Seroconversion stages COVID19 into distinct pathophysiological states
Figures
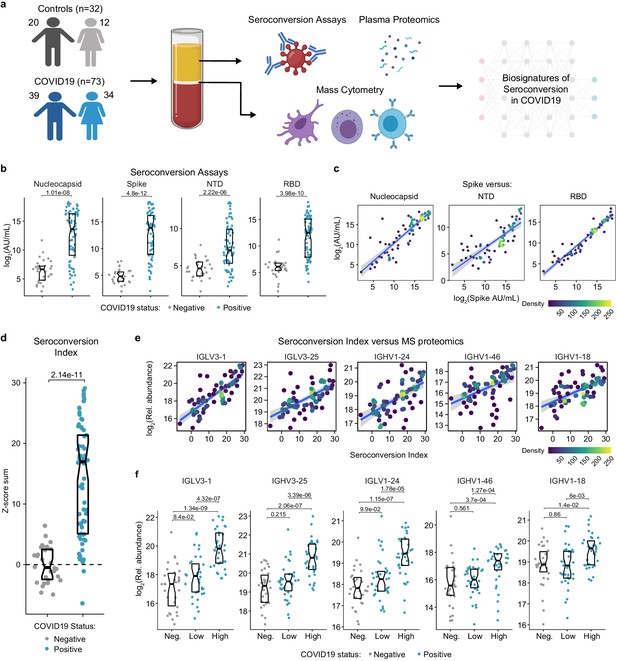
Highly variable seroconversion status among hospitalized COVID19 patients.
(a) Overview of experimental approach. Blood samples from 105 research participants, 73 of them with COVID19, were analyzed by matched multiplex immunoassays for detection of antibodies against SARS-CoV-2, plasma proteomics using mass spectrometry (MS), SOMAscan proteomics, and cytokine profiling using Meso Scale Discovery (MSD) technology. Data was then analyzed to define biosignatures of seroconversion. (b) Multiplex immunoassays were used to measure antibodies against the SARS-CoV-2 nucleocapsid and spike proteins, as well as specific peptides encompassing the N-terminus domain (NTD) and receptor-binding domain (RBD) of the spike protein. Data are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers above brackets are p-values for Mann–Whitney tests. (c) Scatter plots showing correlations between antibodies against the full-length spike protein versus antibodies against the nucleocapsid, NTD, and RBD domains. Points are colored by density; lines represent linear model fit with 95% confidence interval. (d) Seroconversion indices were calculated for each research participant by summing the Z-scores for each of the four seroconversion assays. Z-scores were calculated from the adjusted concentration values for each epitope in each sample, based on the mean and standard deviation of COVID19-negative samples. (e) Scatter plots displaying the top five correlations between seroconversion indices and proteins detected in the MS proteomics data set among COVID19 patients. Points are colored by density; lines represent linear model fit with 95% confidence interval. (f) Sina plots showing values for the top five proteins correlated with seroconversion comparing the control cohort (Negative, Neg.) to COVID19 patients divided into seroconversion low and high status. Data are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers above brackets are q-values for Mann–Whitney tests. See also Figure 1—figure supplement 1.
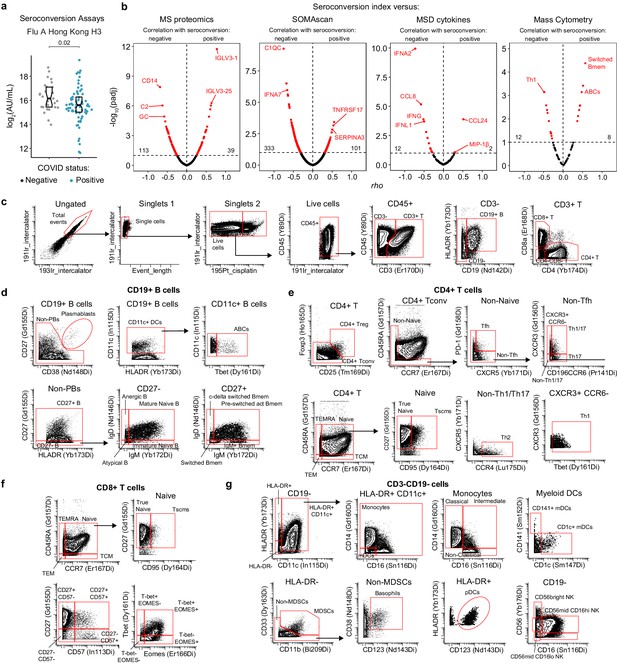
Biosignatures of seroconversion among hospitalized COVID19 patients.
