The gut microbiota is a transmissible determinant of skeletal maturation
Figures
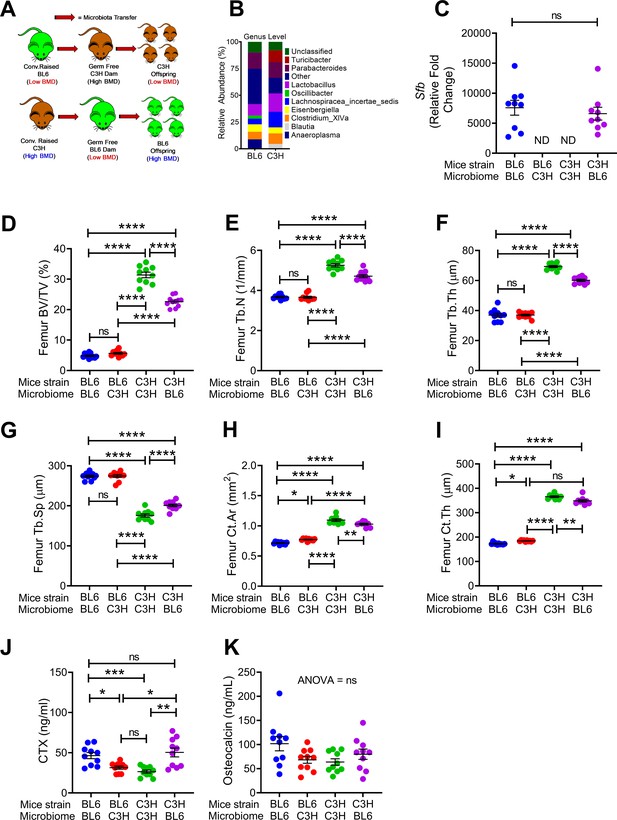
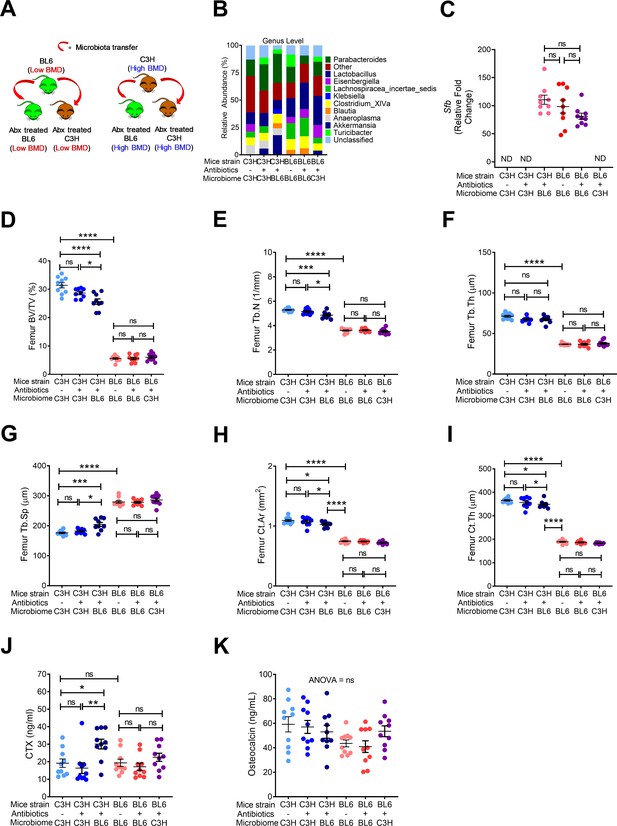
Nongenomic effects of gut microbiome on bone volume, structure and turnover are hereditarily transferred.
(A) Diagram of the experimental outline. Fecal material from conventional C57BL/6 (low BMD) or C3H/HeN (high BMD) mice was transferred to germ-free mating pairs of the other strain. Bone structure and turnover were analyzed in 16-week-old female mice of the F1 generation to determine the influence of the donor microbiota on post-natal skeletal development. (B) Fecal microbiome composition in C57BL/6 (BL6) mice and C3H/HeN (C3H) mice. (C) Quantitative PCR (qPCR) analysis of Sfb and total bacterial 16S rRNA genes in fecal samples. (D) Femoral trabecular bone volume fraction (BV/TV). (E) Trabecular number (Tb.N). (F) Trabecular thickness (Tb.Th). (G) Trabecular separation (Tb.Sp). (H) Cortical Area (Ct.Ar). (I) Cortical thickness (Ct.Th). (J) Serum levels of CTX, a marker of bone resorption. (K) Serum levels of osteocalcin (OCN), marker of bone formation. n = 9–12 mice per group. Data were expressed as mean ± SEM. All data were normally distributed according to the Shapiro-Wilk normality test. Data were analyzed by 2-way ANOVA and post hoc tests applying the Bonferroni correction for multiple comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 compared to indicated groups.
