A live attenuated-vaccine model confers cross-protective immunity against different species of the Leptospira genus
Figures
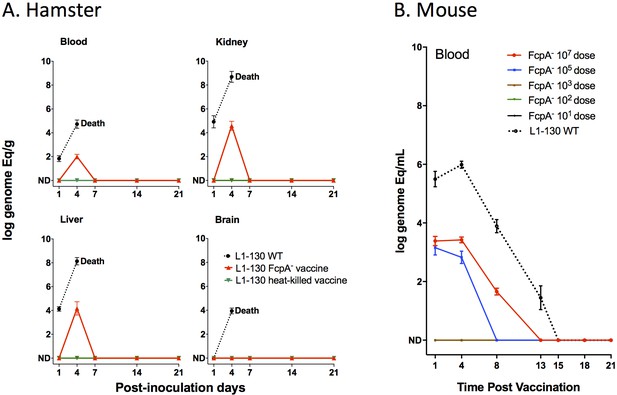
Dissemination of L1-130 fcpA- mutant in animal tissues.
(A) Kinetics of infection of L1-130 WT, L1-130 fcpA- vaccine, and L1-130 heat-killed vaccine in blood, kidney, liver, and brain of hamsters after inoculation with 107 bacteria. All animals infected with WT strain died between 5 and 6 days post-infection; (B) Kinetics of infection of L1-130 WT (107 leptospires) and L1-130 fcpA- attenuated-vaccine (dose range from 107 to 101 leptospires) in blood of mouse. Results are expressed by logarithmic genome equivalent per gram or milliliter of tissues with mean and standard deviation. All doses were inoculated by subcutaneous route in both models.
-
Figure 1—source data 1
Raw data for dissemination experiments in hamster and mouse.
- https://cdn.elifesciences.org/articles/64166/elife-64166-fig1-data1-v1.xls
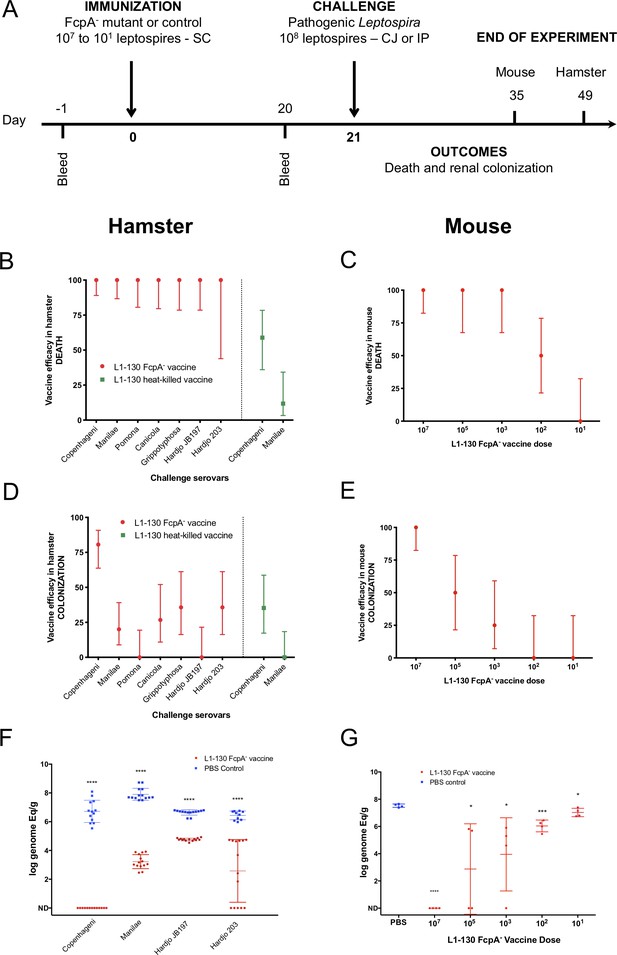
Efficacy of L1-130 fcpA- attenuated-vaccine model.
