CFI1/NET1 phospho-null mutants (A41398, A41399, A41400, A41401, A41404, A41405, A41397, A41402 and A41403; n = 95, 103, 146, 59, 102, 113, 128, 114, and 102 cells respectively). Cells were grown at 25°C in SC medium + 2% glucose and imaged every 5 min for 5 hr. Release of Cdc14 from the nucleolus was quantified as in Figure 6A. Each dot represents a single cell and the solid lines represent the median. ****p<0.0001; **p<0.01; *p<0.05 by two-sided Wilcoxon rank sum test. (C) Representative images showing Cdc14 nucleolar release kinetics for cells harboring wild-type CFI1/NET1 (A41387) or CFI1/NET1 phospho-mutants with all 91 sites (A41404) or only sites in zone 2 (A41399) mutated to alanine. Yellow arrows highlight localization of Cdc14 at dSPB prior to anaphase.
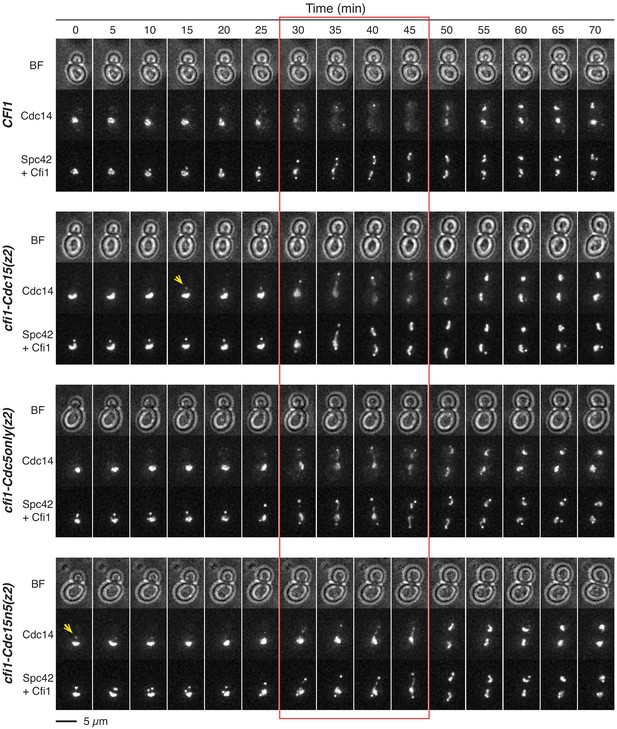
The mitotic exit network (MEN) and CDC5 promote release of Cdc14 from the nucleolus by phosphorylating Cfi1/Net1.
Representative images showing Cdc14 nucleolar release kinetics for cells harboring wild-type CFI1/NET1 (A41387) or CFI1/NET1 phospho-mutants for Cdc15 sites (A41587), Cdc5 sites (A441588), and Cdc15 and Cdc5 sites (A41589). Red box highlights anaphase when Cdc14 is fully released in WT cells. Yellow arrows highlight localization of Cdc14 at daughter spindle pole body (dSPB) prior to anaphase.
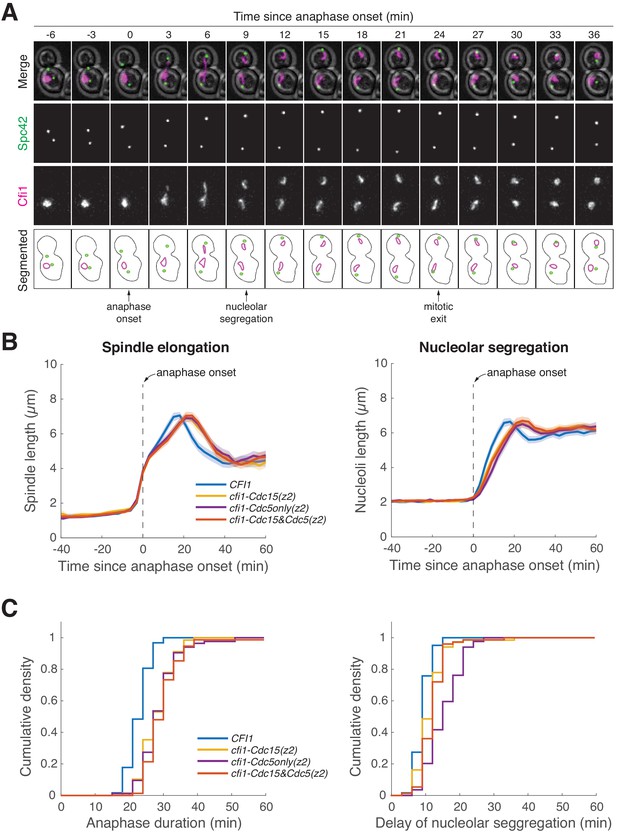
Cfi1/Net1 phospho-mutants delay mitotic exit and nucleolar segregation.