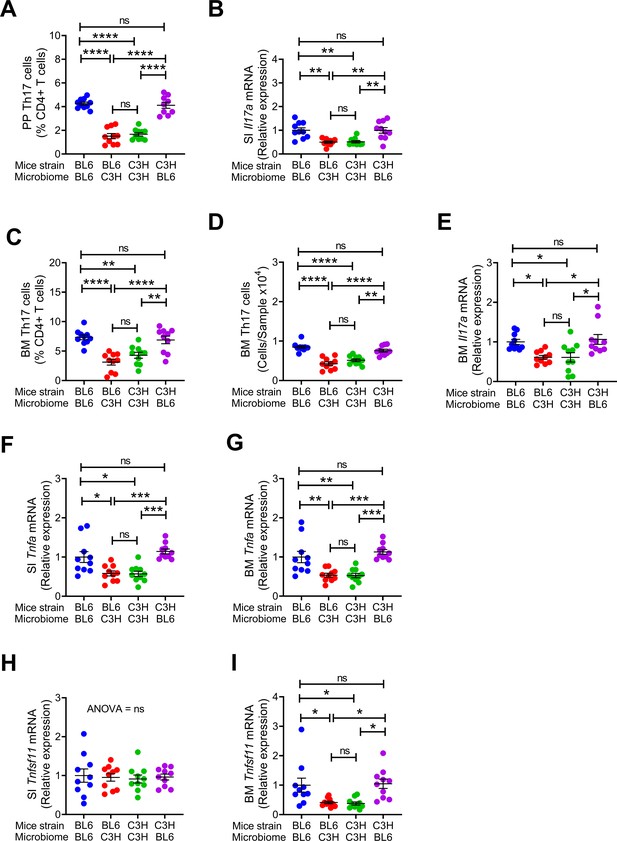
Effects of maternal fecal microbime on the frequency of BM and PP Th17 cells and transcript levels of inflammatory cytokines in the offspring.
(A) Relative frequency of Th17 cells (IL-17+ CD4+ T cells) in PP. (B) SI Il17a mRNA levels. (C,D) Relative and absolute frequency of BM Th17 cells. (E) BM Il17a mRNA levels. (F) SI Tnfa mRNA levels. (G) BM Tnfa mRNA levels. (H) SI Tnfsf11 mRNA levels. (I) BM Tnfsf11 mRNA levels. n = 10 mice per group. Data were expressed as mean ± SEM. All data were normally distributed according to the Shapiro-Wilk normality test. Data were analyzed by 2-way ANOVA and post hoc tests applying the Bonferroni correction for multiple comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 compared to indicated groups.
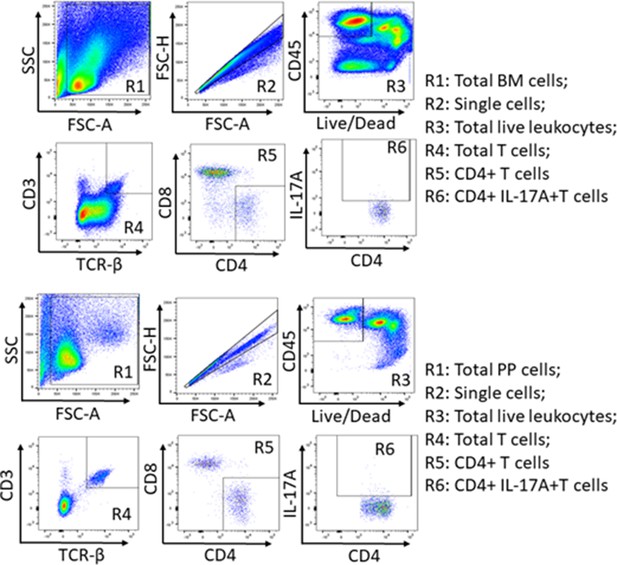
Gating strategy used to identify BM (Panel A) and Payer’s patches (Panel B) Th17 cells.