Animals were vaccinated with a dose of 107 leptospires (hamsters) or a range of doses from 107 to 101 leptospires (mice) by subcutaneous (SC) route. Animals were bled the day before immunization (day −1) and day 20 post-immunization (A). Hamsters were challenged by conjunctival route with either the homologous strain or different heterologous strains. Mice were challenged by intraperitoneal route with the heterologous serovar Manilae of L. interrogans. By combining all vaccine experiments performed, efficacy of the vaccine against death and colonization was evaluated for hamsters (B and D) and mice (C and E) and represented by percentage and 95% CI based on frequency of outcomes compared to PBS-immunized animals. Hamster experiment are showing the results after vaccination with the fcpA- attenuated-vaccine (red) and heat-killed vaccine (blue). Bacterial load in the kidney was measured by qPCR in hamsters (F) and mice (G) and compared between PBS-immunized animals (blue) and animals immunized with fcpA- attenuated-mutant (red). Results are expressed in logarithmic genome equivalents per gram of renal tissue with mean and standard deviation. Asterisk symbols represent statistical significance calculated by t-test: *p<0.01, ***p<0.0001. See also Supplementary files 2 and 3.
-
Figure 2—source data 1
Raw data for qPCR experiments in hamster and mouse.
- https://cdn.elifesciences.org/articles/64166/elife-64166-fig2-data1-v1.xls
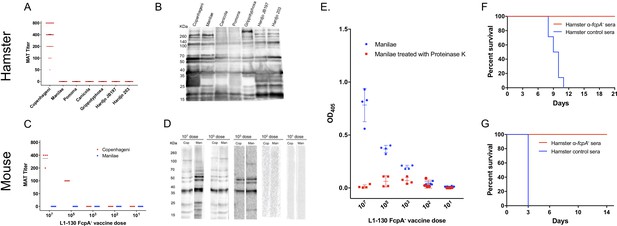
Immunogenicity and correlates of immunity for L1-130 fcpA- attenuated-vaccine model.
Individual sera of hamsters and mice were obtained after 20 days post-vaccination by a subcutaneous (SC) dose of 107 leptospires (hamsters) or a range of doses from 107 to 101 leptospires (mice) of the attenuated-vaccine. Microscopic agglutination test (MAT) (A and C) and western blot (B and D) were performed adopting as antigen all the strains used for challenged in both hamster and mice, respectively. Mice sera was additionally tested using an ELISA assay (E) adopting whole-cell extract of serovar Manilae with (red) and without (blue) Proteinase K treatment as antigen. Furthermore, a pool of hamster immune-sera vaccinated with a dose of 107 leptospires of fcpA- attenuated-vaccine was used for passive transfer experiments. 2 mL or 0.5 mL of sera was passively transfer to naïve hamsters (F) or mice (G), respectively, followed by challenge with a dose of 108 leptospires of heterologous serovar Manilae by conjunctival (CJ) or intraperitoneal (IP) route, respectively. Results are expressed in a survival curve of animals passively transferred with fcpA- anti-sera (red) and control hamster sera (blue).
-
Figure 3—source data 1
Raw data for microscopic agglutination test (MAT) and passive transfer experiments in hamster and mouse, and ELISA data in mouse.
- https://cdn.elifesciences.org/articles/64166/elife-64166-fig3-data1-v1.xls
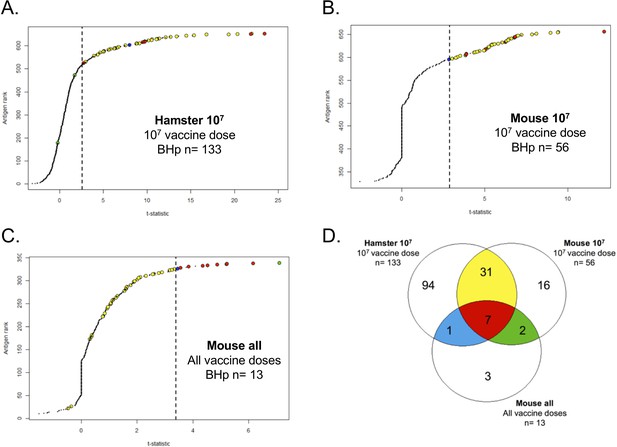
Proteome array analysis of immune-sera against L1-130 fcpA- attenuated-vaccine.
