A unified platform to manage, share, and archive morphological and functional data in insect neuroscience
Figures
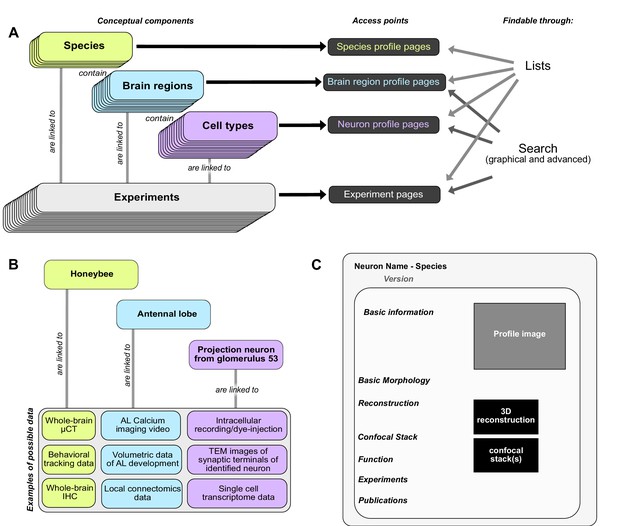
Basic concepts behind the Insect Brain Database.
(A) Organizational layers of the database. Elements in each layer are represented on their respective profile pages, which can be located either through lists or search interfaces. (B) Examples for information that can be deposited on each level of the database, illustrating how diverse data is automatically associated with hierarchical metadata. (C) Schematic illustration of a neuron profile page. Detailed anatomical and functional data is available on this page, from where experiments associated with this cell type are also linked. Similar pages exist for species and brain regions. Abbreviations: AL, antennal lobe; IHC, immunohistochemistry; TEM, transmission electron microscopy; μCT, micro computed tomography.
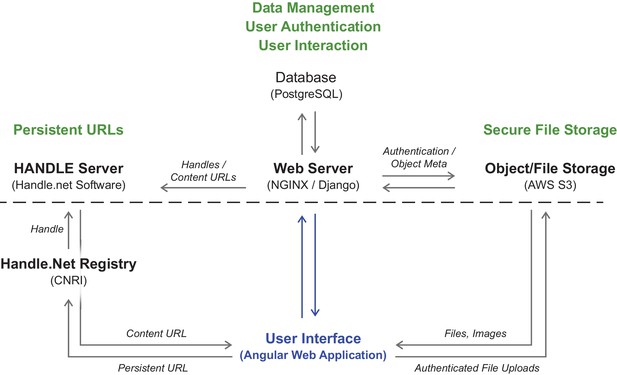
Technical overview of database layout.
Illustration of the interactions between the user interface, the web server, database, data storage and handles management.
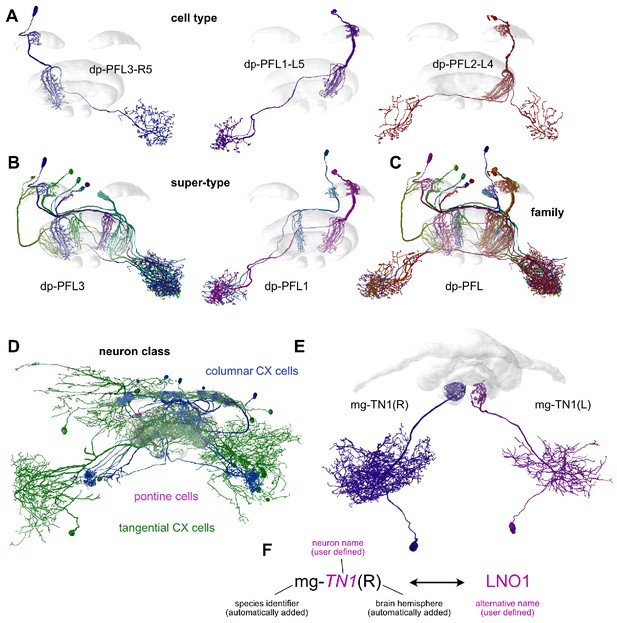
Definition of cell types.
