Matrix-trapped viruses can prevent invasion of bacterial biofilms by colonizing cells
Figures
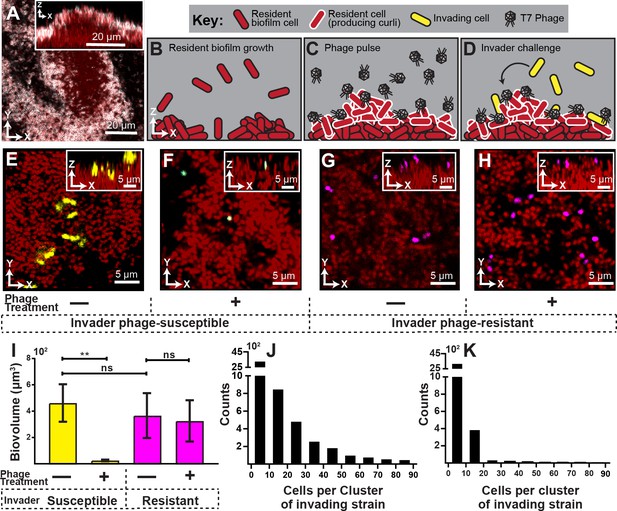
Visualization and quantification of biofilm invasion with or without phage exposure.
(A) Visualization of E. coli biofilm (red) with stained curli matrix (white), including one x-y optical section (main image) and z-projection (inset). (B–D) Illustration of invasion assay procedure. (B) Resident biofilm-producing cells (red) were allowed to grow for 62 hr prior to phage exposure. (C) Inlet tubing was then swapped for 1 hr to new tubing and syringe containing a concentrated phage suspension (2x108 PFU/mL). (D) Resident biofilms were then challenged with isogenic E. coli expressing a different fluorescent protein (yellow); this was performed by swapping for 3 hr to new tubing and syringe containing high density E. coli culture (OD600 = 6.0). Biofilms were imaged 10 hr following this step. (E) Invading cells (yellow) can successfully attach to the periphery of resident biofilm (red) in the absence of phages. (F) Invading cells fail to colonize when biofilms are pretreated with phages, which become trapped in the biofilm matrix. The phage-encoded reporter (cyan) indicates invading cells that have become phage-infected. (G,H) An E. coli mutant that does not permit phage amplification (ΔtrxA, magenta) invades equally well in control and phage pre-treatment conditions. (I) Quantification of image data shown in E-H; average invading biovolume per field of view (150 µm x 150 µm x 15 μm; length x width x height). Error bars represent SEM. Pairwise comparisons were performed with Mann-Whitney Signed Ranks tests with Bonferroni correction (n = 4-6 biological replicates; ** denotes p<0.05). (J–K) Invading cell cluster size distributions for phage-susceptible cells invading biofilms without (J) or with (K) phage pre-treatment (n = 4-6 biological replicates).
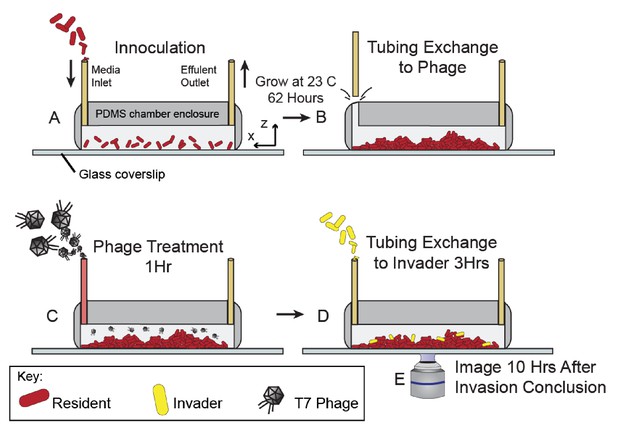
Method for phage pretreatment experiments.
(A) E. coli resident strain was inoculated into microfluidic devices at OD600 = 0.2. Cells were allotted an hour for surface attachment before a media flush was performed (40 µL/min for 40 s). Biofilms were then grown at room temperature for approximately 62 hr. (B) Inlet tubing was carefully removed from devices and replaced with tubing connected to new syringes containing phages. (C) Phages (2x109 PFU/mL) were flowed into the chamber for 1 hr (0.1 µL/min for 1 hr). (D) Following phage pretreatment, isogenic E. coli invaders (OD600 = 6.0) were introduced into the chamber, again through a tubing swap. (E) Tubing was finally returned to sterile media and the device was allowed to incubate for 10 hr prior to imaging.