Using statistical modeling we calculated the t-statistics value for each individual antigen used in the proteome array (660 for hamster and 330 for mice) based on three groups: the contrast between vaccinated and unvaccinated hamsters (A) or mice (B) using a vaccine dose of 107 leptospires; the dose–response relationship for each antigen on mice (C) vaccinated with a range of doses from 107 to 101 leptospires of the attenuated-vaccine. Results are ranked based on individual t-statistics values for each antigen, and the dashed line represents the selection point for the antigens based on Bhp-test. The Venn-diagram (D) shows the relationship of all the 154 antigens identified in the three groups. The subgroups of antigens selected for further characterization are highlighted in color. See Figure 5.
-
Figure 4—source data 1
Raw data of proteome array experiments in hamster and mouse.
- https://cdn.elifesciences.org/articles/64166/elife-64166-fig4-data1-v1.xls
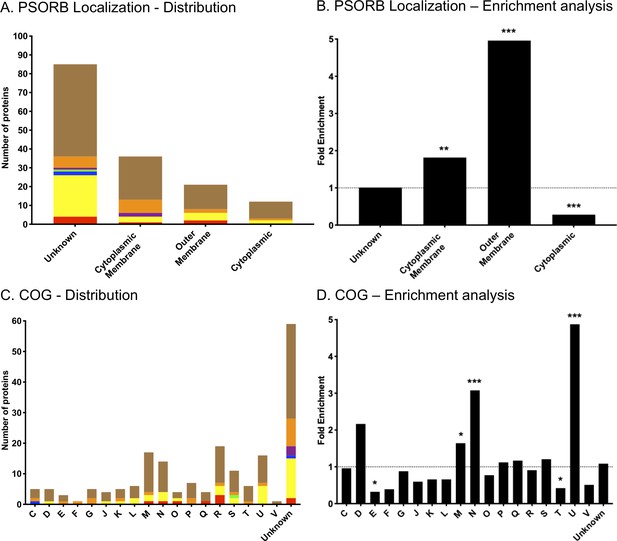
In-silico analysis of protein targets.
Using PSORB information and Genoscope database, we classified the 154 protein targets identified in this study by their putative localization in the cell (A) and their clusters of orthologous group (COG) classification (C). The groups of proteins are classified as follows: proteins identified in all three groups analyzed (7, red); proteins identified in both hamster and mouse vaccinated with a dose of 107 leptospires of the attenuated-vaccine (31, yellow); proteins identified between the group of mice immunized with different doses and the group of mice immunized with a dose of 107 leptospires (2, green); proteins identified between the group of mice immunized with different doses and the group of hamsters immunized with a dose of 107 leptospires (1, blue); proteins identified only in the group of mice immunized with different doses (3, purple); proteins identified only in the group of mice immunized with a dose of 107 leptospires (16, orange); and proteins identified only in the group of hamsters immunized with a dose of 107 leptospires (94, brown). We also performed enrichment analysis of the reactive antigens compared to the whole proteome of Leptospira, based on the PSORB localization (B) and COG (D). Statistical results are represented by *(p<0.05), **(p<0.001), and ***(p<0.0001). See Supplementary file 4.
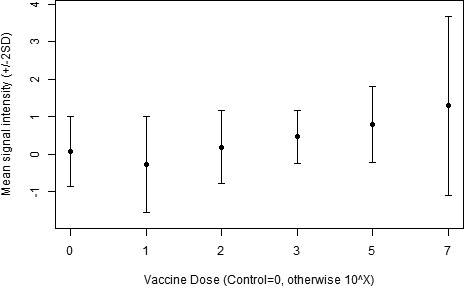
Mouse dose–response relationship.