(A) Tracking anaphase progression and nucleolar segregation using Spc42-eGFP and Cfi1/Net1-mScarlet-I as markers, respectively. A41436 cells were grown at 25°C in SC medium + 2% glucose and imaged every 3 min for 4 hr. (B) Quantification of spindle elongation (spindle length was estimated by measuring the distance between the two spindle pole bodies [SPBs]) and nucleolar segregation (nucleoli length was estimated by measuring the length of the major axis of the nucleolar mass) for CFI1/NET1 mutants using the SPB marker Spc42-eGFP and the nucleolar marker Cfi1/Net1-mScarlet-I (A41436, A41590, A41591, and A41592; same dataset as in Figure 6C). Single cell traces were aligned to anaphase onset (spindle length >3 μm) and averaged. Solid lines represent the average. Shaded areas represent 95% confidence intervals. (C) Cumulative density of anaphase duration (left) and delay of nucleolar segregation (right) for cells in (B). Anaphase duration was defined as the time from anaphase onset (spindle length >3 μm) to mitotic exit (spindle breakdown, determined as relaxation of the distance between SPBs). Delay of nucleolar segregation was defined as the time of nucleolar segregation (clear separation of two nucleolar masses) relative to anaphase onset.
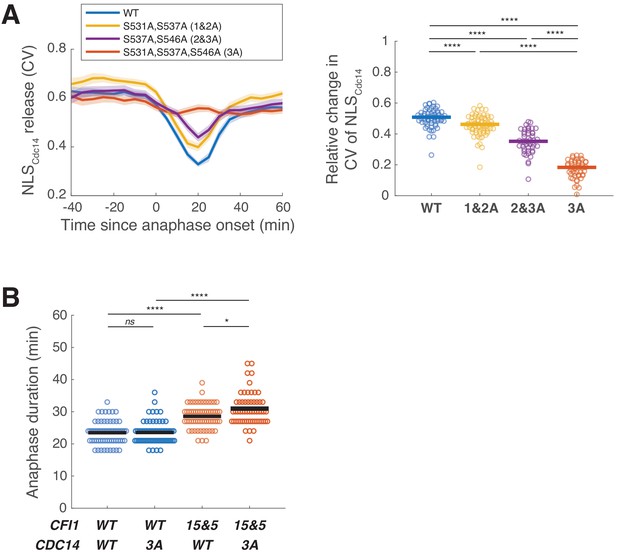
Phosphorylation of the Cdc14 nuclear localization signal (NLS) does not play a major role in promoting mitotic exit.
(A) Profiles (left) and relative degrees (right) of nuclear release of wild-type (A41584, n = 69 cells) and mutant NLSCdc14 reporters where all three potential Dbf2-Mob1 target sites (A41585, n = 59 cells) or only two out of three sites (A41586 and A41623, n = 77 and 55 cells) were mutated to alanine. Cells were grown at 25°C in SC medium + 2% glucose and imaged every 5 min for 4 hr. Solid lines (right) represent the median. (B) Distribution of anaphase duration for CDC14 phospho-mutants in combination with CFI1 or cfi1-Cdc15&Cdc5(z2) measured using the spindle pole body (SPB) marker Spc42-eGFP (A41436, A41707, A41592, and A41708; n = 64, 65, 61, and 58 cells, respectively). Cells were grown at 25°C in SC medium + 2% glucose and imaged every 3 min for 4 hr. Anaphase duration was defined as the time from anaphase onset (spindle length >3 μm) to mitotic exit (spindle breakdown). Solid lines represent the mean. ****p<0.0001; *p<0.05; ns, not significant (p>0.05) by two-sided Wilcoxon rank sum test.

Mutating both CDK and Cdc5 sites in Cfi1/Net1 results in severe delays in mitotic exit and nucleolar segregation.