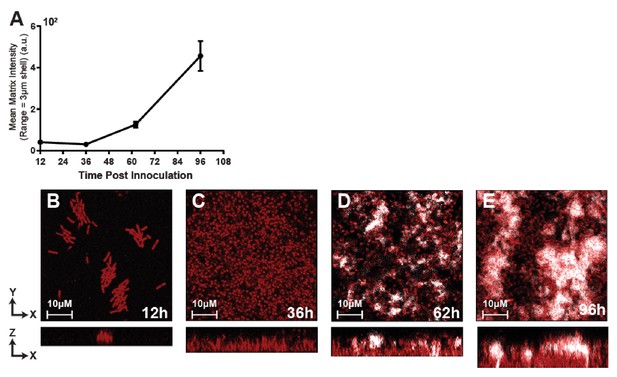
Time course imaging of E. coli biofilms documenting the course of curli matrix protein accumulation.
Biofilms were imaged at 12, 36, 62, and 96 hr post inoculation and initiation of medium flow. Curli production was visualized using a strain of E. coli producing csgA-6xHis, allowing for immunostaining of curli protein by addition of anti-His antibodies to the influent medium. Consistent with prior work (Vidakovic et al., 2018), appreciable curli accumulation sufficient to block T7 phage diffusion into the biofilm interior was not consistently observed until ~60 hr after the start of biofilm growth. Biofilms of this age were therefore chosen as the base resident biofilms to which phages were added prior to challenge with new invading stains of E. coli (see main text). Localized matrix intensity was calculated as for Figure 3 in the main text (n = 3-4 replicates per time point; error bars denote SEM).
-
Figure 1—figure supplement 2—source data 1
- https://cdn.elifesciences.org/articles/65355/elife-65355-fig1-figsupp2-data1-v2.xlsx
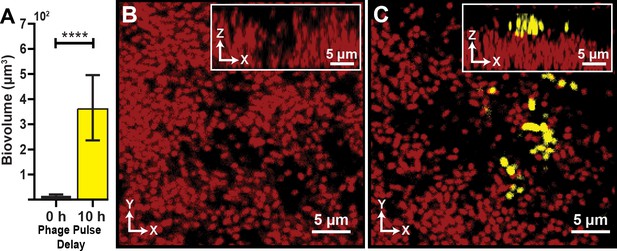
Visualization and quantification of colonization success with phage exposure post-colonization.
(A) Average invading biovolume per field of view (150 µm x 150 µm x 15 μm; length x width x height). Error bars represent SEM. Pairwise comparisons were performed with a Mann-Whitney Signed Ranks test (n = 7-11 biological replicates; **** denotes p<0.00005). (B) Invading cells (yellow, though absent in B) are killed when phages are introduced immediately after their arrival. Resident biofilm cells are shown in red. (C) Invading cells are not killed when phages are introduced 10 hr after their arrival.
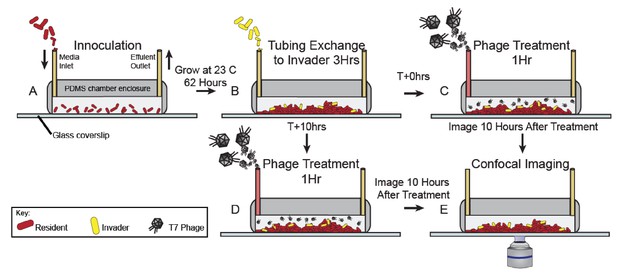
Method for phage posttreatment experiments.
(A) E. coli resident strain was inoculated into microfluidic devices at OD600 = 0.2. Cells were allotted an hour for surface attachment before a media flush was performed (40 µL/min for 40 s). Biofilms were then grown at room temperature for 62 hr. (B) Invading cells were then introduced into all chambers (OD600 = 6.0) for 3 hr. After the invasion, individual chambers were separated into two groups: Chambers in group C received phage treatment (2x108 PFU/mL, 0.1 µL/min for 1 hr) immediately following the invasion step. These chambers were imaged 10 hr after the conclusion of the phage treatment (E). Chambers in group D were incubated at room temperature for 10 hr before receiving phage treatment. Ten hours following the completion of the phage treatment, chambers in group D were imaged.
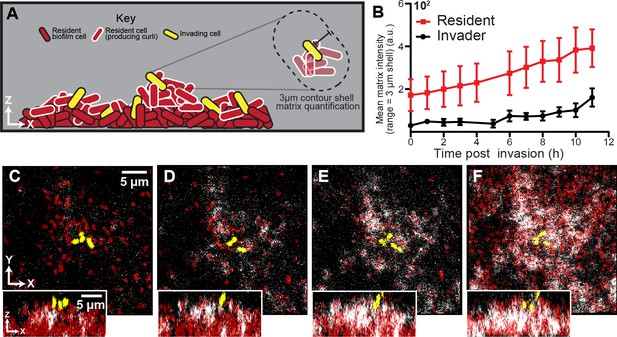
Spatial and temporal dynamics of curli fiber localization around resident and invading cells.
