Applications of genetic-epigenetic tissue mapping for plasma DNA in prenatal testing, transplantation and oncology
Figures
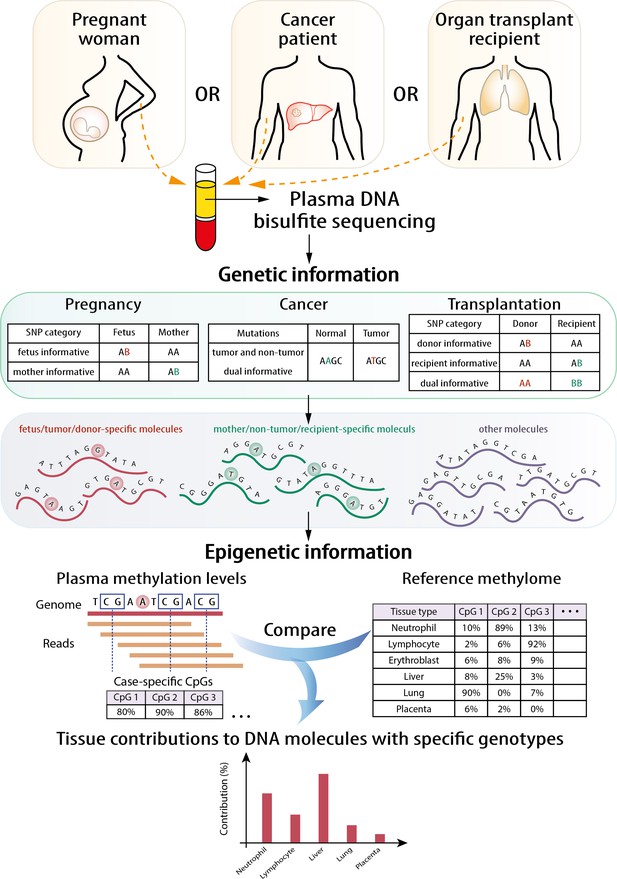
Schematic illustration of the principle of genetic-epigenetic tissue mapping (GETMap) analysis.
The paired individuals (e.g., fetus/mother, organ donor/recipient, and tumor/normal tissue) are genotyped to identify single nucleotide polymorphism (SNP) alleles specific for one of them. After bisulfite sequencing, plasma DNA molecules carrying individual-specific alleles and at least one CpG site are identified. The plasma DNA methylome is compared with the methylation profiles of reference tissues to determine the tissue composition of the subset of plasma DNA molecules derived from a particular individual.
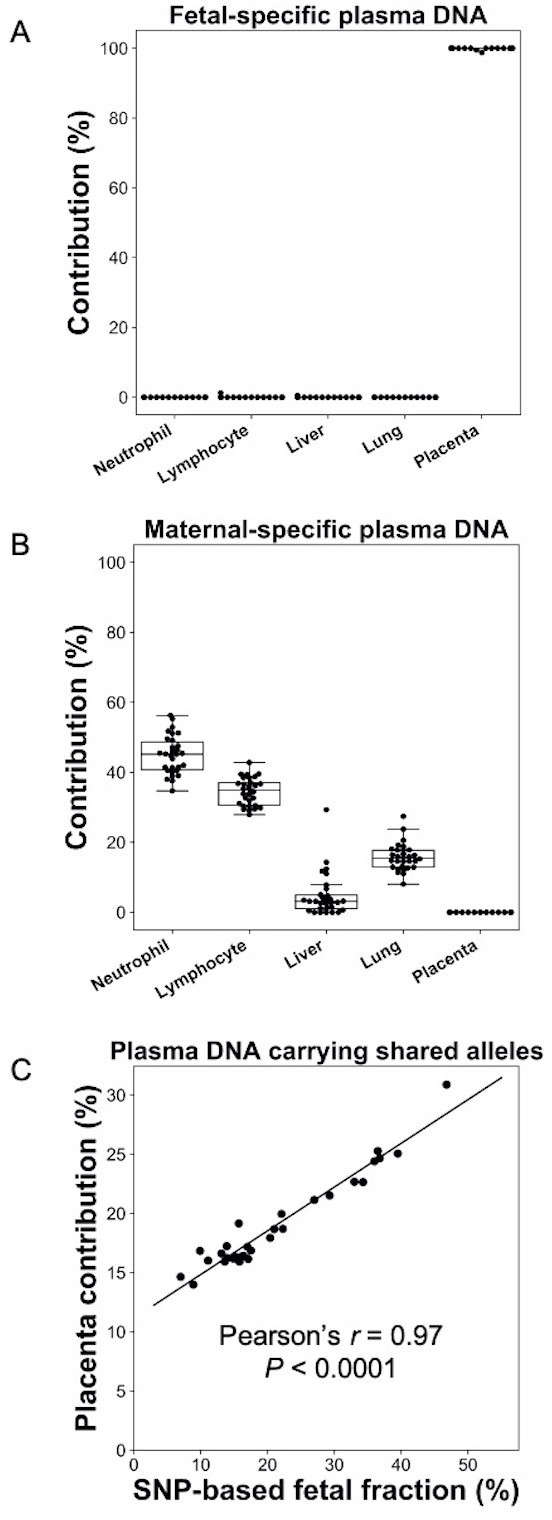
Percentage contributions of different cell types to maternal plasma DNA carrying (A) fetal-specific alleles and (B) maternal-specific alleles in 30 pregnant women.