(a) Meso Scale Discovery (MSD) assays show no elevation in levels of circulating antibodies against the Flu A Hong Kong H3 virus strain among COVID19 patients. Data are presented as modified Sina plots with boxes indicating median and interquartile range. Number above brackets is p-value for Mann–Whitney test. (b) Volcano plots for Spearman correlations between seroconversion indices and circulating plasma proteins detected by mass spectrometry (MS), SOMAscan assays, MSD assays, as well as immune cell subsets detected by mass cytometry (MC) among all live cells. X axes show Spearman rho values. Y axes show −log10 p-values adjusted with Benjamini–Hochberg method. Dashed vertical line indicates rho = 0. Dashed horizontal line indicates the statistical cut off of false discovery rate (FDR) = 10 (q = 0.1). (c–g) Representation of gating strategy employed during MC analysis of peripheral immune cell lineages. In (c), single live cells were gated for CD45+ staining followed by gating into T cells (CD3+), B cells (CD3- CD19+), and CD4+ and CD8+ T cells subsets. In (d), B cells were further gated into the indicated subsets. In (e) and (f), CD4+ and CD8+ T cell lineages were further characterized in the indicated subsets. Panel (g) shows gating for the indicated myeloid subsets and natural killer (NK) cells.
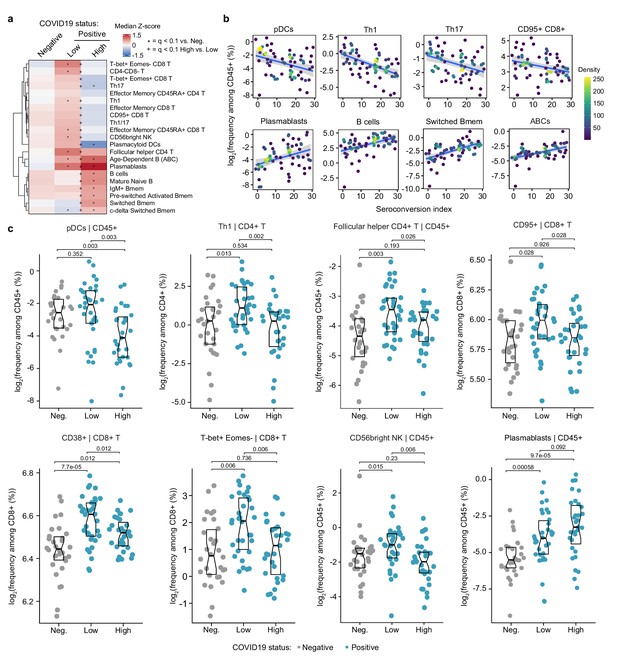
Seroconversion associates with significant changes in peripheral immune cell frequencies.
(a) Heatmap representing changes in the frequency of immune cell subsets that are significantly correlated, either positively or negatively with seroconversion status. Values displayed are median Z-scores, derived from cell frequencies among all CD45+ cells, for each cell subset for controls (negative, Neg.) versus COVID19 patients divided into seroconversion low (Low) and high (High) status. Z-scores were calculated from the adjusted frequency values for each cell type in each sample, based on the mean and standard deviation of COVID19-negative samples. Asterisks indicate a significant difference relative to the control COVID19-negative group, and the + symbols indicate a significant difference between sero-low and sero-high groups after multiple hypothesis correction (q < 0.1, Mann–Whitney test). (b) Scatter plots for indicated immune cell types significantly correlated with seroconversion indices among COVID19 patients. Points are colored by density; lines represent linear model fit with 95% confidence interval. (c) Sina plots showing values for indicated immune cell types significantly correlated with seroconversion indices among COVID19 patients. The parent cell lineage is indicated in the header and Y axis label for each plot. Data are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers above brackets are q-values for Mann–Whitney tests. See also Figure 2—figure supplement 1.