Cell types are defined as all neurons that are morphologically indistinguishable. In many cases, a cell type comprises an individual neuron, while in other cases, several neurons with identical morphologies comprise a cell type. For highly organized brain regions, cell types can be grouped into higher order categories. (A). Three cell types of the Monarch butterfly central complex (CX), which differ in their innervated columns, overall morphology, detailed location of dendrites and axonal branching patterns. (B). Cell types that only differ in the innervated CX column are grouped into a super-type. (C). Several super-types can be grouped into a neuron family. (D). Illustration of neuron classes: CX neurons, for example in dung beetles can be grouped in three classes (columnar CX neurons, tangential CX neurons and pontine neurons). (E). Corresponding neurons in different brain hemispheres are treated as separate cell types, but are cross linked in the database. (F). The neuron name is composed of a species prefix, hemisphere suffix (both automatically generated) and a user defined neuron name. Alternative names can be assigned to the neuron, for example to refer to older naming schemes or names from different species. Those names are automatically queried as well, when searching for neuron names. Note that the hemisphere suffix is not used for columnar CX neurons, as hemisphere is already identified via the innervated CX column. Data from Heinze et al., 2013 (A–C), El Jundi et al., 2018 (D), and Stone et al., 2017 (E); all visualizations generated with Insectbraindb.org.
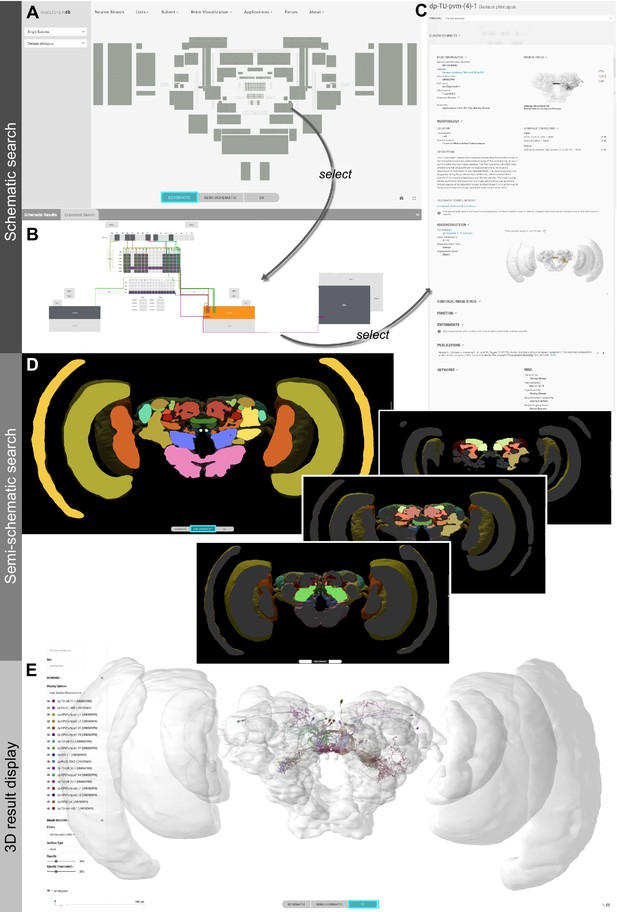
Neuron search in the IBdb.
(A) Screenshot of the schematic brain search interface (Monarch butterfly) in single species mode. Selecting a neuropil will reveal all neurons connected to that neuropil. (B) Schematic wiring diagram view of the search result; orange neuropil was queried. Selecting an individual neuron will reveal the profile page of that cell type. (C) Example of a neuron profile page with anatomical information. Confocal image stack and functional data are not listed in this example. (D) Semi-schematic search interface. The section view is scrollable and allows the user to query individual neuropils for connected neurons by clicking the cross section. The inserts show the results view at three levels of the brain. Neuropils connected to the queried neuropil are highlighted. Switching to the schematic view will then show the neurons as wiring diagram. Switching to the 3D mode will show registered neurons in 3D. (E) The 3D results viewer allows one to view all neurons registered into a common reference frame; example from Monarch butterfly (data from Heinze and Reppert, 2012) . The user can continuously switch between the three modes (schematic, semi-schematic, 3D). All visualizations generated with Insectbraindb.org.