(A) Cdc14 nucleolar release kinetics in cells harboring wild-type CFI1/NET1 (A41387, n = 39 cells) or CFI1/NET1 phospho-mutants for CDK sites combined with Cdc5 sites in zone 2 (A41691, n = 79 cells). Cells were grown at 25°C in SC medium + 2% glucose and imaged every 5 min for 5 hr. Single cell traces were aligned to the frame where the daughter spindle pole body (dSPB) entered the bud and averaged. Solid lines represent the average. Shaded areas represent 95% confidence intervals. Each dot represents a single cell. Solid lines represent the median. ****p<0.0001 by two-sided Wilcoxon rank sum test. (B) Distribution of anaphase duration and relative delay of nucleolar segregation for different CFI1/NET1 phospho-mutants (A41436, A41692, and A41694; n = 58, 55, and 44 cells respectively) measured using the SPB marker Spc42-eGFP and the nucleolar marker Cfi1/Net1-mScarlet-I. Cells were grown at 25°C in SC medium + 2% glucose and imaged every 3 min for 4 hr. Each dot represents a single cell. Solid lines represent the median. ****p<0.0001 by two-sided Wilcoxon rank sum test.
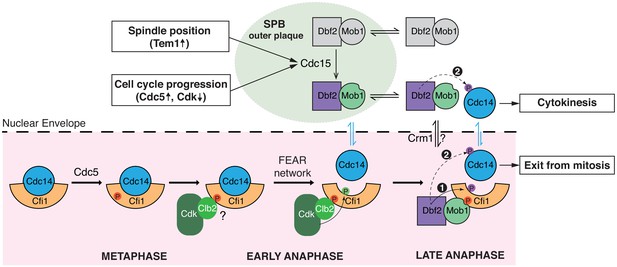
A model for Cdc14 activation and mitotic exit in budding yeast.
In metaphase, Cdc5 phosphorylates Cfi1/Net1 in the nucleolus to prepare for Cdc14 release/activation in anaphase. Upon anaphase onset, the Cdc fourteen early anaphase release (FEAR) network promotes phosphorylation of Cfi1/Net1 by Clb2-Cdk1 which results in transient release of Cdc14 from the nucleolus. In the meantime, the mitotic exit etwork (MEN) kinase Cdc15 is activated by integrating inputs from both spindle position (via Tem1) and cell cycle progression (via Cdc5 and CDK activities). Activated (spindle pole body [SPB]-localized) Cdc15 phosphorylates the SPB outer plaque protein Nud1 which creates a dynamic docking site for the MEN terminal kinase complex Dbf2-Mob1 and facilitates phosphorylation and activation of Dbf2-Mob1 by Cdc15. Activated Dbf2-Mob1 gains access to the nucleus and is targeted to the nucleolus by interacting with Cdc5-primed Cfi1/Net1. Nucleolar Dbf2-Mob1 then phosphorylates Cfi1/Net1, keeping Cdc14 dissociated from its nucleolar inhibitor to trigger exit from mitosis. In addition, active Dbf2-Mob1 in the nucleolus and/or cytoplasm phosphorylates Cdc14 at its nuclear localization signal (NLS) resulting in cytoplasmic retention of Cdc14 to facilitate cytokinesis.
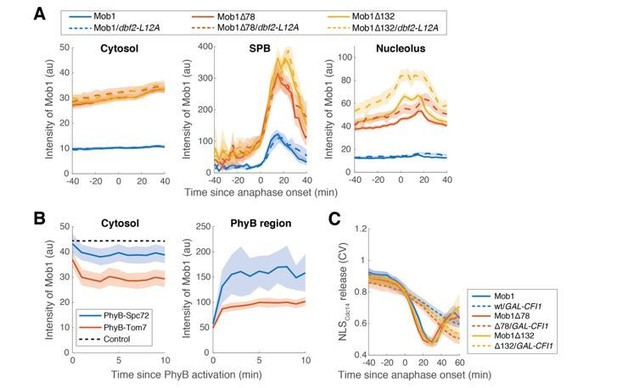
Changes in the nucleolar localization of Dbf2-Mob1 do not affect its SPB enrichment.