(C) Correlation between percentage contribution of the placenta to maternal plasma DNA molecules carrying alleles shared by the fetus and mother and single nucleotide polymorphism (SNP)-based fetal DNA fraction.
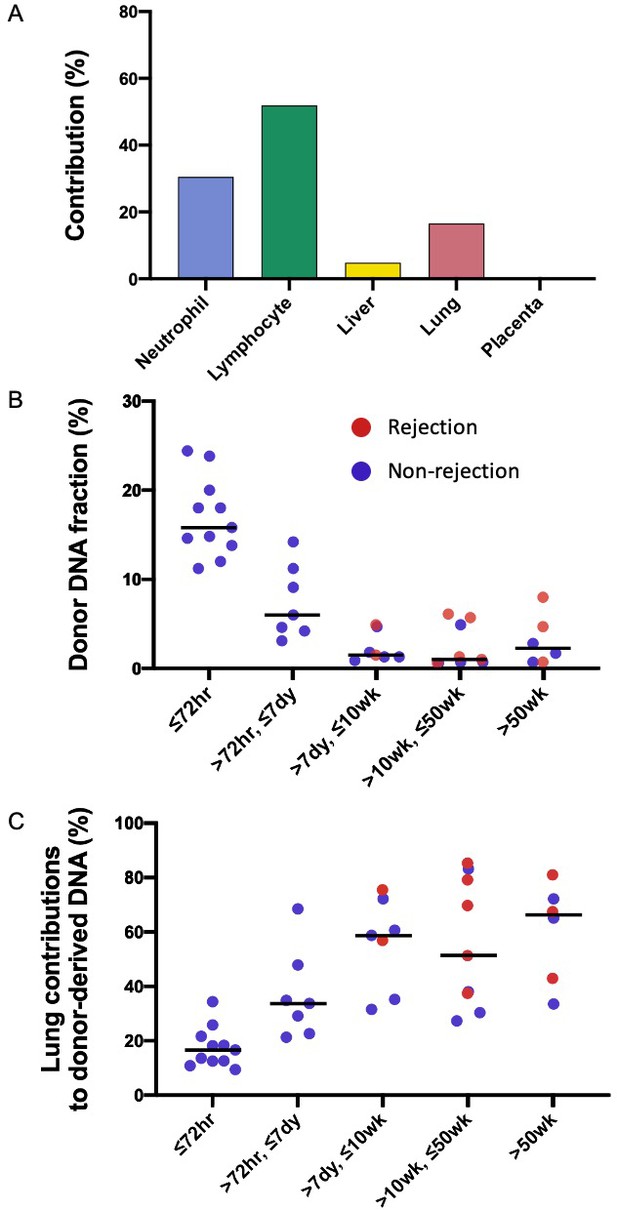
Genetic-epigenetic tissue mapping (GETMap) analysis on donor-derived plasma DNA molecules in lung-transplant recipients.
(A) The median percentage contributions of different cell types to plasma DNA carrying donor-specific alleles in patients with lung transplantation at 72 hr post-transplant. (B) Fractional concentrations of donor-derived DNA and (C) percentage contributions of the lung to plasma DNA carrying donor-specific alleles in patients with lung transplantation.
