Type VI secretion system killing by commensal Neisseria is influenced by expression of type four pili
Figures
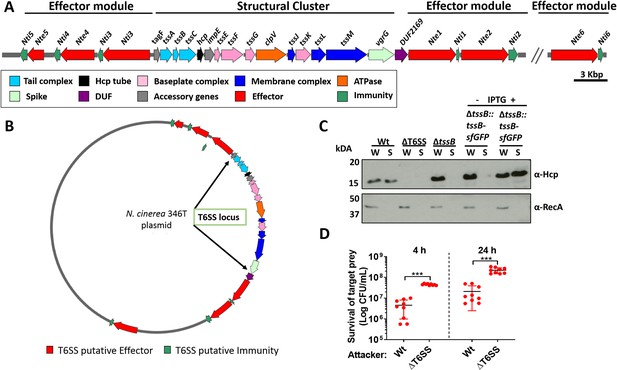
N. cinerea expresses a functional T6SS.
(A) Schematic representation of T6SS genes in N. cinerea 346T. Canonical tss nomenclature was used for genes in the T6SS cluster. (B) Map of the T6SS-associated genes encoded by the N. cinerea 346T plasmid. See also Figure 1—figure supplement 1. (C) Expression and secretion of Hcp by wild-type N. cinerea 346T (Wt) and the tssB mutant (ΔtssB). Hcp protein was detected in the whole cell lysates (W) and supernatants (S) by western blot analysis. For strain ΔtssB::tssBsfGFP, bacteria were grown in the presence (+) or absence (-) of 1 mM IPTG; molecular weight marker shown in kDa. RecA is only detected in whole cell lysates. (D) Survival of the prey, N. cinerea 27178A, after 4 and 24 h co-incubation with wild-type N. cinerea 346T or the T6SS mutant (ΔT6SS) at approximately 10:1 ratio, attacker:prey. The mean ± SD of three independent experiments is shown: ***p < 0.0001 using unpaired two-tailed Student’s t-test.
-
Figure 1—source data 1
Western Blot of N. cinerea Hcp secretion and expression.
- https://cdn.elifesciences.org/articles/63755/elife-63755-fig1-data1-v1.docx
-
Figure 1—source data 2
Survival of N. cinerea 27178A (prey) after 4 and 24 h competition with wild-type N. cinerea 346T or the T6SS mutant.
- https://cdn.elifesciences.org/articles/63755/elife-63755-fig1-data2-v1.xlsx
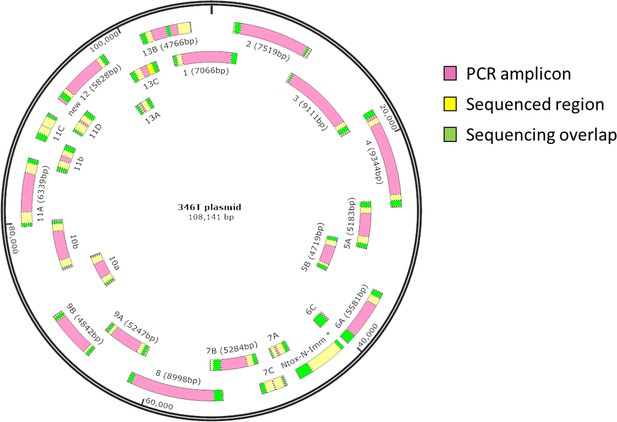
The N. cinerea 346T T6SS is encoded on a plasmid.
Overlapping PCR and sequencing confirms extra-chromosomally closed circular DNA fragment. A total of 25 PCR fragments (pink bars) were amplified from N. cinerea 346T gDNA to confirm the plasmid predicted by PacBio whole-genome sequencing. Yellow shows regions which were sequenced, and green indicates the overlapping amplified regions.
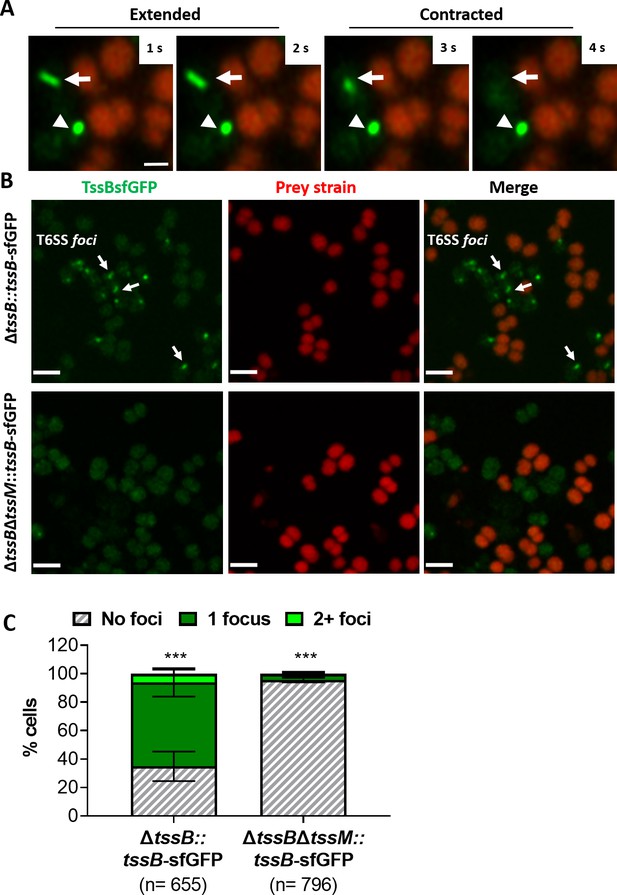
Visualisation of T6SS activity in N. cinerea.
(A) Assembly and contraction of the T6SS in N. cinerea; white arrows indicate contracting T6SSs. Time-lapse images of N. cinerea 346TΔtssB::tssBsfGFP (green) and prey N. cinerea 27178A_ sfCherry (red); the arrowhead shows a non-dynamic focus, scale bar, 1 µm. See also Figure 2—video 1. (B) Representative images of N. cinerea strains with the TssB::sfGFP fusion with (upper panels) or without (lower panels) TssM. Loss of fluorescent foci upon deletion of tssM indicates that foci correspond to active T6SSs. The scale bar represents 2 µm. (C) Quantification of TssB-sfGFP foci in different strains. T6SS foci were quantified using ‘analyse particle’ (Fiji) followed by manual inspection. For each strain, at least two images from gel pads were obtained on two independent occasions. Percentage of cells with 0, 1, or 2+ foci are shown and n = number of cells analysed. Data shown are mean ± SD of two independent experiments: ***p<0.0001 using two-way ANOVA test for multiple comparison. See also Figure 2—video 2.
-
Figure 2—source data 1
Quantification of TssB-sfGFP foci by live-microscopy.