Following red blood cell lysis, single cell suspensions were prepared from Payer’s patches and BM and stained with antibodies to the indicated antigens and live/dead cell dye. Gated regions are numbered from R1 to R6. The figure shows one representative gating of flow cytometric plot.
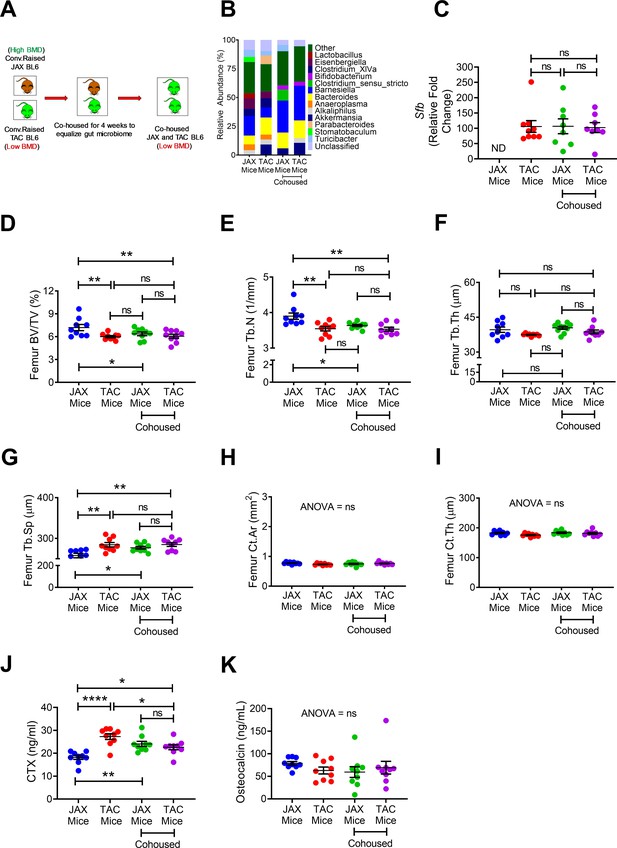
Effects of co-housing on bone volume, structure and turnover.
(A) Diagram of the experimental outline. 10-week-old female JAX and TAC mice were housed separately, or co-housed for 4 weeks. (B) Effects of co-housing on the composition and frequency of microbiome. (C) Quantitative PCR analysis of Sfb and total bacterial 16S rRNA genes in fecal samples. (D) Femoral trabecular bone volume fraction (BV/TV). (E) Trabecular number (Tb.N). (F) Trabecular thickness (Tb.Th). (G) Trabecular separation (Tb.Sp). (H) Cortical Area (Ct.Ar). (I) Cortical thickness (Ct.Th). (J) Serum levels of CTX. (K) Serum levels of OCN. n = 8–9 mice per group. Data were expressed as mean ± SEM. All data were normally distributed according to the Shapiro-Wilk normality test. Data were analyzed by two-way ANOVA and post hoc tests applying the Bonferroni correction for multiple comparisons. *p<0.05, **p<0.01, ****p<0.0001 compared to indicated groups.
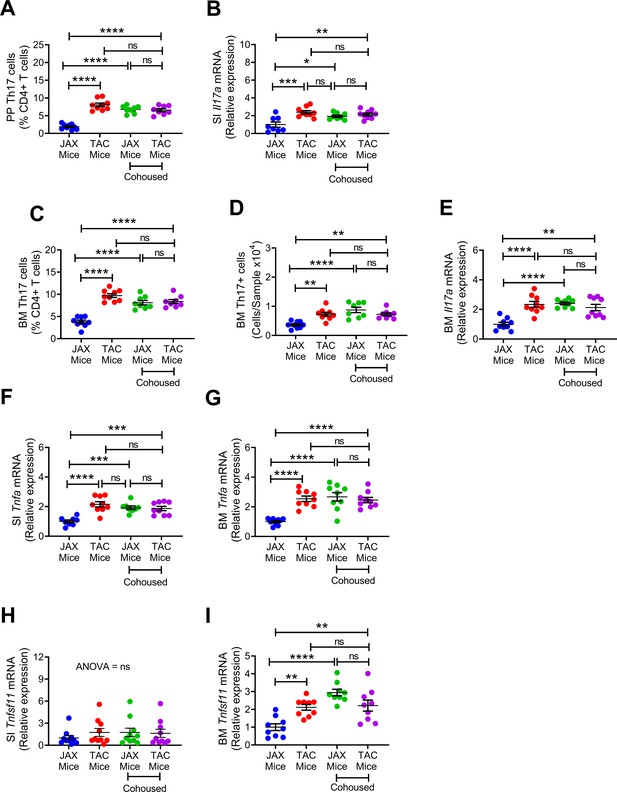
Effects of cohousing on the frequency of BM and PP Th17 cells and transcript levels of inflammatory cytokines.