(A) Illustration of matrix quantification method. Localized curli matrix was quantified by measuring CsgA-His immunofluorescence in 3 µm shells surrounding individual segmented cell volumes. (B) Quantification of matrix localization surrounding the resident and invading strain populations during an 11 hr time course after invading cells arrive (n = 3-4 biological replicates per time point, errors bars denote SEM). (C–F) Representative images displaying resident cells (red) producing labeled curli matrix (white); invading cells are shown in yellow. Images in (C-F) were acquired respectively at 1 , 5, 7, and 10 hr following the arrival of invading cells.
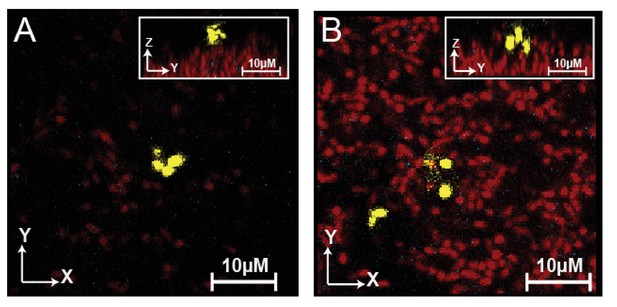
Invading cells do not produce curli matrix.
(A) Representative image of invading cells (yellow) attached to resident biofilm (red) 3 hr following the conclusion of the invasion. Invading cells encode 6xHis labeled curli monomers. No matrix was detectable by immunostaining (some noise from anti-His antibody background is visible as white/cyan dots in these images). (B) Representative image of invading cells ten hours after invasion. Again, invaders were not producing curli matrix.
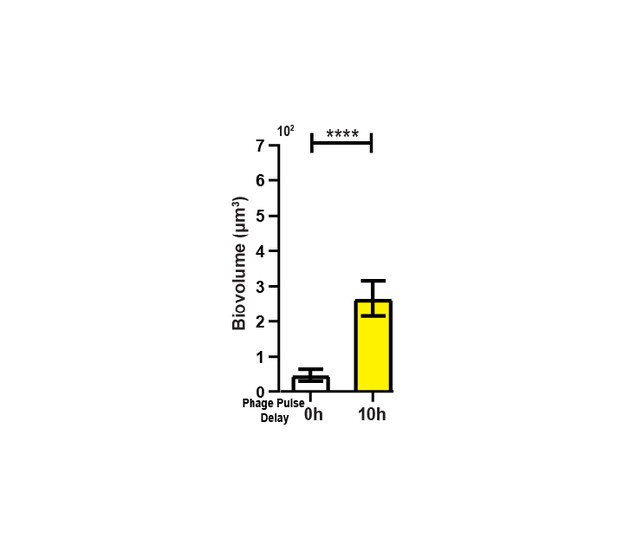
Quantification of invasion success of E. coli lacking the CsgB baseplate required for curli polymerization, with phage exposure post-invasion.
(A) Average invading biomass per 150 x 150 x 15μm (L, W, H) view field. Error bars represent standard error of the mean. Pairwise comparisons were performed with Mann-Whitney Signed Ranks tests with Bonferroni correction (n=7; **** denotes p<0.00005).
-
Figure 3—figure supplement 2—source data 1
- https://cdn.elifesciences.org/articles/65355/elife-65355-fig3-figsupp2-data1-v2.xlsx
Tables
Strains, plasmids, and oligos used in this study.
| Strain | Relevant markers/Genotype | Source | ||
|---|---|---|---|---|
| E. coli | ||||
| CNE 689 | AR3110 wild type Ptac-mKo-κ inserted at attB site | This study | ||
| CNE 762 | AR3110 wild type Ptac-mKate2 inserted at attB site | This study | ||
| CNE 760 | AR3110 wild type Ptac-mTFP inserted at attB site | This study | ||
| CNE 284 | AR3110 ΔcsgB::scar, Ptac-mruby inserted at attB site | (19) | ||
| CNE 773 | AR3110 CsgA with C-terminal 6x His Tag, Ptac-mKo-κ inserted at attB site | This study, (19) | ||
| CNE 691 | AR3110 ΔtrxA::scar, Ptac-mKo-κ inserted at attB site | This study | ||
| CNE 198 | AR3110 wild type | (19) | ||
| T7 Phage | ||||
| CNX 06 | T7 with sfgfp under phi 10 promotor control | (19) | ||
| Plasmid | Origin, marker | Comments | Templates, primers | Source |
| pCN754 | pR6Kγ, Kan | Housing plasmid/template for Ptac-mTFP insert | CNO 198, CNO 199, pCN 752, pCN 664 | This study |
| pCN755 | pR6Kγ, Kan | Housing plasmid/template for Ptac-mKate2 insert | CNO 198, CNO 199, pCN 753, pCN 664 | This study |
| pCN664 | pR6Kγ, Kan | Housing plasmid/template for Ptac-mKo-κ insert | CNO 198 CNO 199 | This study |
| pNUT1336 | pR6Kγ,Kan | pUC housing IDT-synthesized double mKO optimized for E. coli AR3110 | This study | |
| pCN753 | pR6Kγ, Kan | pUC housing IDT-synthesized double mTFP optimized for E. coli AR3110 | This study | |