- https://cdn.elifesciences.org/articles/63755/elife-63755-fig2-data1-v1.xlsx
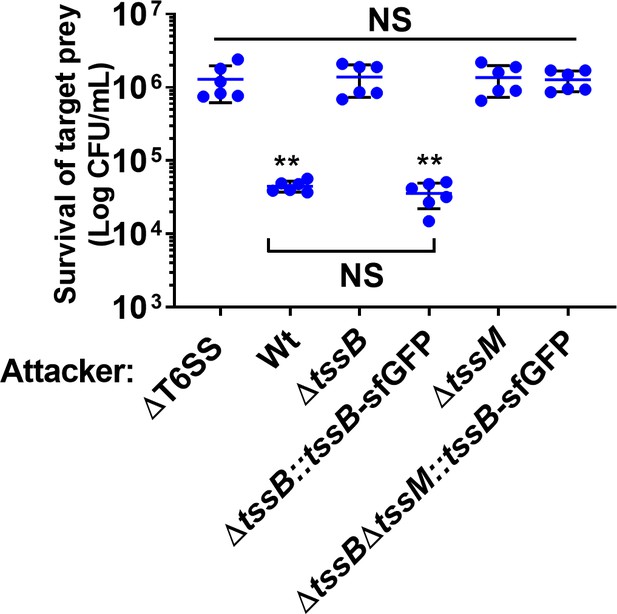
N. cinerea T6SS with a TssB C-terminal sfGFP fusion is functional and activity is lost upon deletion of tssM.
Competition assay measuring the recovery of prey (N. meningitidis 8013) after 4 hr co-incubation with wild-type N. cinerea 346T (Wt) and specified mutants at ratio of 100:1 (attacker:prey). Data shown are the mean ± SD of two independent experiments: NS, not significant, **p < 0.005 using one-way ANOVA test for multiple comparisons.
Visualisation of N. cinerea T6SS contraction.
N. cinerea 346TΔtssB::tssBsfGFP (green) and prey cells N. cinerea 27178A_sfCherry (red) were mixed at a ratio of 1:1 and spotted on a 1% agarose PBS pad supplemented with 0.1 mM IPTG. The cells were imaged for 12 s with a rate of 1 image per second.
Visualisation of N. cinerea T6SS foci.
N. cinerea 346TΔtssB::tssBsfGFP (green) or N. cinerea 346TΔtssBΔtssM::tssBsfGFP (green) and prey cells N. cinerea 27178A_sfCherry (red) were mixed at a ratio of 1:1 and spotted on a PBS 1% agarose pad supplemented with 0.1 mM IPTG. The cells were imaged for 200 s with a rate of one image per 10 s.
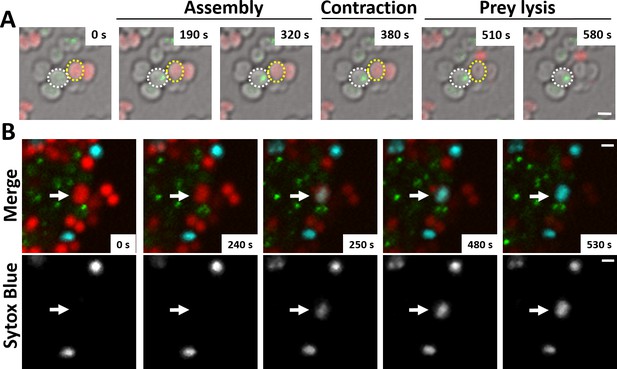
N. cinerea T6SS induces lysis in prey bacteria.
(A) Assembly of T6SSs and prey lysis. Time-lapse series of merged images with phase contrast, N. cinerea 346T ΔtssB+tssBsfGFP (green), and N. cinerea 27178A sfCherry (red); scale bar, 1 µm. (B) Top row shows merged images of GFP (green, indicating T6SS assembly/contraction), mCherry (red, prey strain), and SYTOX Blue (cyan, showing membrane permeabilisation) channels. The bottom row arrows highlight a prey cell losing membrane integrity (increase in SYTOX Blue staining inside cells) arrows. Representative image from two biological repeats. Scale bars represent 1 µm. See also Figure 3—video 1.
N. cinerea T6SS elicits prey lysis.
N. cinerea 346TΔtssB::tssBsfGFP (green) and N. cinerea 27178A_sfCherry (red) were mixed at a ratio of 1:1 and spotted onto agarose pads supplemented with 0.1 mM IPTG and the cell-impermeable DNA stain SYTOX Blue (0.5 μM) as an indicator for loss of membrane integrity. The cells were imaged for 10 min with an image taken every 10 s.
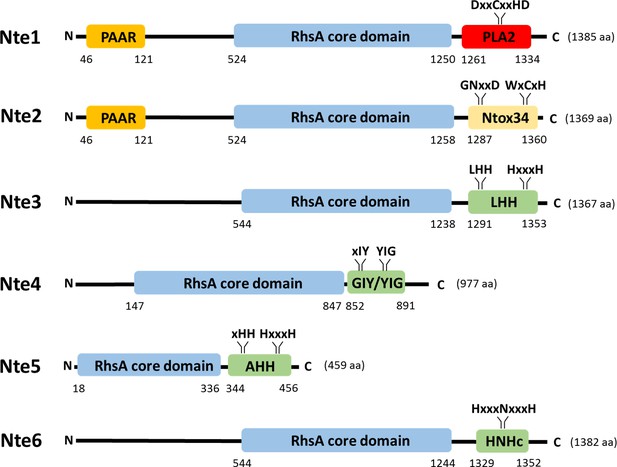
Predicted domain organisation of N. cinerea 346T T6SS effectors.
Schematic representation of bioinformatically identified effectors in N. cinerea 346T. The domain organisation of the putative effectors is shown, with PAAR motifs indicated in orange, Rhs domains in blue, endonuclease motifs (Tox-LHH pfam14411; Tox-GIY/YIG cd00719; Tox-AHH pfam14412; and Tox-HNHc cd00085) in green, RNase (Ntox34, pfam15606) motif in yellow and the phospholipase (PLA2_like, cd00618) domain in red. The conserved domains annotation was retrieved from the NCBI database.
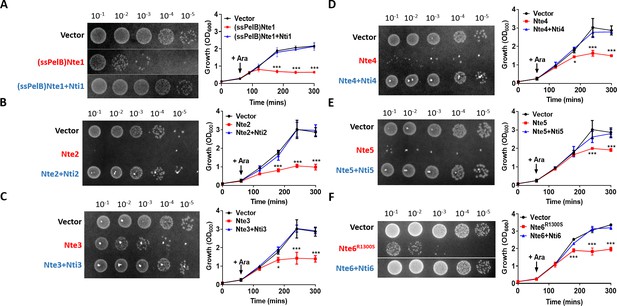
Putative N. cinerea T6SS effectors are toxic in E. coli.
(A) Arabinose (Ara)-induced expression of T6SS effector Nte1 in periplasm of E. coli leads to reduction in CFU and OD at 600 nm (OD600). Co-expression of putative immunity Nti1 restores growth to levels of strain with empty vector (pBAD33). See also Figure 5—figure supplement 1. (B-E) Cytoplasmic expression of putative effectors Nte2-5 without cognate immunity reduces growth and survival of E. coli. (F) Expression of Nte6R1300S reduces viability and growth when expressed in E. coli. Expression of Nti6 with Nte6 does not impact growth. In (A-F) number of CFU at 120 min post-induction are shown. Data shown are the mean ± SD of three independent experiments: NS, not significant, ***p<0.0001, *p<0.05 using two-way ANOVA test for multiple comparison. Images of colonies for Nte1 and Nte6 are composite as strains were spotted to different areas of the same plates.
-
Figure 5—source data 1
Growth of E. coli strains expressing putative N. cinerea 346T effector/immunity .
- https://cdn.elifesciences.org/articles/63755/elife-63755-fig5-data1-v1.xlsx
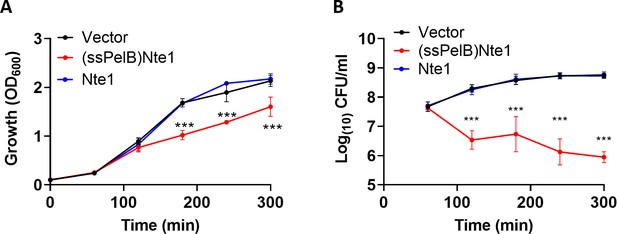
N cinerea putative T6SS effector Nte1 requires a PelB signal sequence for toxicity in E. coli.