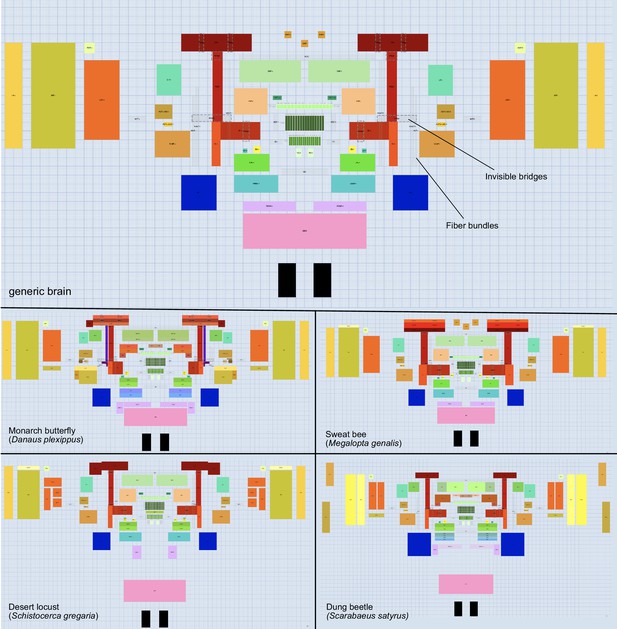
Schematic outline of generic brain and variations across species.
The brain builder module enables to generate the schematic brain models required for the graphical search functions. It allows the user to draw brain regions (tagged as either super-region, neuropil, or sub-region) as well as fiber bundles and invisible bridges. The latter provide shortcuts for neurons in the wiring diagram view (schematic search results), preventing overly convoluted neuron paths. The top panel shows the generic brain that is used as the template for all other species and which combines brain regions and fiber bundles shared by the majority of insect species. The bottom four panels illustrate the modifications of this template needed to account for species-specific features of four insect species examples.
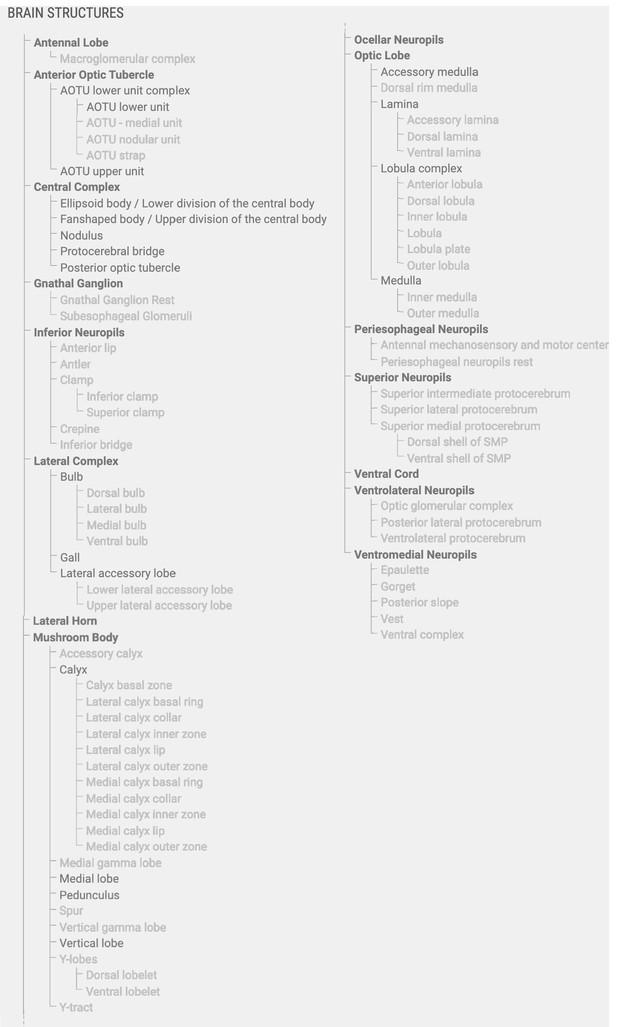
Brain structures in the database and their hierarchical organization.