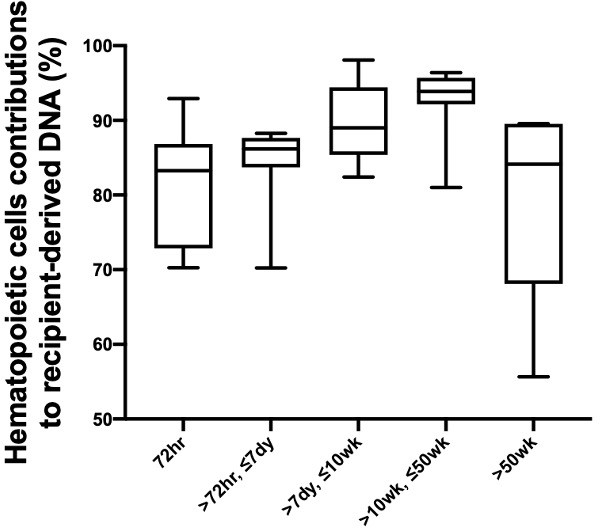
Percentage contributions of hematopoietic cells to the plasma DNA carrying recipient-specific alleles in patients with lung transplantation.
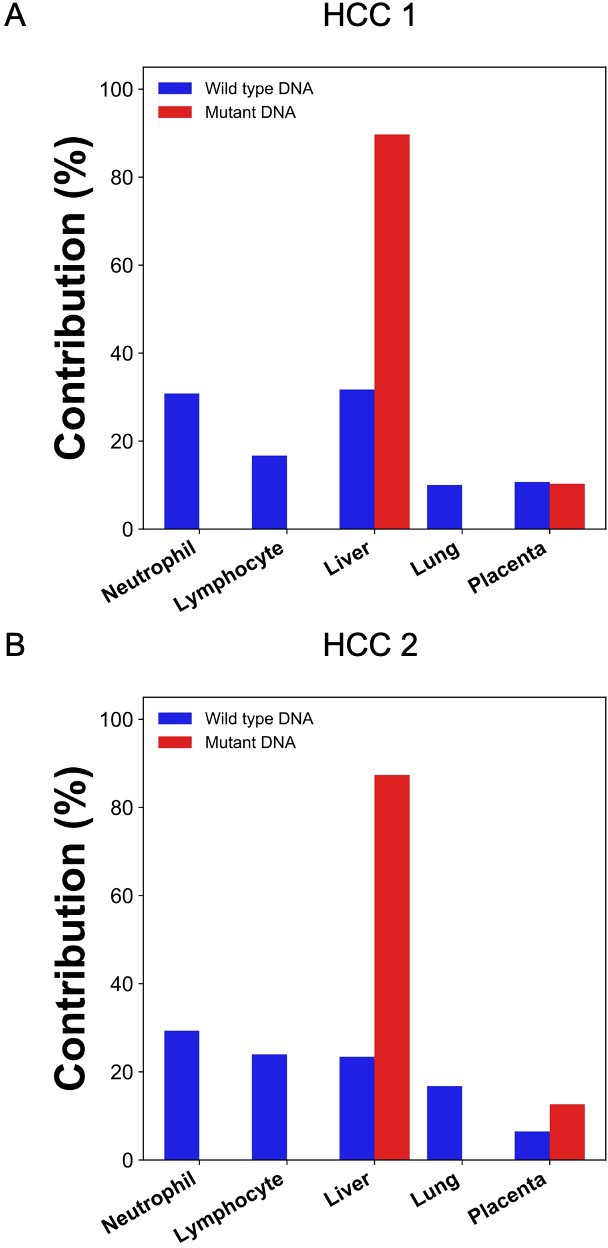
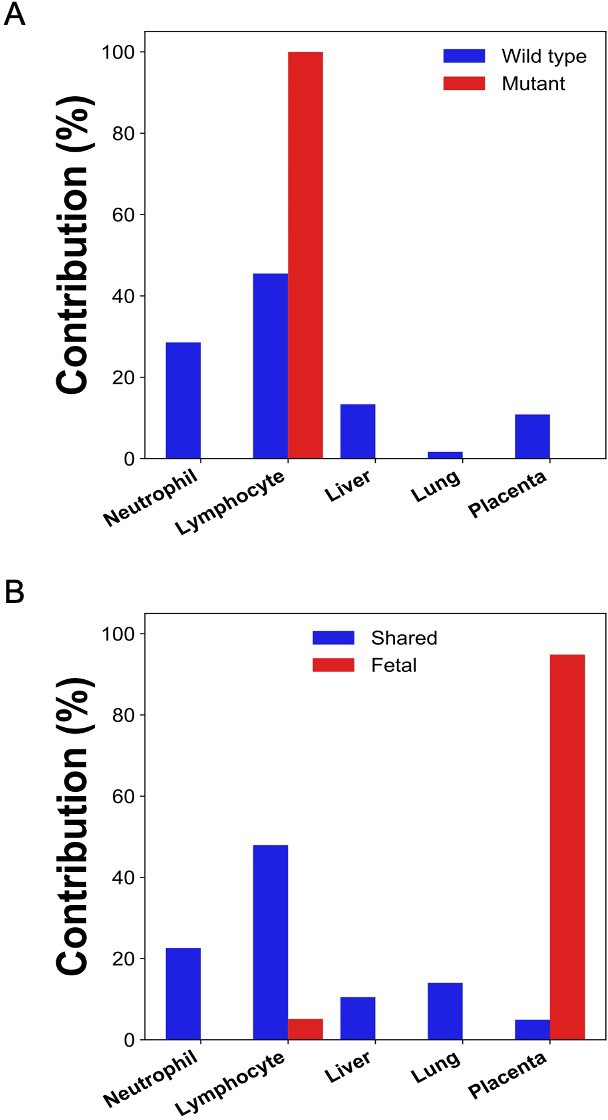
Percentage contributions of different tissues to plasma DNA with tumor-specific and wildtype alleles in two hepatocellular cancer (HCC) patients.