(A) Toxicity of Nte1 with or without the PelB signal sequence (ssPelB) following expression in E. coli. Expression was induced with L-arabinose at 60 min and bacterial growth was monitored by measuring the OD600 of cultures. A reduction on OD600 was only observed when Nte1 was expressed with ssPelB. (B) Samples collected after addition of arabinose were plated to media containing glucose to repress toxin expression and enumerated. Data shown are mean ± SD of three independent experiments: NS, not significant, ***p<0.0001, *p<0.05 using two-way ANOVA test for multiple comparison.
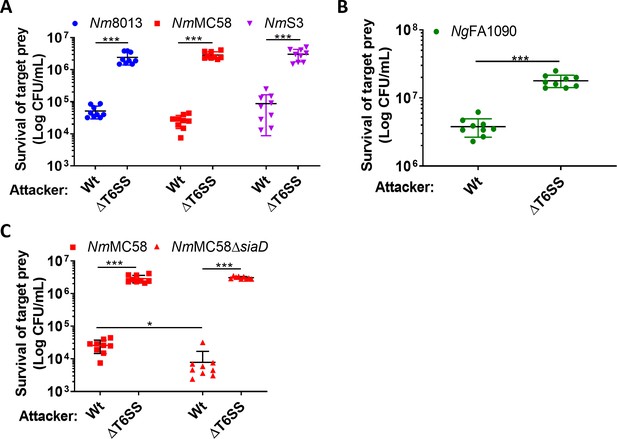
N. cinerea T6SS is active against pathogenic N. meningitidis and N. gonorrhoeae.
(A) Recovery of wild-type N. meningitidis (Nm8013, NmMC58, NmS3) after 4 hr co-incubation with N. cinerea 346T wild-type (Wt) or the T6SS mutant (ΔT6SS) at approx. 100:1 attacker:prey ratio. (B) Recovery of wild-type N. gonorrhoeae (FA1090) after 4 hr co-incubation with N. cinerea 346T wild-type (Wt) or the T6SS mutant (ΔT6SS) at approximately 10:1 attacker:prey ratio. (C) Unencapsulated N. meningitidis (NmMC58ΔsiaD) is more susceptible to T6SS-mediated killing than wild-type N. meningitidis. Recovery of NmMC58 or the capsule-null mutant (NmMC58ΔsiaD) after 4 hr co-culture with N. cinerea 346T (Wt) or a T6SS-deficient mutant (ΔT6SS) at ratio of approximately 100:1, attacker:prey. Data shown are the mean ± SD of three independent experiments: NS, not significant, ***p < 0.0001, **p < 0.001 using unpaired two-tailed Student’s t-test for pairwise comparison (B and C) or one-way ANOVA test for multiple comparison (A).
-
Figure 6—source data 1
Survival of N. meningitidis strains after 4 hr co-incubation with N. cinerea 346T wild-type or the T6SS mutant.
- https://cdn.elifesciences.org/articles/63755/elife-63755-fig6-data1-v1.xlsx
-
Figure 6—source data 2
Survival of N. gonorrhoeae FA1090 strain after 4 hr co-incubation with N. cinerea 346T wild-type or the T6SS mutant.
- https://cdn.elifesciences.org/articles/63755/elife-63755-fig6-data2-v1.xlsx
-
Figure 6—source data 3
Survival of N. meningitidis MC58 or the capsule-null mutant strain after 4 hr co-incubation with N. cinerea 346T wild-type or the T6SS mutant.
- https://cdn.elifesciences.org/articles/63755/elife-63755-fig6-data3-v1.xlsx
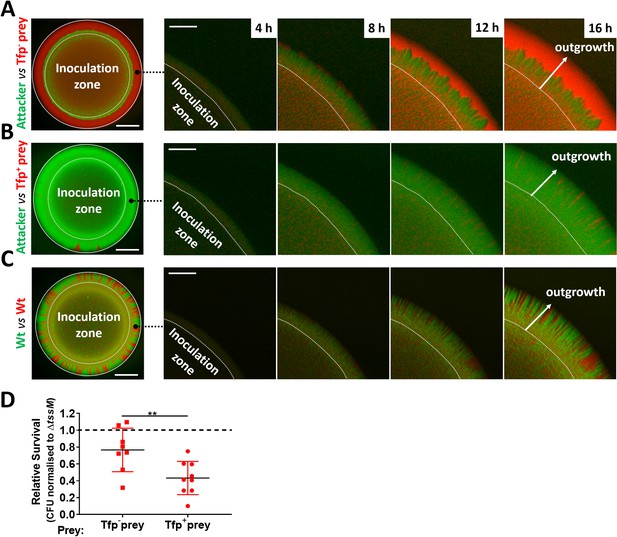
Attacker and prey piliation promotes T6SS killing.
(A) Fluorescence microscopy images taken at specific times after inoculation of mixed (1:1 ratio) bacterial colonies. A T6SS-susceptible, non-piliated prey strain (346TΔnte/i3-5ΔpilE1/2_sfCherry, red) migrates to the expanding edge of the colony over time, segregating from the T6SS+ attacker strain (N. cinerea 346T_gfp, green) and dominating the expanding population. See also Figure 7—video 1, (B) The same susceptible prey strain but expressing pili does not segregate, and after 24 hr is outcompeted by the piliated T6SS+ attacker. See also Figure 7—video 2. (C) The non-T6SS-susceptible, piliated prey strain (346T_sfCherry, red) and piliated attacker strain (346T_sfGfp, green) do not segregate, but due to immunity against T6SS attack, no dominance is observed. Images of colonies are representative of three independent experiments. See also Figure 7—video 3. Scale bar, 500 µm. Expanding colony edge images are stills at indicated times from time-lapse imaging performed on one occasion. Scale bar 100 µm. Flow cytometry data are presented in Figure 7—figure supplement 2. (D) The influence of piliation on T6SS killing. Recovery of non-piliated and piliated prey strains after 24 hr co-culture with N. cinerea 346T (Wt) and a tssM-deficient mutant (ΔtssM) at ratio of approx. 10:1, attacker:prey. Relative survival is defined as the fold change in recovery of prey following incubation with wild-type attacker N. cinerea compared to N. cinerea ΔtssM. Data shown are the mean ± SD of three independent experiments: **p < 0.01 using unpaired two-tailed Student’s t-test for pairwise comparison. See also Figure 7—figure supplement 3.
-
Figure 7—source data 1
Survival of non-piliated and piliated prey strains after 24 hr co-culture with N. cinerea 346T and a tssM-deficient mutant.
- https://cdn.elifesciences.org/articles/63755/elife-63755-fig7-data1-v1.xlsx
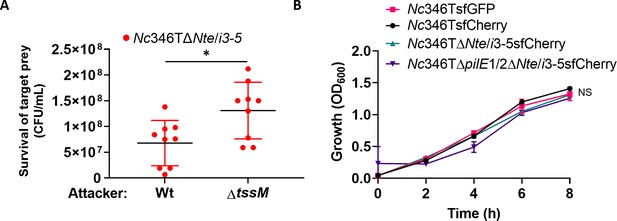
N. cinerea 346TΔnte/i3-5 prey is susceptible to T6SS-killing by wild-type N. cinerea 346T and fluorophore expressing mutants have comparable growth.