| pCN752 | pR6Kγ, Kan | pUC housing IDT-synthesized double mKate2 optimized for E. coli AR3110 | This study | |
| Primer name | Sequence (designed using Snapgene) | Description | ||
| CNO 198 | ACAACTTTTTGTCTTTTTACCTTCCCGTTTCGCTCAAGT TAGTATttgacaattaatcatcggctcg | Universal primer to amplify new E. coli Ptac_FP construct, with attB integration tail | ||
| CNO 199 | TCCGGGCTATGAAATAGAAAAATGAATCCGTTGAAGCC TGCTTTTcatgggaattagccatggtcc | Universal primer to amplify new E. coli Ptac_FP construct, with attB integration tail | ||
| CNO 138 | ACAACGAAACCAACACGCCAGGCTTA TTCCTGTGGAGTTATATgtgtaggctggagctgcttc | Forward primer to amplify pKD3 FRT-Cm-FRT with homology to trxA up flank | ||
| CNO 139 | GCGTCCAGTTTTTAGCGACGGGGCACCCGAACATG AAATTCCCCcatatgaatatcctccttagt | Reverse primer to amplify pKD3 FRT-Cm-FRT with homology to trxA down flank | ||
| CNO 146 | GAATGGGCGTACAGTTATGAAAC | Forward primer to check sequence of trxA deletion in E. coli, 200 bp upstream | ||
| CNO 147 | TGCCTGGTCACAGGAGAGT | Reverse primer to check sequence of trxA deletion in E. coli, 200 bp downstrm | ||
| CNO 179 | GTGGATTGGGAACCGAGCA | Sequencing primer for pCN 664 | ||
| CNO 180 | GGAGATCCCAGACTACTTCAAAC | Sequencing primer for pCN 664 | ||
| CNO 181 | gtcaagaccgacctgtcc | Sequencing primer for pCN 664 | ||
| CNO 182 | ggacatagcgttggctacc | Sequencing primer for pCN 664 | ||
| CNO 183 | caccaatttcatattgctgtaagtg | Sequencing primer for pCN 664 | ||
| CNO 223 | TCTTTCACTTCCAGGTTAATGGTG | Sequencing primer for pCN 754 | ||
| CNO 224 | CGTTTGTGATTGAAGGCGAAG | Sequencing primer for pCN 754 | ||
| CNO 225 | GTGTATGAAAGCGCGGTGG | Sequencing primer for pCN 754 | ||
| CNO 226 | GGAACGTATGTACGTTCGTGAC | Sequencing primer for pCN 754 | ||
| CNO 229 | CATAATCCACCTCCTTTACTGGTC | Sequencing primer for pCN 755 | ||
| CNO 230 | AAACCGTATGAAGGCACCC | Sequencing primer for pCN 755 | ||
| CNO 231 | AAAGAAACCTATGTGGAACAGCAT | Sequencing primer for pCN 755 | ||
| CNO 232 | CGAATGGTCCGGTTATGC | Sequencing primer for pCN 755 | ||
| CNO 173 | TTTGGATCCTCTAAGCTTCATcctag | Forward primer for amplification of IDT 2x-mKO E. coli optimized (kde1336 template) | ||
| CNO 174 | ctccagcctacactttGAATTCtttTCTAGAAAGGAGCTCatg | Reverse primer for kde1336 template with overlap to FRT-Kan from pKD4 | ||
| CNO 175 | GAATTCaaagtgtaggctggagctgc | Forward primer for FRT-Kan amplification from pKD4 template with overlap to IDT 2x-mKO from pNUT1336 | ||
| CNO 176 | ggaagaaatagcgcatgggaattagccatggtcc | Reverse primer for FRT-Kan from pKD4, with overlap to pSC101 from kde970 | ||
| CNO 177 | ctaattcccatgcgctatttcttccagaattgc | Forward primer for pSC101 from kde970 with overlap to FRT_Kan from pKD4 | ||
| CNO 178 | aaaGGATCCattggtgagaatccaagcactag | Reverse primer for amplifying pSC01 from kde970, with BamHI site for ligating to upstream fragment from pNUT1336 | ||
| CNO 189 | aaaGGATCCttgacaattaatcatcggctcgtataatgcctaggc CTAAGCTTCATcctaggGACAc | introduction of Ptac (no lacO) into new FP construct vector for E. coli | ||
| CNO 190 | GCTCGCGGTAATTTTTTCGG | for sequencing of Ptac inserted with CNO 189 |