(A) Raw intensity measurements for the experiments shown in Figure 3E. (B) Quantification of Mob1 intensity in the cytosol or PhyB anchored region upon PhyB activation for the optogenetics experiments shown in Figure 3—figure supplement 1B. (C) Kinetics of MEN activation (as measured by the release of NLSCdc14 reporter) for the experiments shown in Figure 2F with GAL-CFI1. For all graphs, solid lines represent the average of single cell traces aligned to the event described in the axis. Shaded areas represent 95% confidence intervals.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (S. cerevisiae) | See Supplementary file 1 | |||
| Strain, strain background (S. cerevisiae) | W303 | https://www.yeastgenome.org/strain/w303 | ||
| Genetic reagent (S. cerevisiae) | See Supplementary file 1 | |||
| Recombinant DNA reagent | See Supplementary file 2 | |||
| Strain, strain background (E. coli) | DH5α | New England Biolabs | Cat# C2987U | Chemical competent cells |
| Antibody | Anti-GFP [JL-8] (Mouse monoclonal) | Takara Bio | Cat# 632381; RRID:AB_2313808 | WB (1:1000) |
| Antibody | Anti-Myc [9E10] (Mouse monoclonal) | Abcam | Cat# ab32; RRID:AB_303599 | WB (1:500) |
| Antibody | Anti-V5 (Mouse monoclonal) | Invitrogen | Cat# R960-25; RRID:AB_255656 | WB (1:2000) |
| Antibody | Anti-Kar2 (Rabbit polyclonal) | Gift from Mark Rose | N/A | WB (1:200,000) |
| Antibody | HRP-conjugated anti-mouse IgG (Sheep monoclonal) | GE | Cat# NA9310; RRID:AB_772193 | (1:10,000) |
| Antibody | HRP-conjugated anti-rabbit IgG (Donkey monoclonal) | GE | Cat# NA934; RRID:AB_772206 | (1:10,000) |
| Antibody | Anti-tubulin [YOL1/34] (Rat monoclonal) | Abcam | Cat# Ab6161; RRID:AB_305329 | IF (1:100) |
| Antibody | FITC-anti-Rat IgG (Donkey polyclonal) | Jackson ImmunoResearch | Cat# 712-095-153; RRID:AB_2340652 | (1:50) |
| Peptide, recombinant protein | Streptavidin | Sigma-Aldrich | Cat# 189730 | |
| Peptide, recombinant protein | α-factor | The Koch Institute Swanson Biotechnology Center – Biopolymers Core Facility | N/A | |
| Commercial assay or kit | Gibson Assembly Master Mix | New England Biolabs | Cat# E2611S | |
| Commercial assay or kit | Q5 Site-Directed Mutagenesis Kit | New England Biolabs | Cat# E05545 | |
| Commercial assay or kit | Bradford protein assay | BioRad | Cat# 5000006 | |
| Commercial assay or kit | BCA protein assay | Pierce | Cat# 23227 | |
| Commercial assay or kit | MyOne Streptavidin C1 dynabeads | Thermo Fisher Scientific | Cat# 65001 | |
| Chemical compound, drug | Nocodazole | Sigma-Aldrich | Cat# M1404 | |
| Chemical compound, drug | Biotin | Sigma-Aldrich | Cat# B4639 | |
| Chemical compound, drug | 1-NA-PP1 | Cayman Chemical | Cat# 10954 | |
| Chemical compound, drug | CMK | MedChem Express | Cat# HY-52101 | |
| Chemical compound, drug | Phycocyanobilin (PCB) | Santa Cruz Biotechnology | Cat# sc-396921 | |
| Software, algorithm | FIJI (ImageJ) | Schindelin et al., 2012 | RRID:SCR_002285 | |
| Software, algorithm | MATLAB_R2018b | Mathworks, Inc (2018) | RRID:SCR_001622 | |
| Software, algorithm | SnapGene v4.3 | https://www.snapgene.com | RRID:SCR_015052 | |
| Software, algorithm | Custom MATLAB codes | This paper | https://github.com/snow-zhou/Dbf2-Mob1 (copy archived at swh:1:rev:edb372c2e4ddf8eb2278536a7fa580abaa60acf1) | |
| Other | Mini Bead Beater | Biospec Products | N/A |
Additional files
-
Supplementary file 1
Yeast strains used in this study.
- https://cdn.elifesciences.org/articles/63645/elife-63645-supp1-v1.xlsx
-
Supplementary file 2
Plasmids used in this study.
- https://cdn.elifesciences.org/articles/63645/elife-63645-supp2-v1.xlsx
-
Supplementary file 3
Summary of TurboID labeling experiments.
- https://cdn.elifesciences.org/articles/63645/elife-63645-supp3-v1.xlsx
-
Supplementary file 4
Phosphorylation of Cfi1/Net1.
- https://cdn.elifesciences.org/articles/63645/elife-63645-supp4-v1.xlsx
-
Supplementary file 5
Summary of phosphoproteomics results for inhibiting Cdc15 and Cdc5 in anaphase cells and Cdc5 in metaphase cells.
- https://cdn.elifesciences.org/articles/63645/elife-63645-supp5-v1.xlsx
-
Supplementary file 6
Complete list of phosphopeptides for each phosphorylation site detected in the DIA-MS experiments.
- https://cdn.elifesciences.org/articles/63645/elife-63645-supp6-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63645/elife-63645-transrepform-v1.docx