Analysis showing an association between the different doses of the attenuated L1-130 fcpA- attenuated-vaccine in mice and the mean signal response intensity against all different proteins.
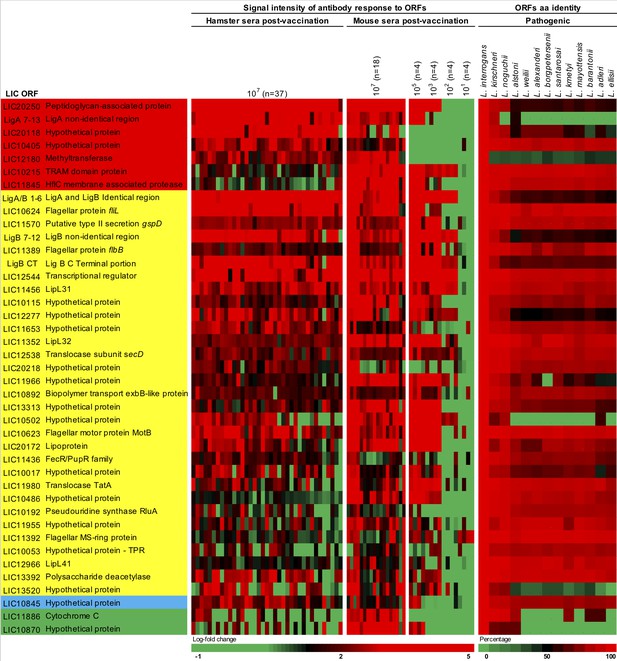
Heat-map of 41 seroreactive proteins recognized by hamsters and mice immunized with attenuated L1-130 fcpA- attenuated-vaccine.
Proteins were selected based on the groups depicted on Figure 4 and Supplementary file 4: present in all three groups of analysis (red), present in both hamster and mice immunized with 107 leptospires (yellow), present in both hamsters immunized with 107 leptospires and mice immunized with a dose range (blue), and present in both mice immunized with 107 leptospires and mice immunized with a dose range (green). The proteins are identified by their L. interrogans serovar Copenhageni ORF number and the heat-map shows the signal intensity of antibody response (based on log-fold change) in all animals vaccinated with the fcpA- mutant used for this analysis (37 hamsters and 34 mice). Right panel shows amino acid sequence identity of respective ORFs among a representative of all pathogenic Leptospira species.
-
Figure 5—source data 1
Raw data of amino acid identity of 41 selected protein targets among different Leptospira species.
- https://cdn.elifesciences.org/articles/64166/elife-64166-fig5-data1-v1.xls
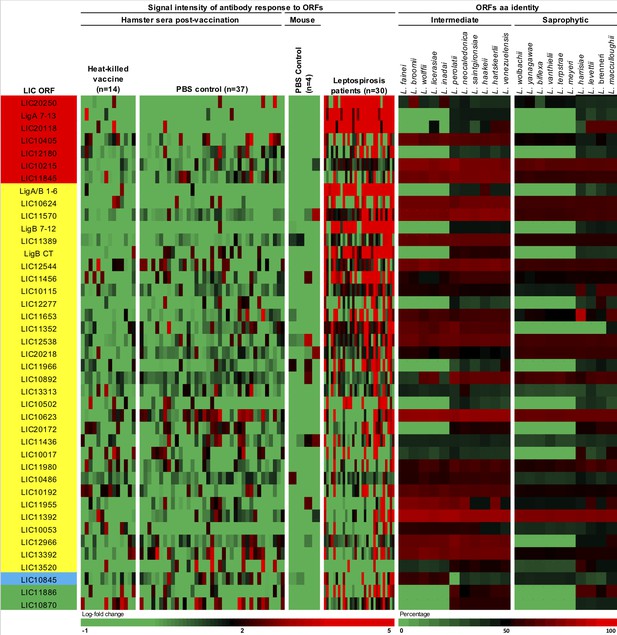
Complementary heat-map of 41 seroreactive proteins recognized by hamsters and mice immunized with attenuated L1-130 fcpA- attenuated-vaccine.