Brain structures included in the insect brain database. All regions shown in black font are included in the generic insect brain that serves as a template for all other species and as a fall-back option for cross-species search. Bold regions are super-regions, which consist of neuropils, which in turn can be divided into sub-regions. Note that neuropils and subregions can be species-specific (gray font) and no single species contains all listed structures.
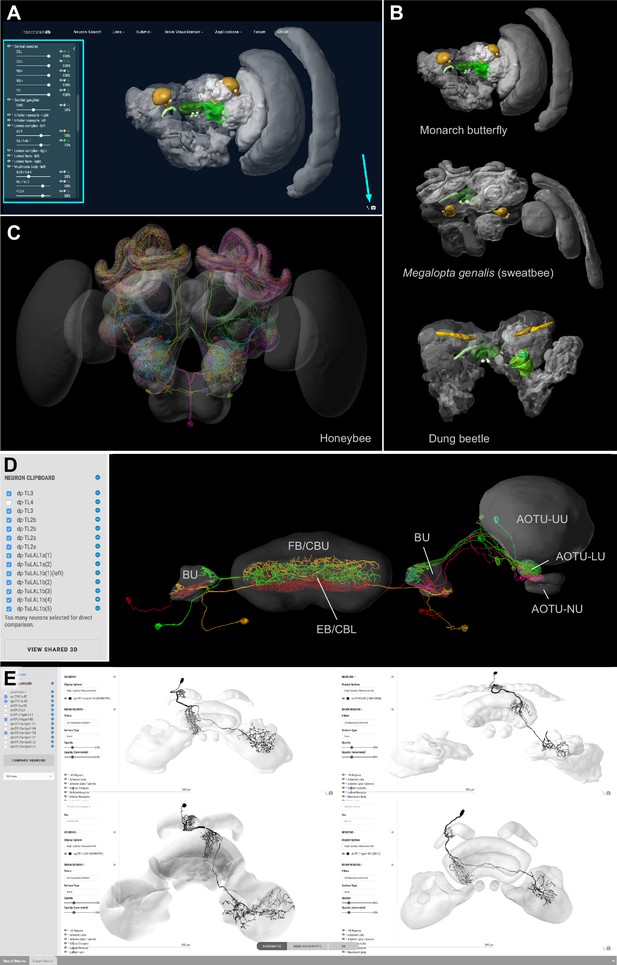
Visualization tools and applications.
(A) Screenshot illustrating the functionality of the 3D viewer in the insect brain database. Cyan arrow: Screenshot button. Cyan panel: Tools for adjusting appearance of neuropils. (B) Examples of neuropil images generated with the IBdb 3D viewer, illustrating navigation relevant neuropils in three insect species (Monarch butterfly [Heinze and Reppert, 2012], sweat bee Megalopta genalis [Stone et al., 2017], dung beetle [Immonen et al., 2017]). (C) Neurons associated with the antennal lobe of the honeybee, generated with the IBdb 3D neuron viewer (data from Rybak, 2012). (D) Elements in the neuron clipboard (left) can be arbitrarily combined and displayed in the 3D viewer to highlight neural pathways and circuits. Shown are two parallel input pathways from the anterior optic tubercle to the ellipsoid body of the central complex in the Monarch butterfly (data from Heinze et al., 2013). (E) Side-by-side neuron viewer. Screenshot showing comparison of 3D skeletons of CPU1 (PFL) neurons from four species (top left: Monarch butterfly [Heinze et al., 2013]; bottom left: desert locust [El Jundi et al., 2009b]; top right: Dung beetle [el Jundi et al., 2015]; bottom right: Bogong moth [de Vries et al., 2017]). All visualizations generated with Insectbraindb.org.