(A) Competition assay measuring the recovery of the indicated N. cinerea 346TΔNte/i3-5 mutant strain after 4 hr of co-incubation wild-type N. cinerea 346T (Wt) or a tssM-deficient mutant (ΔtssM) at ratio of 10:1 (attacker: prey). Data shown are the mean ± SD of three independent experiments performed in triplicate: *p<0.05 using unpaired two-tailed Student’s t-test. (B) N. cinerea 346T WT and mutant strains expressing fluorophores were grown in liquid BHI media for 8 hr at 37°C with 5% CO2. Growth was monitored by measuring the OD600 of cultures. Data are representative of two independent experiments: NS, not significant, using two-way ANOVA test for multiple comparison.
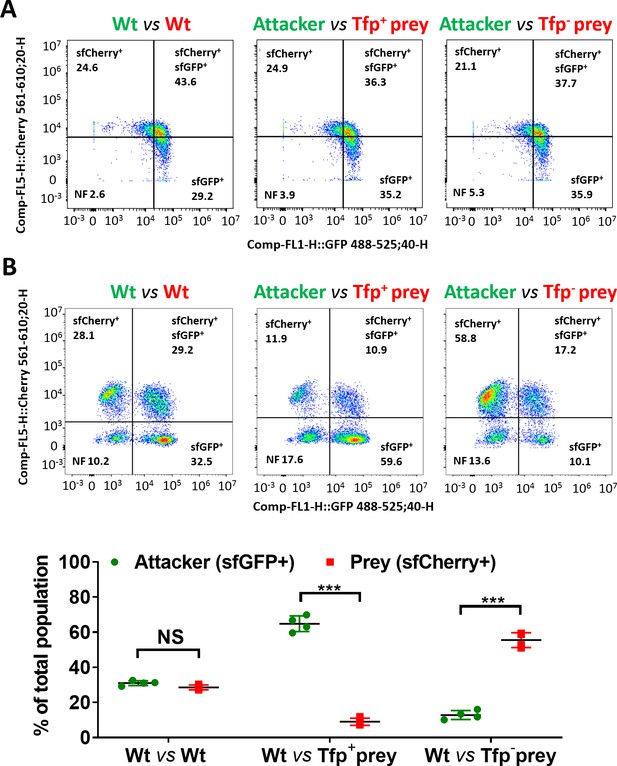
Flow cytometry analysis of relative proportion of attacker and prey strains.
(A) Representative FACS plots corresponding to analysis of fixed input suspensions of indicated attacker and prey strains mixed at a 1:1 ratio. Plots revealed a population of dual labelled (sfCherry+sfGFP+) particles which likely correspond to associated sfGFP+ and sfCherry+ cells, although additional experiments are required to confirm this. Data from two experiments indicated no significant difference in the % of total population between sfGFP+ and sfCherry+ strains. (B) Total bacteria in mixed colonies grown on agar plates for 24 hr (Figure 7A-C) were collected, fixed, and analysed by Flow cytometry. Plots are representative of two independent experiments and show relative proportion of sfCherry+ (prey), sfGFP+ (attacker), non-fluorescent (NF), and dual-labelled populations (sfCherry+sfGFP+) populations. To determine whether the dual labelled population could affect interpretation of the relative overall proportions of attacker (sfGFP+) and prey (sfCherry+) cells, the fluorescence of this population was analysed further. The Median fluorescence Intensity (MFI) for sfGFP and sfCherry of this population was determined. MFI values were normalised relative to the MFI of the sfGFP or sfCherry single populations respectively, to allow a comparison of the red and green fluorescence of this dual labelled population. This revealed no significant difference for all samples except for the Wt attacker vs Tfp- prey, in which we observed a slight increase towards the dominant population, that is, MFIsfCherry>MFIsfGFP of the sfGFP+sfCherry+ population. Therefore, dual fluorescence populations were excluded from analysis and graphs indicate the percentage of total population for sfGFP+ and sfCherry+ populations as mean ± SD of two independent experiments with duplicate samples. When WT:WT are mixed (both strains are T6SS+ and Tfp+) both were detected in approximately equal proportions (red 346T_sfCherry 29 ± 1%, green 346T_gfp 31 ± 1%, p=0.046). In the context of piliated attacker and prey strains, the attacker cells constitute the majority of the population (red prey 346TΔNte/i3-5 9 ± 2%, green attacker 346T_gfp 65 ± 5%, p<0.0001); however, when non-piliated, the prey strain (red) dominates, despite being susceptible to T6SS-mediated attack (red prey 346TΔNte/i3-5ΔpilE1/2_sfCherry 56 ± 4%, green attacker 346T_gfp 13 ± 3%; p<0.0001). NS, not significant, ***p < 0.0001 using unpaired two-tailed Student’s t-test.
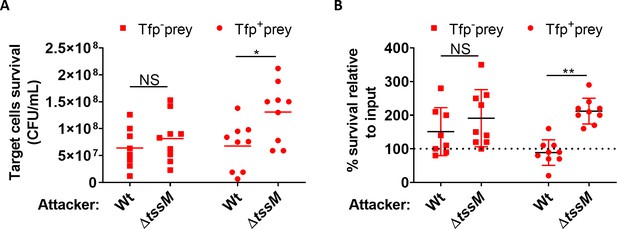
CFU and % survival data for Tfp+/- strains.
(A) Data shown in Figure 7D presented as total recovery in CFU/ml of non-piliated (346TΔnte/i3-5ΔpilE1/2 as Tfp-prey) and piliated prey (346TΔnte/i3-5 as Tfp+prey) strains after 24 hr co-culture with N. cinerea 346T (Wt) and a tssM-deficient mutant (ΔtssM) at ratio of 10:1, attacker:prey. (B) Data shown in Figure 7D presented as percentage survival of prey (Tfp-prey and Tfp+prey) relative to input levels of each prey strain following incubation with wild-type attacker N. cinerea. Data shown are the mean ± SD of three independent experiments: NS, not significant *p<0.05 and **p<0.01 using or two-way ANOVA test for multiple comparison.
Growing edge of colonies with a piliated attacker N. cinerea 346T_gfp, (green) and non-piliated prey 346TΔnte/i3-5ΔpilE1/2_sfCherry (red).
Strains were spotted at a ratio of 1:1. Colonies were imaged every 2 hr between 4 and 16 hr post inoculation. Over time a population of non-piliated prey segregates to the edge, escape T6SS assault and dominates the growing colony.
Growing edge of colonies with a piliated attacker N. cinerea 346T_gfp, (green) and piliated prey 346TΔnte/i3-5_sfCherry (red).
Strains were spotted at a ratio of 1:1. Colonies were imaged every 2 hr between 4 and 16 hr post inoculation. Over time the T6SS expressing strain dominates the colony.
Growing edge of colonies with two wild-type strains.
N. cinerea 346T_gfp (green) and N. cinerea 346T_sfCherry (red). Strains were spotted at a ratio of 1:1. Colonies were imaged between 4 and 16 hr post inoculation. Images obtained every 2 hr. No dominance of either strain is observed.