Proteins were selected based on the groups depicted on Figure 4 and Supplementary file 4: present in all three groups of analysis (red), present in both hamster and mice immunized with 107 leptospires (yellow), present in both hamsters immunized with 107 leptospires and mice immunized with a dose range (blue), and present in both mice immunized with 107 leptospires and mice immunized with a dose range (green). The proteins are identified by their L. interrogans serovar Copenhageni ORF number and the heat-map shows the signal intensity of antibody response (based on log-fold change) in all control animals used for this analysis (14 hamsters vaccinated with heat-killed vaccine, 37 PBS control hamsters, and 4 PBS control mice). The heat-map also shows the result for 30 leptospirosis patients. Right panel shows amino acid sequence identity of respective ORFs among a representative of all intermediate and saprophytic Leptospira species.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Leptospira interrogans) | Fiocruz L1-130 fcpA- mutant | Wunder et al., 2016a doi:10.1111/mmi.13403 | L. interrogans serovar Copenhageni strain Fiocruz L1-130 fcpA- | Fiocruz L1-130 fcpA- mutant |
| Antibody | Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson Immuno Research | Cat# 115-035-166 RRID:AB_2338511 | ELISA (1:50,000) WB (1:100,000) |
| Antibody | Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson Immuno Research | Cat# 107-035-142 RRID:AB_2337454 | WB (1:100,000) |
| Antibody | Hamster α-fcpA- sera (Hamster polyclonal) | This paper | See Material and methods WB (1:100) | |
| Antibody | Hamster control sera (Hamster polyclonal) | This paper | See Material and methods WB (1:100) | |
| Chemical compound, drug | SureBlue Reserve TMB 1-Component Microwell Peroxidase Substrate | SeraCare | Cat. #: 5120–0081 | |
| Chemical compound, drug | Sulfuric Acid (H2SO4) | Sigma-Aldrich | Cat. #: 258105 | |
| Chemical compound, drug | Proteinase K | Thermo Fisher | Cat. #: EO0491 | |
| Chemical compound, drug | Platinum Quantitative PCR Supermix-UDG | Thermo Fisher | Cat. #: 11730017 | |
| Software, algorithm | GraphPad Prism software | GraphPad Software | RRID:SCR_015807 | Version 8.0.0 |
| Software, algorithm | RStudio software | RStudio | RRID:SCR_000432 | Version 1.0.153 |
Additional files
-
Supplementary file 1
Leptospira strains used in this study for challenge after vaccination with L1-130 fcpA- mutant.
- https://cdn.elifesciences.org/articles/64166/elife-64166-supp1-v1.docx
-
Supplementary file 2
Efficacy of the immunization with a dose of 107 leptospires of the attenuated L1-130 fcpA- mutant in hamsters followed by challenge with 108 leptospires with homologous or heterologous strains by conjunctival route.
- https://cdn.elifesciences.org/articles/64166/elife-64166-supp2-v1.docx
-
Supplementary file 3
Efficacy of the immunization with different doses of the attenuated L1-130 fcpA- mutant in mice followed by challenge with 108 leptospires of heterologous strain by intraperitoneal route.
- https://cdn.elifesciences.org/articles/64166/elife-64166-supp3-v1.docx
-
Supplementary file 4
Complete list of 154 protein targets identified by the proteome array as correlates of immunity for the attenuated-vaccine model.
- https://cdn.elifesciences.org/articles/64166/elife-64166-supp4-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64166/elife-64166-transrepform-v1.docx