(A) Relative frequency of Th17 cells (IL-17+ CD4+ T cells) in PP. (B) SI Il17a mRNA levels. (C,D) Relative and absolute frequency of BM Th17 cells. (E) BM ll17a mRNA levels. (F) SI Tnfa (TNF) mRNA levels. (G) BM Tnfa mRNA levels. (H) SI Tnfsf11 mRNA levels. (I) BM Tnfsf11 mRNA levels. n = 8–9 mice per group. Data were expressed as mean ± SEM. All data were normally distributed according to the Shapiro-Wilk normality test. Data were analyzed by 2-way ANOVA and post hoc tests applying the Bonferroni correction for multiple comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 compared to indicated groups.
Effects of FMT on bone volume, structure and turnover.
(A) Diagram of the experimental outline. (B) Fecal microbiome composition in C57BL/6 (BL6) mice and C3H/HeN (C3H) mice. (C) Quantitative PCR analysis of Sfb and total bacterial 16S rRNA genes in fecal samples. (D) Femoral trabecular bone volume fraction (BV/TV). (E) Trabecular number (Tb.N). (F) Trabecular thickness (Tb.Th). (G) Trabecular separation (Tb.Sp). (H) Cortical Area (Ct.Ar). (I) Cortical thickness (Ct.Th). (J) Serum levels of CTX. (K) Serum levels of OCN. n = 9–10 mice per group. Data were expressed as mean ± SEM. All data were normally distributed according to the Shapiro-Wilk normality test. Data were analyzed by 2-way ANOVA and post hoc tests applying the Bonferroni correction for multiple comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 compared to indicated groups.
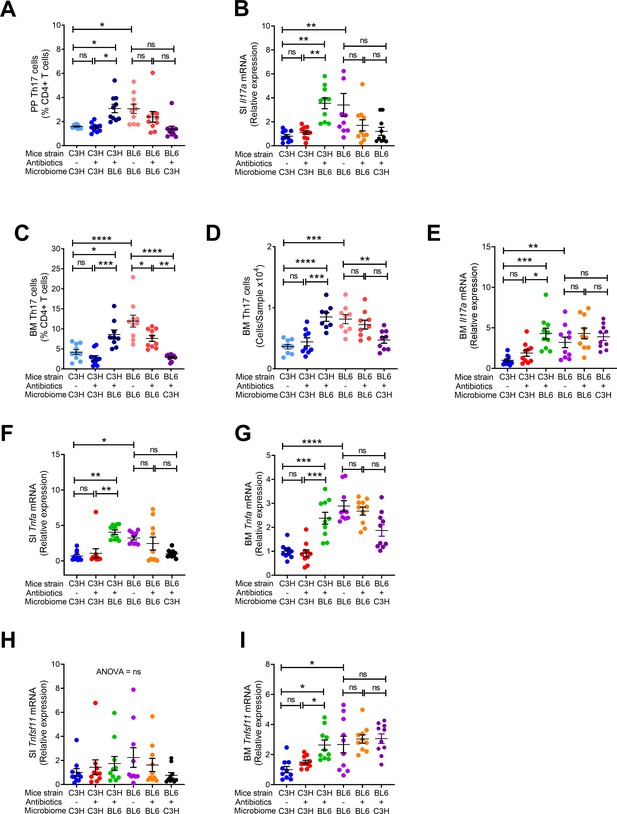
Effects of FMT on the frequency of BM and PP Th17 cells and transcript levels of inflammatory cytokines.