The tumor-specific mutations were deduced from the tumor tissues.
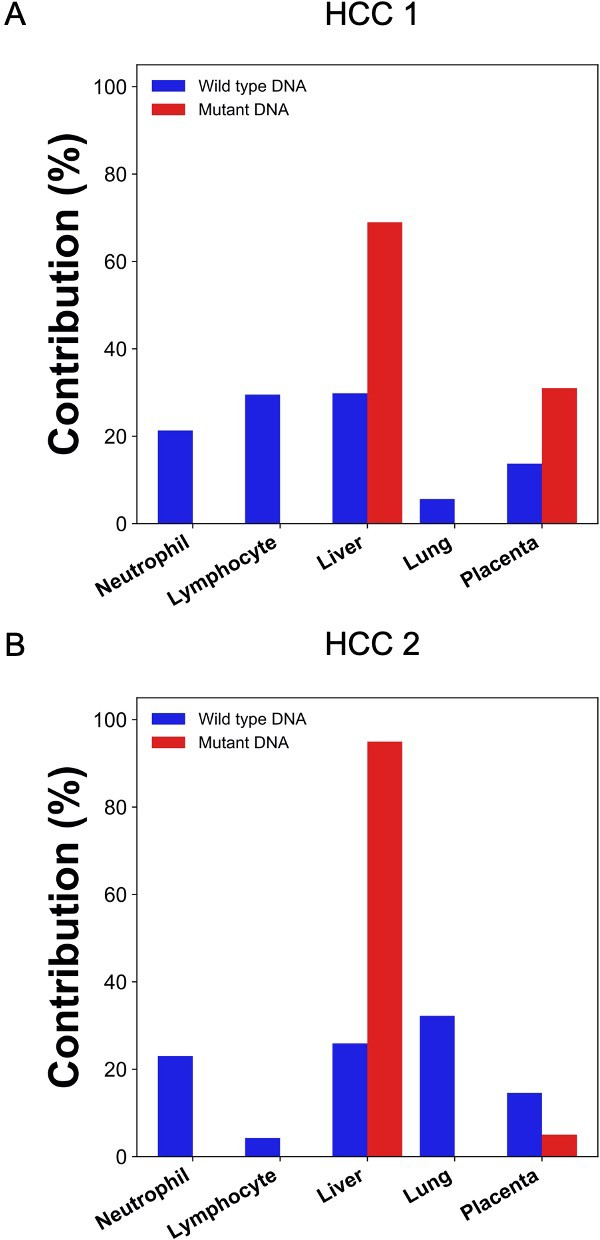
Percentage contributions of different tissues to plasma DNA with tumor-specific and wildtype alleles in two hepatocellular cancer (HCC) patients.
The tumor-specific mutations were deduced directly from the plasma.