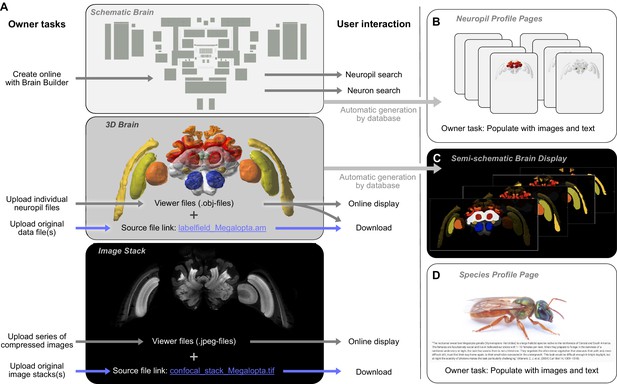
Contributing a species to the IBdb.
(A) Three main elements have to be created for each new species: the schematic brain, the 3D brain, and an image stack. The schematic brain is generated directly on the IBdb website using the 'Brain Builder', while the other two elements are uploaded. For each, both source files and viewer files are needed. Viewer files are used for online display, while source files can be downloaded by users. (B) Neuropil profile pages are automatically generated when creating the schematic brain. They have to be populated with images and texts by the user. (C) The semi-schematic brain is automatically generated based on the provided 3D brain. (D) The species profile page must be populated with images, texts and a bibliography to provide context for the species. Visualizations in A,B,c generated with Insectbraindb.org, Megalopta genalis data from Stone et al., 2017.
Photograph in D reproduced with permission from Ajay Narendra.
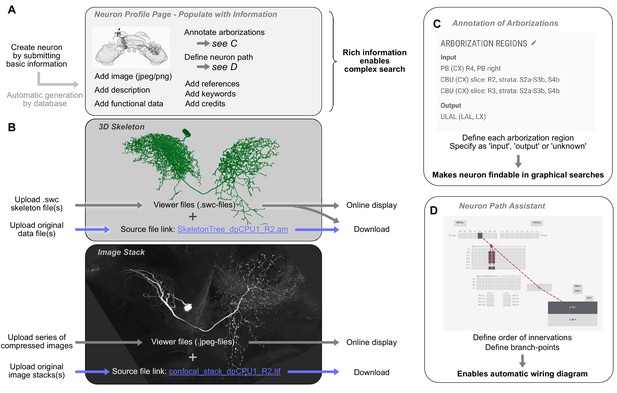
Contributing a neuron type to the IBdb.
(A) A new neuron type can be created by submitting a neuron form containing basic information. This generates a neuron profile page that then has to be populated with information by the owner. Each entry is findable by the expanded search function. (B) 3D-skeletons are added as swc-files (online display) and source files (download). Confocal image stacks are uploaded as jpeg series for online display and as original data files for download. (C) All arborization regions of the neuron must be defined (at the level specified in the species' schematic brain) and labeled as either input, output, or unknown polarity. (D) To enable automatic drawing of wiring diagrams in schematic search results, the order of innervation of neuropils and the branch-points of the neuron must be defined using the path assistant. All visualizations generated with Insectbraindb.org.
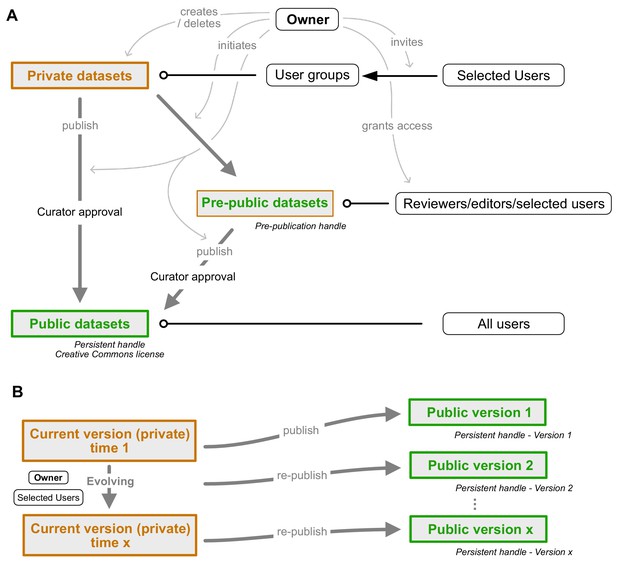
Dataset publication concept.