(A) Relative frequency of Th17 cells (IL-17+ CD4+ T cells) in PP. (B) SI Il17a mRNA levels. (C,D) Relative and absolute frequency of BM Th17 cells. (E) BM Il17a mRNA levels. (F) SI Tnfa (TNF) mRNA levels. (G) BM Tnfa mRNA levels. (H) SI Tnfsf11 mRNA levels. (I) BM Tnfsf11 mRNA levels. n = 9–10 mice per group. Data were expressed as mean ± SEM. All data were normally distributed according to the Shapiro-Wilk normality test. Data were analyzed by 2-way ANOVA and post hoc tests applying the Bonferroni correction for multiple comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 compared to indicated groups.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (M. musculus) | C57BL/6J | Jackson Laboratory | Cat# JAX:000664, RRID:IMSR_JAX:000664 | Mice for in vivo experiments |
| strain, strain background (M. musculus) | C57BL/6NTac | Taconic biosciences | Cat# TAC:b6, RRID:IMSR_TAC:b6 | Mice for in vivo experiments |
| strain, strain background (M. musculus) | C3H/HeNTac | Taconic biosciences | Cat# TAC:c3h, RRID:IMSR_TAC:c3h | Mice for in vivo experiments |
| Chemical compound, drug | Ampicillin | Sigma-Aldrich | Cat# A9393 | For Microbiota depletion |
| Chemical compound, drug | Vancomycin | Selleckchem | Cat# S2575 | For Microbiota depletion |
| Chemical compound, drug | Neomycin Sulfate | Sigma-Aldrich | Cat# 1062540100 | For Microbiota depletion |
| Chemical compound, drug | Metronidazole | Sigma-Aldrich | Cat# M3761 | For Microbiota depletion |
| Chemical compound, drug | Trizol | Life Technologies | Cat# 15596018 | Cells Sample collection for total RNA isolation |
| Chemical compound, drug | Zombie NIR | Biolegend | Cat# 423105 | FACS (0.1 ul per test) |
| Chemical compound, drug | Cell Activation Cocktail (without Brefeldin A) | Biolegend | Cat# 423301 | Cell Activation Cocktail |
| Chemical compound, drug | Monensin Solution | Biolegend | Cat# 420701 | Monensin for cell activation |
| Chemical compound, drug | Fixation Buffer | Invitrogen | Cat# 2178648 | For cell fixation |
| Chemical compound, drug | Permeabilization Buffer | Invitrogen | Cat# 2229113 | Cell Permeabilization Buffer |
| Antibody | Anti-Mouse CD16/32 (clone 93) (Mouse monoclonal) | Biolegend | Cat# 101302, RRID:AB_312801 | FACS (1 ul per test) |
| Antibody | BV 510-CD45 (clone 30-F11) (Mouse monoclonal) | Biolegend | Cat# 103138, RRID:AB_2563061 | FACS (1 ul per test) |
| Antibody | BV 421-TCRβ (clone H57-597) (Mouse monoclonal) | Biolegend | Cat# 109230, RRID:AB_2562562 | FACS (1 ul per test) |
| Antibody | AF 700-CD3 (clone 17A2) (Mouse monoclonal) | Biolegend | Cat# 100216, RRID:AB_493697 | FACS (1 ul per test) |
| Antibody | PerCP/Cy5.5-CD4 (clone RM4-5) (Mouse monoclonal) | Biolegend | Cat# 100540, RRID:AB_893326 | FACS (1 ul per test) |
| Antibody | BV 711-CD8 (clone 53–6.7) (Mouse monoclonal) | BD Biosciences | Cat# 100748, RRID:AB_2562100 | FACS (1 ul per test) |
| Antibody | Anti-mouse PE-IL-17A (clone eBio17B7) (Mouse monoclonal) | Thermo Fisher Scientific | Cat# 12-7177-81, RRID:AB_763582 | FACS (1 ul per test) |
| sequenced-based reagent | cDNA Synthesis kit | Invitrogen | Cat# 18080–051 | For RT-PCR |
| sequenced-based reagent | SYBR GREEN | Applied Biosciences | Cat# 4367659 | For RT-PCR |
| commercial assay or kit | Osteocalcin | Immunodiagnostic systems Ltd. | Cat# AC-12F1 | For the measurement of serum levels of Osteocalcin |
| commercial assay or kit | CTX | Immunodiagnostic systems Ltd. | Cat# AC-06F1 | For the measurement of serum levels of CTX |
| commercial assay or kit | DNA Stool Qiagen kit | Qiagen | Cat# 51604 | For the 16S and SFB PCR |
| software, algorithm | Flow cytometry | LSR II system | BD Biosciences | |
| software, algorithm | FlowJo software | Tree Star, Inc | NA | |
| software, algorithm | MicroCT-40 scanner | Scanco | NA | |
| software, algorithm | QIIME | Open Source | NA | |
| software, algorithm | GraphPad Prism 8 | GraphPad Software | NA |