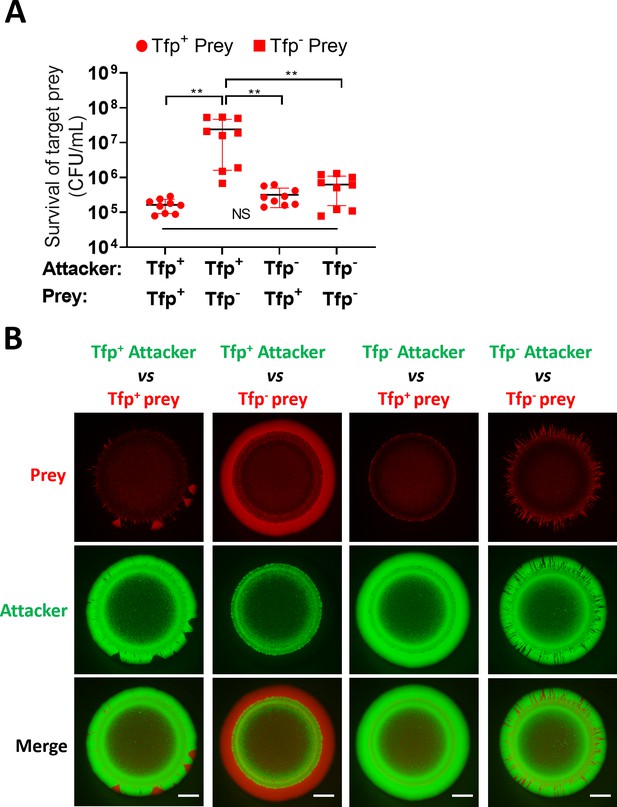
Tfp loss influences prey survival.
(A) Role of Tfp in the attacker and prey population during competition. Recovery of non-piliated (346TΔnte/i3-5ΔpilE1/2_sfCherry) and piliated prey (346TΔnte/i3-5_sfCherry) strains after 24 hr co-culture with piliated N. cinerea 346T (346T_sfGfp) and non-piliated attacker (346TΔpilE1/2_sfGfp) strains at ratio of approx. 10:1, attacker:prey. Data shown are the mean ± SD of three independent experiments: NS, not significant, **p<0.001 using one-way ANOVA test for multiple comparison. See also Figure 8—figure supplement 1. (B) Fluorescence microscopy images taken at 24 hr after inoculation of mixed (1:1 ratio) bacterial colonies. A T6SS-susceptible, piliated prey strain (Tfp+ prey, 346TΔnte/i3-5_sfCherry, red) does not segregate, and after 24 hr is outcompeted by the piliated T6SS+ attacker (Tfp+ attacker, 346T_sfGfp, green). The same prey, but non-piliated (Tfp- prey, 346TΔnte/i3-5ΔpilE1/2_sfCherry, red), segregates from the piliated T6SS+attacker strain (Tfp+ attacker, 346T_sfGfp, green) and dominates the edge of the colony. When the prey is piliated (Tfp+ prey, 346TΔnte/i3-5_sfCherry, red) and attacker is non-piliated (Tfp- attacker, 346TΔpilE1/2_sfGfp, green), the non-piliated attacker population segregates to the edge and dominates the outer region of the colony. In a mixed population with a non-piliated prey (Tfp- prey, 346TΔnte/i3-5ΔpilE1/2_sfCherry, red) and a non-piliated attacker (Tfp- attacker, 346TΔpilE1/2_sfGfp, green), the prey does not segregate from attacker and attacker and prey form expanding sectors in the region of outgrowth at the colony edge. Images of colonies are representative of three independent experiments. Scale bar, 500 µm. See also Figure 8—figure supplement 2.
-
Figure 8—source data 1
Survival of non-piliated and piliated prey strains after 24 hr co-culture with piliated N. cinerea 346T and non-piliated attacker strains.
- https://cdn.elifesciences.org/articles/63755/elife-63755-fig8-data1-v1.xlsx
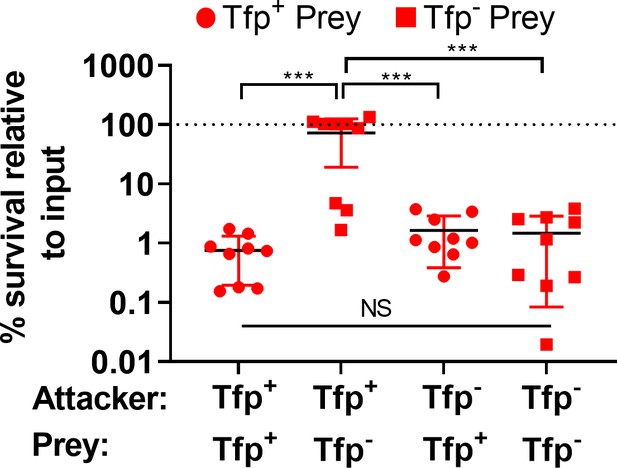
% survival data for prey strains +/-Tfp.
Data shown in Figure 8A presented as percentage of prey (Tfp-prey and Tfp+prey) relative to input, following incubation with attacker N. cinerea (Tfp- attacker and Tfp+ attacker). Data shown are the mean ± SD of three independent experiments: NS, not significant ***p < 0.0001 using one-way ANOVA test for multiple comparison.
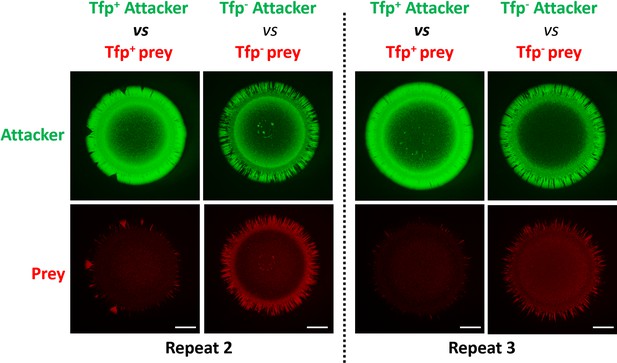
Fluorescence microscopy images of colonies of piliated (Tfp+/Tfp+) or non-piliated (Tfp-/Tfp-) attacker and prey strains.
Additional examples of fluorescence microscopy images taken at 24 hr after inoculation of mixed (1:1 ratio) bacterial colonies. Gfp-expressing attacker, with Tfp (Tfp+, 346T_sfGfp, green) or without Tfp (Tfp-, 346T_sfGfpΔpilE1/2, green) and sfCherry-expressing T6SS susceptible prey strains with Tfp (346TΔnte/i3-5_sfCherry, red) or without Tfp (346TΔnte/i3-5ΔpilE1/2_sfCherry, red) were mixed as indicated.

The deletion of all four putative endonuclease effectors impairs T6SS function and Hcp secretion.