(A) Interconnection of private, public, and pre-public datasets. Private datasets can be viewed and edited by members of user groups with access granted to a particular dataset. Pre-public datasets can be viewed by anyone in possession of the pre-publication handle (distributed by the data owner, e.g. within a manuscript). Public datasets can be accessed by all users. Publication of data cannot be undone as persistent handles are generated. Gray arrows indicate control actions employed by the dataset owner. (B) Re-publication strategy of evolving datasets. A current version of each public dataset remains present in the owner's private database mode and can be edited at wish. Once sufficient updates have accumulated, the dataset can be re-published. A new version of the persistent handle is assigned and the now public dataset (version 2) becomes locked. Datasets can be edited by the owner and anyone who has been granted permission to edit by the owner.
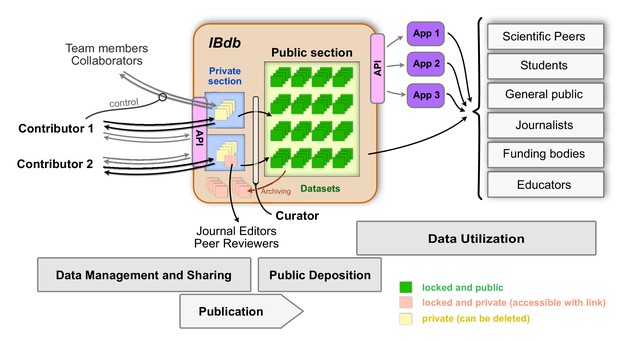
The Insect Brain Database and the possible interactions between users and deposited data.
The private sections of the database are accessible to only the owner of the data, and datasets within this section can be shared with team members and collaborators. Up- and download of these data are possible either directly or via an application programming interface (API). As these datasets are unlocked, they can continuously be updated and also be deleted. Upon publication and curator approval, datasets become locked (persistent) and are deposited in the public section of the database. As an intermediate step, datasets can be pre-published (locked but private) and made available to journal editors and peer reviewers when including datasets in manuscripts of journal articles. Data in the public section of the database are accessible directly for all interested users (relevant user groups are shown on the right). Additionally, an API also allows automated access of public data, which can therefore be used by third party applications (illustrated as ’App 1–3’) for generating specific user experiences with additional capabilities, for instance in the context of teaching. To remove obsolete datasets from the public domain, they can be archived, a process that preserves persistence but prevents datasets from being findable without the explicit handle.
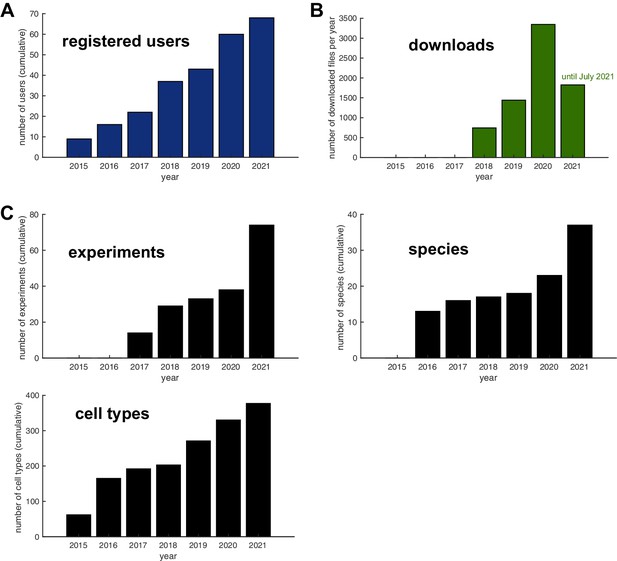
Database usage statistics.
Usage statistics of the IBdb since its initial launch in 2015. (A) Number of registered user accounts (cumulative). (B) Number of downloaded files for each year since tracking of download was implemented in 2018. Note that for 2021 only data until July could be included before submission of this publication. (C) Number of deposited datasets, for Experiments, Species, and Neuron types for each year (cumulative). Note that usage of the database has grown steadily during the development phase since its launch in 2015 and that most substantial growth in data deposition has occurred in 2020 and 2021, that is, since the deposition of the preprint version of this publication.