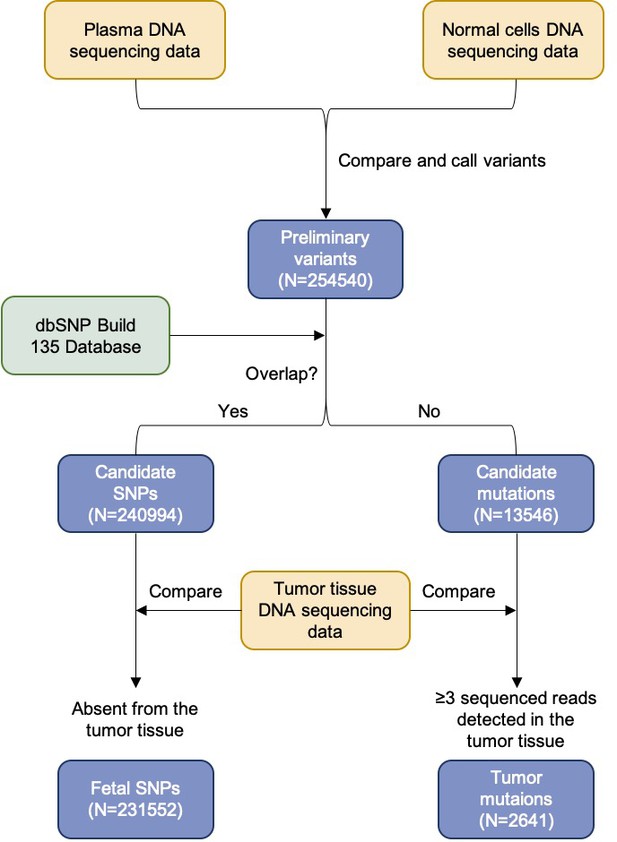
Flowchart of the steps for identifying the fetal-specific alleles and cancer mutations in the pregnant woman with lymphoma.
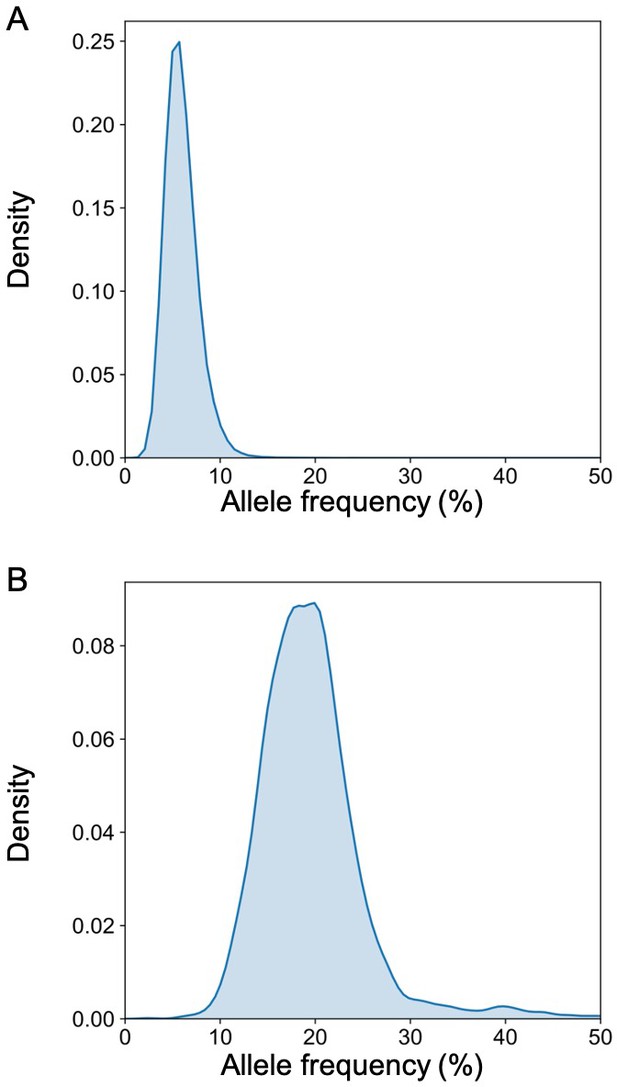
The distribution of the allele frequency of (A) the fetal-specific alleles and (B) the mutant alleles in the plasma of the pregnant woman with lymphoma.
Tables
Results of deconvolution of bisulfite sequencing data from reference tissues for scenarios of (A) pregnancy, (B) lung transplantation, and (C) liver cancer.