(A) Recovery of wild-type N. meningitidis 8013 after 4 h co-incubation with N. cinerea 346T wild-type (Wt) and the T6SS mutant (ΔT6SS) or strains lacking specific effectors at a 100:1 attacker:prey ratio. Data shown are the mean ± SD of three independent experiments: NS, not significant, ***p < 0.0001, one-way ANOVA test for multiple comparison. (B) Western blot detection of Hcp in whole cell lysates (W) and supernatants (S) from bacteria grown in liquid BHI media for 4-5 h. WT (wild-type N. cinerea 346T), ΔTssB (strain with deletion of tssB), Δ4Nte (strain lacking nte/nti 3,4,5,6).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Goat polyclonal anti-rabbit IgG–HRP | Santa Cruz | sc-2004 | Target rabbit IgG antibodies WB (1:5000) |
| Antibody | Rabbit polyclonal anti-RecA | Abcam | ab63797 | Target bacterial RecA protein WB (1:5000) |
| Antibody | Rabbit polyclonal anti-Hcp sera | This paper | Antibody raised to target full-length N. cinerea 346T Hcp protein WB (1:10000) | |
| Chemical compound, drug | SYTOX Blue | Thermo Fisher Scientific | S34857 | SYTOX Blue is a high-affinity nucleic acid stain that does not penetrate uncompromised cell membranes |
| Software, algorithm | FiJi | Schindelin et al., 2012 DOI:10.1038/nmeth.2019 | https://fiji.sc RRID:SCR_002285 | |
| Software, algorithm | Graphpad Prism7 | San Diego, CA | https://www.graphpad.com/ RRID:SCR_002798 | |
| Software, algorithm | FlowJo v10 | Becton Dickinson Company | https://www.flowjo.com/ RRID:SCR_008520 | |
| Strain, strain background (Neisseria cinerea) | CCUG346T (346T) | Bennett et al., 2012 DOI:10.1099/mic.0.056077-0 | wild-type N. cinerea | |
| Strain, strain background (Neisseria cinerea) | CCUG27178A (27178A) | Bennett et al., 2012 DOI:10.1099/mic.0.056077-0 | wild-type N. cinerea | |
| Strain, strain background (Neisseria cinerea) | 346T_sfGFP | Wörmann et al., 2016 DOI: 10.1099/mic.0.000248 | 346T with chromosomally integrated sfGfp; EryR | |
| Strain, strain background (Neisseria cinerea) | 346T_sfGFPΔpilE1/2 | Wörmann et al., 2016 DOI: 10.1099/mic.0.000248 | 346T with pilE1 and pilE2 deleted by insertion mutagenesis, and chromosomally integrated sfGfp; EryR and KanR | |
| Strain, strain background (Neisseria cinerea) | 346T_sfCherry | This paper | 346T with chromosomally integrated sfCherry; EryR | |
| Strain, strain background (Neisseria cinerea) | 27178A_sfCherry | This paper | 27178 with chromosomally integrated sfCherry; SpecR | |
| Strain, strain background (Neisseria cinerea) | 346TΔT6SS | This paper | 346T with insertion-deletion of tssC – vgrG region; EryR | |
| Strain, strain background (Neisseria cinerea) | 346TΔtssB | This paper | 346T with insertion-deletion of tssB; EryR | |
| Strain, strain background (Neisseria cinerea) | 346TΔtssB::tssBsfGFP | This paper | 346T with insertion-deletion of native tssB and ectopic chromosomal insertion of tssB-sfGFP fusion; SpecR EryR | |
| Strain, strain background (Neisseria cinerea) | 346TΔtssM | This paper | 346T with insertion-deletion of tssM; TetR | |
| Strain, strain background (Neisseria cinerea) | 346TΔtssBΔtssM::tssB-sfGFP | This paper | 346T with insertion-deletion of native tssB and tssM and ectopic chromosomal insertion of tssB-sfGFP fusion; SpecR EryR TetR | |
| Strain, strain background (Neisseria cinerea) | 346TΔnte3Δnte4Δnte5 | This paper | immunity genes; EryR | deletion mutagenesis, nte3-nte5 locus deletion including respective immunity genes; EryR |
| Strain, strain background (Neisseria cinerea) | 346TΔnte/i3-5_sfCherry | This paper | 346T with insertion-deletion of nte/i3-5 region and ectopic chromosomal insertion of sfCherry; SpecR EryR | |
| Strain, strain background (Neisseria cinerea) | 346TΔnte6 | This paper | deletion mutagenesis, nte6 deficient; SpecR | |
| Strain, strain background (Neisseria cinerea) | 346TΔnte3Δnte4Δnte5Δnte6 | This paper | deletion mutagenesis, nte3-nte5 locus deletion including respective immunity genes plus nte6 deletion; EryR SpecR | |
| Strain, strain background (Neisseria cinerea) | 346TΔnte/i3-5ΔpilE1/2_sfCherry | This paper | 346T with insertion-deletion of nte/i3-5 region; ectopic chromosomal insertion of sfCherry; insertion-deletion of pilE1 and pilE2; kanR,SpecR EryR | |
| Strain, strain background (Neisseria meningitidis) | 8013 | Rusniok et al., 2009 DOI: 10.1186/gb-2009-10-10-r110 | N. meningitidis wild-type | |
| Strain, strain background (Neisseria meningitidis) | MC58 | Tettelin et al., 2000 DOI: 10.1126/science.287.5459.1809. | N. meningitidis wild-type | |
| Strain, strain background (Neisseria meningitidis) | S3 | Uria et al., 2008 DOI: 10.1084/jem.20072577 | N. meningitidis wild-type | |
| Strain, strain background (Neisseria meningitidis) | MC58ΔsiaD | Virji et al., 1995 DOI:10.1111/j.1365-2958.1995.mmi_18040741.x | deletion mutagenesis, NEIS0051; KanR | |
| Strain, strain background (Neisseria gonorrhoeae) | FA1090 pGCC4 | Mehr and Seifert, 1997 DOI:10.1046/j.1365-2958.1997.2971660.x | FA1090 with chromosomally integrated plasmid pGCC4; EryR | |
| Strain, strain background (Escherichia coli) | Dh5α | Lab collection | DH5α is an E. coli strain used for general cloning applications. | |
| Strain, strain background (Escherichia coli) | Dh5α pNCC1-Spec | This paper | Dh5α with pNCC1SpecR plasmid | |
| Strain, strain background (Escherichia coli) | Dh5α pNCC1-Spec-sfGFP | This paper | Dh5α with pNCC1-Spec with sfGFP insert; | |
| Strain, strain background (Escherichia coli) | Dh5α pNCC101-Spec-sfCherry | Lab collection | DH5α with plasmid pNCC101+sfCherry insert. SpecR | |
| Strain, strain background (Escherichia coli) | Dh5α pUC19 | Lab collection | pUC19 vector RRID:Addgene_50005 | E. coli DH5α strain harbouring pUC19 for general cloning applications. |
| Strain, strain background (Escherichia coli) | Dh5α pUC19::ΔtssB | This paper | DH5α with pUC19::ΔtssB deletion construct; CarbR EryR | |
| Strain, strain background (Escherichia coli) | Dh5α pUC19::ΔtssM | This paper | DH5α with pUC19::ΔtssM deletion construct; CarbR TetR | |
| Strain, strain background (Escherichia coli) | Dh5α pUC19::ΔT6SS | This paper | DH5α with pUC19::ΔtssC-vgrG locus deletion construct; CarbR EryR | |
| Strain, strain background (Escherichia coli) | Dh5αpUC19:: Δnte3Δnte4Δnte5 | This paper | DH5α with pUC19::Δnte3Δnte4Δnte5 region including respective immunity genes deletion construct; CarbR EryR | |
| Strain, strain background (Escherichia coli) | Dh5α pUC19:: Δnte6 | This paper | nte6 deletion construct; CarbR SpecR | |
| Strain, strain background (Escherichia coli) | B834 pET28a | Lab collection | pET28a Novagen Cat. No. 69864–3 | B834 with pET28a IPTG-inducible expression vector, KanR |
| Strain, strain background (Escherichia coli) | Dh5α pET28a-His-3C-Hcp | This paper | Dh5α with pET28a vector for IPTG inducible expression of Nc 346T Hcp with N-terminal cleavable HIS tag. KanR | |
| Strain, strain background (Escherichia coli) | B834 pET28a-His-3C-Hcp | This paper | B834 expression strain, with pET28a vector for IPTG inducible expression of Nc 346T Hcp with N-terminal cleavable HIS tag. KanR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33 | Lab collection | pBAD33 RRID:Addgene_36267 | Dh5α with pBAD33 vector for Arabinose-inducible expression, CmR |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::(ssPelB)Nte1-His | This paper | Dh5α with pBAD33 encoding Nte1 with N-terminal PelB leader peptide and C-terminal his-tag under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33:: (ssPelB)Nte1+Nti1 | This paper | Dh5α with pBAD33 encoding Nte1 with N-terminal PelB leader peptide and C-terminal his-tag plus Nti, under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte1-His | This paper | Dh5α with pBAD33 encoding Nte1 with N-terminal his-tag under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte2 | This paper | Dh5α with pBAD33 encoding Nte2 under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte2+Nti2 | This paper | Dh5α with pBAD33 encoding Nte2+Nti2 under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte3 | This paper | Dh5α with pBAD33 encoding Nte3 under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte3+Nti3 | This paper | Dh5α with pBAD33 encoding Nte3+Nti3 under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte4 | This paper | Dh5α with pBAD33 encoding Nte4 under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte4+Nti4 | This paper | Dh5α with pBAD33 encoding Nte4+Nti4 under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte5 | This paper | Dh5α with pBAD33 encoding Nte5 under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte5+Nti5 | This paper | Dh5α with pBAD33 encoding Nte5+Nti5 under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte6R1300S | This paper | Dh5α with pBAD33 encoding Nte6R1300S under arabinose-inducible promoter control; CmR | |
| Strain, strain background (Escherichia coli) | Dh5α pBAD33::Nte6+Nti6 | This paper | Dh5α with pBAD33 encoding Nte6+Nit6 under arabinose-inducible promoter control; CmR | |
| Sequence-based reagent | T6SSdel-1 | This paper | 5’-CGAAAAGTG CCACCTGACGTATGACTGAAAAGCAATTAGATATC | Deletion of tssC-vgrG locus |
| Sequence-based reagent | T6SSdel-2 | This paper | 5’-GTTAAATTTAAGGATAAGAAACGTGGCAG | Deletion of tssC-vgrG locus |
| Sequence-based reagent | T6SSdel-3 | This paper | 5’-TTTCTTATCC TTAAATTTAACGATCACTCATCATG | Deletion of tssC-vgrG locus |
| Sequence-based reagent | T6SSdel-4 | This paper | 5’-ACTCAAACATTTACTTATTAAATAATTTATAGCTATTGAAAAG | Deletion of tssC-vgrG locus |
| Sequence-based reagent | T6SSdel-5 | This paper | 5’-TTAATAAGTAAATGTTTGAGTTGCAGAACTTTAC | Deletion of tssC-vgrG locus |
| Sequence-based reagent | T6SSdel-6 | This paper | 5’-GATAATAATGGTTTCTTAGACGTGCCGTTCCAATAGGCCATAG | Deletion of tssC-vgrG locus |
| Sequence-based reagent | T6SSdel-conf-F | This paper | 5’-CCTAAAGCG GCTTCCAAAGACG | Confirmation of tssC-vgrG locus deletion |
| Sequence-based reagent | T6SSdel-conf-R | This paper | 5’-CCATGCCGG TAAAGGTCAGT | Confirmation of tssC-vgrG locus deletion |
| Sequence-based reagent | TssBdel-1 | This paper | 5’-GATCCTCTA GAGTCGACCTGCAGGCATGCACTTACCCTGATCCACAAAGCC | Deletion of tssB |
| Sequence-based reagent | TssBdel-2 | This paper | 5’-ATTCAATGACCTTTAAATGATAAAAGTTGT | Deletion of tssB |
| Sequence-based reagent | TssBdel-3 | This paper | 5’-ACAACTTTTATCATTTAAAGGTCATTGAATATGAACGAGAAAAATATAAAACACAGTC | Deletion of tssB |
| Sequence-based reagent | TssBdel-4 | This paper | 5’-TTACTTATTA AATAATTTATAGCTATTGAAAAGAGATAAGAATTG | Deletion of tssB |
| Sequence-based reagent | TssBdel-5 | This paper | 5’-TATAAATTATTTAATAAGTAAGCTTCCAAAGACGAGCAGTAA | Deletion of tssB |
| Sequence-based reagent | TssBdel-6 | This paper | 5’-CAGGAAACA GCTATGACCATGATTACGCCTAAGTTGCGGGCAACTTCTT | Deletion of tssB |
| Sequence-based reagent | TssBdel-conf-F | This paper | 5’-ATAGAAACCTACTTTTTCGGAAAGC | Confirmation of tssB deletion |
| Sequence-based reagent | TssBdel-conf-R | This paper | 5’-TTACTTATTA AATAATTTATAGCTATTGAAAAGAGATAAGAATTG | Confirmation of tssB deletion |
| Sequence-based reagent | TssMdel-1 | This paper | 5’-GATCCTCTA GAGTCGACCTGCAGGCATGCAACCCTGTCTTGGCTAGAGTC | Deletion of tssM |
| Sequence-based reagent | TssMdel-2 | This paper | 5’-ATTTGTTTTT CCGTATCAATCCAATTTCA | Deletion of tssM |
| Sequence-based reagent | TssMdel-3 | This paper | 5’-ATTGGATTGATACGGAAAAACAAATATGAAAATTATTAATATTGGAGTTTTAGCTCATGTT | Deletion of tssM |
| Sequence-based reagent | TssMdel-4 | This paper | 5’-CTAAGTTATTTTATTGAACATATATCGTACTTTATCTATCCG | Deletion of tssM |
| Sequence-based reagent | TssMdel-5 | This paper | 5’-AAGTACGATATATGTTCAATAAAATAACTTAGAATAAATTAAGGAATTTTCAGTGCATTTGAAG | Deletion of tssM |
| Sequence-based reagent | TssMdel-6 | This paper | 5’-CAGGAAACA GCTATGACCATGATTACGCCGGCAATATCTAGAACGGATTTATCG | Deletion of tssM |
| Sequence-based reagent | TssMdel-Conf-F | This paper | 5’-AGGACTTCC AAGATAGAAGTACGG | Confirmation of tssM deletion |
| Sequence-based reagent | TssMdel-Conf-R | This paper | 5’-AAAGCCCCT TGTACGATAGC | Confirmation of tssM deletion |
| Sequence-based reagent | Nte345del-1 | This paper | 5’-GATCCTCTA GAGTCGACCTGCAGGCATGCAGACCTTCATGCTGACTAGTGAT | Deletion of Nte3-Nte5 locus |
| Sequence-based reagent | Nte345del-2 | This paper | 5’-GAAGTGTTG GATGAACTTTTTCTATG | Deletion of Nte3-Nte5 locus |
| Sequence-based reagent | Nte345del-3 | This paper | 5’-CATAGAAAAAGTTCATCCAACACTTCTTAAATTTAACGATCACTCATCATGT | Deletion of Nte3-Nte5 locus |
| Sequence-based reagent | Nte345del-4 | This paper | 5’-TTACTTATTA AATAATTTATAGCTATTG | Deletion of Nte3-Nte5 locus |
| Sequence-based reagent | Nte345del-5 | This paper | 5’-CAATAGCTAT AAATTATTTAATAAGTAAAATAAGAAACTGTAAACACAGTGTG | Deletion of Nte3-Nte5 locus |