The underlined numbers represent the percentage of contribution accurately assigned to the respective tissues by genetic-epigenetic tissue mapping (GETMap).
| (A) | Tissue contribution as determined by GETMap analysis | |||||
|---|---|---|---|---|---|---|
| Neutrophils | Lymphocytes | Liver | Lung | Placenta | ||
| Reference tissue used for the simulation | Neutrophils | 96.78 | 2.01 | 0.59 | 0.33 | 0.29 |
| Lymphocytes | 0.52 | 98.30 | 0.41 | 0.20 | 0.58 | |
| Liver | 0.31 | 0.64 | 98.36 | 0.27 | 0.42 | |
| Lung | 0.24 | 0.66 | 0.35 | 98.36 | 0.39 | |
| Placenta | 0.13 | 0.05 | 0.00 | 0.09 | 99.73 | |
| (B) | Tissue contribution as determined by GETMap analysis | |||||
| Neutrophils | Lymphocytes | Liver | Lung | Placenta | ||
| Reference tissue used for the simulation | Neutrophils | 98.21 | 0.77 | 0.42 | 0.43 | 0.17 |
| Lymphocytes | 0.48 | 98.70 | 0.20 | 0.31 | 0.31 | |
| Liver | 0.32 | 0.19 | 99.25 | 0.11 | 0.13 | |
| Lung | 0.21 | 0.09 | 0.22 | 99.39 | 0.09 | |
| Placenta | 0.00 | 0.09 | 0.08 | 0.05 | 99.78 | |
| (C) | Tissue contribution as determined by GETMap analysis | |||||
| Neutrophils | Lymphocytes | Liver | Lung | Placenta | ||
| Reference tissue used for the simulation | Neutrophils | 96.08 | 2.23 | 0.32 | 0.37 | 1.00 |
| Lymphocytes | 0.94 | 95.46 | 0.79 | 2.06 | 0.75 | |
| Liver | 0.50 | 0.44 | 96.67 | 1.48 | 0.91 | |
| Lung | 0.90 | 1.71 | 0.80 | 96.08 | 0.51 | |
| Placenta | 0.49 | 0.13 | 0.77 | 0.34 | 98.27 | |
The demographic profiles of lung transplant recipients.
| Case number | Recipient age | Recipient gender | Donor age | Donor gender | Diagnosis for transplant | Single/ double lung | Cause of death | Time of sample collection post-transplant |
|---|---|---|---|---|---|---|---|---|
| 1 | 34 | M | 32 | M | Cystic fibrosis | Double | Alive | 72 hr |
| 2 | 59 | F | 27 | F | Interstitial lung disease | Double | Alive | 72 hr |
| 3 | 53 | M | 20 | M | Interstitial lung disease | Double | Alive | 72 hr |
| 4 | 63 | M | 16 | F | Interstitial lung disease | Double | Alive | 72 hr, 6 dy |
| 5 | 55 | F | 36 | F | Interstitial lung disease | Double | Alive | 72 hr, 7 dy |
| 6 | 66 | M | 48 | F | Interstitial lung disease | Single | Alive | 72 hr, 4 wk |
| 7 | 66 | F | 18 | M | Chronic obstructive pulmonary disease | Single | Alive | 72 hr, 7 dy, 5 wk, 20 wk, 25 wk, 157 wk |
| 8 | 32 | F | 39 | M | Cystic fibrosis | Double | Alive | 72 hr, 7 dy, 8 wk, 38 wk, 77 wk, 129 wk |
| 9 | 67 | F | 53 | F | Sarcoidosis | Double | Respiratory failure | 72 hr, 7 dy, 6 wk, 13 wk, 22 wk |
| 10 | 44 | M | 35 | F | Retransplant | Double | Alive | 72 hr, 7 dy, 10 dy, 4 wk, 14 wk, 25 wk, 103 wk |
| 11 | 67 | F | 32 | M | Pulmonary arterial hypertension | Single | Alive | 72 hr, 7 dy, 5 wk, 15 wk, 26 wk, 61 wk, 104 wk |
-
*Samples collected when the patient was having a rejection episode were underlined.
Additional files
-
Supplementary file 1
The information of all the samples analyzed in this study, including sequencing depth, number of informative single nucleotide polymorphisms (SNPs), number of informative sequencing fragments, number of informative CpG sites, and number of CpG sites used for deconvolution.
- https://cdn.elifesciences.org/articles/64356/elife-64356-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64356/elife-64356-transrepform-v2.docx