| Sequence-based reagent | Nte345del-6 | This paper | 5’-CAGGAAACA GCTATGACCATGATTACGCCAGTTTAACTGTTCGGAAAGGGTGT | Deletion of Nte3-Nte5 locus |
| Sequence-based reagent | Nte345del-conf-F | This paper | 5’-GTTTTCGTTGGTGAGGACGG | Confirmation of Nte3-Nte5 locus deletion |
| Sequence-based reagent | Nte345del-conf-R | This paper | 5’-CTACTTATAATCCAAATATTTTATTGAACAGAGAAC | Confirmation of Nte3-Nte5 locus deletion |
| Sequence-based reagent | TssBsfGFP1 | This paper | 5’-CATGATTACGAATTCCCGGATTAATTAAAATGTCACGAAACAAATCATCCGG | tssB amplification to fuse with sfGFP and clone into pNCC1-spec |
| Sequence-based reagent | TssBsfGFP2 | This paper | 5’-CTGCTCGTCTTTGGAAGC | tssB amplification to fuse with sfGFP and clone into pNCC1-spec |
| Sequence-based reagent | TssBsfGFP3 | This paper | 5’-GCTTCCAAA GACGAGCAGGCAGCAGCAGGTGGTGGTAGCAAAGGAGAAGAACTTTTCAC | sfGFP amplification and addition of DNA linker to fuse with tssB and clone into pNCC1-spec |
| Sequence-based reagent | TssBsfGFP4 | This paper | 5’-GATCCTCTA GAGTCGACCTGCAGGCATGCTCATTTGTAGAGCTCATCCATGC | sfGFP amplification and addition of DNA linker to fuse with tssB and clone into pNCC1-spec |
| Sequence-based reagent | sfGFP-Prom-F | This paper | 5’-TGACCCGGG TCATTTGTAGAGCTCATCCATGCC | sfGFP amplification from pNCC1-sfGFP to clone into pNCC1-spec |
| Sequence-based reagent | sfGFP-Prom-R | This paper | 5’-TGAAAGCTTTTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCCCAACATGTTACACAATAATGGAGTAATGAACATATGAGCAAAGGAGAAGAACT | sfGFP amplification from pNCC1-sfGFP to clone into pNCC1-spec |
| Sequence-based reagent | pGib-RBS-Nte2-F | This paper | 5’-GATCCTCTA GAGTCGACCTGCAGGCATGCAAAGAAGGAGATATACCATGGCATTCAATAAAATCGCCC | Nte2 amplification and addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nte2-R | This paper | 5’-AAAATCTTCTCTCATCCGCCAAAACAGCCATCATTTTTTCCTATTGTTACATTTATCCT | Nte2 amplification and addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nti2-R | This paper | 5’-AAAATCTTCTCTCATCCGCCAAAACAGCCATTATTCAAATTTCTTTAGCAGTATTTTTCT | Nte2 and Nti2 amplification plus addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nte3-F | This paper | 5’-GATCCTCTA GAGTCGACCTGCAGGCATGCAAAGAAGGAGATATACCATGGCCTCTTTCGGTAAC | Nte3 amplification and addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nte3-R | This paper | 5’-AAAATCTTCTCTCATCCGCCAAAACAGCCATCATTTAATACCTCTTCTTGATAATTCTTT | Nte3 amplification and addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nti3-R | This paper | 5’-AAAATCTTCTCTCATCCGCCAAAACAGCCACTATTCACCCAACAATGTTTCT | Nte3 and Nti3 amplification plus addition of RBS to clone into pBAD33 |
| Sequence-based reagent | PGIB-RBS-NTE4-F | This paper | 5’-GATCCTCTA GAGTCGACCTGCAGGCATGCAAAGAAGGAGATATACCATGGTCGAACACAACCAG | Nte4 amplification and addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nte4-R | This paper | 5’-AAAATCTTCTCTCATCCGCCAAAACAGCCATTAAATTATTGGAAGATTTTTACAACCA | Nte4 amplification and addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nti4-R | This paper | 5’-AAAATCTTCTCTCATCCGCCAAAACAGCCATTACGCTTTTAAATTCCGGTG | Nte4 and Nti4 amplification plus addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nte5-F | This paper | 5’-GATCCTCTA GAGTCGACCTGCAGGCATGCAAAGAAGGAGATATACCATGGGTCGTCTGAAAAGC | Nte5 amplification and addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nte5-R | This paper | 5’-AAAATCTTCTCTCATCCGCCAAAACAGCCACTAATCTAATCGTTTGGGCG | Nte5 amplification and addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nti5-R | This paper | 5’-AAAATCTTCTCTCATCCGCCAAAACAGCCATTAATCCCAATAACTGTCTAAATTGT | Nte5 and Nti5 amplification plus addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nte6-F | This paper | 5’-GATCCTCTA GAGTCGACCTGCAGGCATGCAAAGAAGGAGATATACCATGGCCTCTTTCGGTAAC | Nte6 amplification and addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nte6-R | This paper | 5’-AAAATCTTCTCTCATCCGCCAAAACAGCCACTATTATCTAGGAACAATCTGATTAATTATTCC | Nte6 amplification and addition of RBS to clone into pBAD33 |
| Sequence-based reagent | pGib-RBS-Nti6-R | This paper | 5’-AAAATCTTCTCTCATCCGCCAAAACAGCCATTAAATTTCCTCTAGTTTTTCTTTCATC | Nte6 and Nti6 amplification plus addition of RBS to clone into pBAD33 |
| Sequence-based reagent | CE043-F | This paper | 5’-GGCCGGTCT AGAAAGAAGGAGATATACCATGAAATACCTGCTGCCGACCGCTGCTGCTGGTCTGCTGCTCCTCGC | Addition of 5′ PelB leader peptide and 3′ 6xHIS-tag to PLA2 domain to clone into pBAD33 |
| Sequence-based reagent | CE044-F | This paper | 5’-GGTCTGCTG CTCCTCGCTGCCCAGCCGGCGATGGCCATGGGGGGAAGTAATTTTTATGCGTTTGCA | PLA2 domain amplification and addition of 5′ PelB leader peptide |
| Sequence-based reagent | CE046-R | This paper | 5’-CCGGCCGCA TGCCTAGTGATGGTGATGGTGATGCCTATGATTTTTAGAC | Addition of 3′ 6xHIS-tag to PLA2 domain with or without 5′ PelB leader peptide to clone into pBAD33 |
| Sequence-based reagent | CE047-R | This paper | 5’-GATGCCTAT GATTTTTAGACGTTTTTTTAATTGTTTTATCG | PLA2 domain amplification with or without addition of 5′ PelB leader peptide |
| Sequence-based reagent | CE048-R | This paper | 5’-CCGGCCGC ATGCCTAGTGATGGTGATGGTGATGATTAAGTTTGGATAGTTTGAAAATTTTTTTAAGCTTATATATAAG | PLA2 domain amplification with or without a 5′ PelB leader peptide and amplification of Nti1 adding a 3′ 6xHIS-tag to clone into pBAD33 |
| Sequence-based reagent | CE083-F | This paper | 5’-GGCCGGTC TAGAAAGAAGGAGATATACCATGGGGGGAAGTAATTTTTATGCGTTTGCA | PLA2 domain amplification and addition of 3′ 6xHIS-tag to clone into pBAD33 |
| Sequence-based reagent | MW312 | This paper | 5’- TATAAGGAG GAACATATGGAATACATGTTATAATAACTATAAC | Spectinomycin cassette amplification from pDG1728 to clone into pNCC1 |
| Sequence-based reagent | MW313 | This paper | 5’- GTATTCCATATGTTCCTCCTTATAAAATTAGTATAATTATAG | pNCC1 plasmid backbone amplification |
| Sequence-based reagent | MW314 | This paper | 5’- GCATCCCTTAACGACGTCAATTGAAAAAAGTGTTTCCACC | Spectinomycin cassette amplification from pDG1728 to clone into pNCC1 |
| Sequence-based reagent | MW315 | This paper | 5’-TCAATTGACGTCGTTAAGGGATGCATAAACTGCATCCCTTAAC | pNCC1 plasmid backbone amplification |
Additional files
-
Supplementary file 1
Table of Putative T6SS core components in N. cinerea 346T.
- https://cdn.elifesciences.org/articles/63755/elife-63755-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63755/elife-63755-transrepform-v1.docx