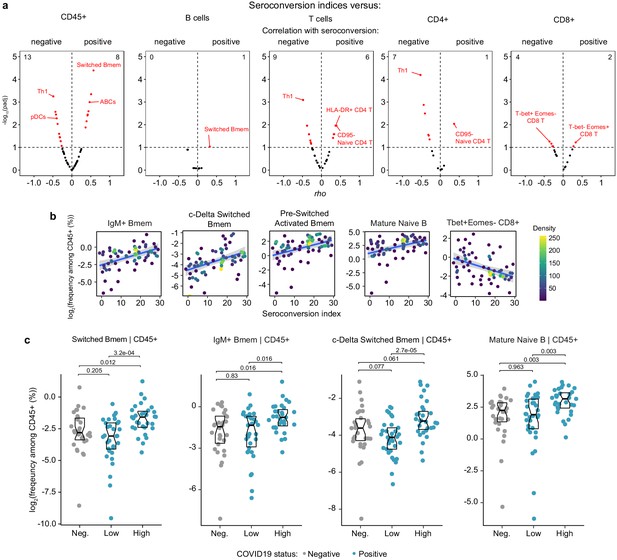
Immune cell signatures of seroconversion.
(a) Volcano plots displaying the correlations between seroconversion indices and circulating levels of immune cell subsets detected by mass cytometry (MC). X axes show Spearman rho values. Y axes show −log10 p-values adjusted with Benjamini–Hochberg method. Dashed vertical line indicates rho = 0. Dashed horizontal line indicates a false discovery rate (FDR) threshold of 10% (q = 0.1). Correlations were calculated for immune cell subsets measured as frequencies among all live CD45+ cells (far left), all B cells, all T cells, CD4+ T cells, and CD8+ T cells (far right). (b) Scatter plots for indicated cell types against seroconversion indices. Values shown are derived from frequency among all CD45+ cells. Points are colored by density; lines represent linear model fit with 95% confidence interval. (c) Sina plots for indicated immune cell types comparing controls (Negative, Neg.) to COVID19 patients divided into seroconversion low (Low) and high (High) status. Data are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers above brackets are q-values for Mann–Whitney tests.
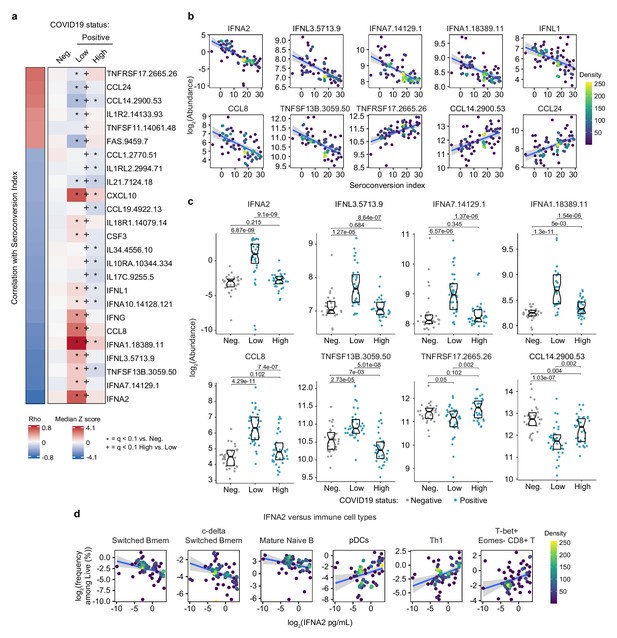
Seroconversion is associated with decreased interferon signaling.
(a) Heatmap displaying changes in circulating levels of immune factors that are significantly correlated, either positively or negatively, with seroconversion status. The left column represents Spearman rho values, while the right columns display median Z-scores for each immune factor for controls (negative, Neg.) versus COVID19 patients divided into seroconversion low (Low) and high (High) status. Factors are ranked from most positively correlated (top, high rho values) to most anti-correlated (bottom, low rho values) with seroconversion index. Z-scores were calculated from the adjusted concentration values for each immune factor in each sample, based on the mean and standard deviation of COVID19-negative samples. Asterisks indicate a significant difference relative to the control COVID19-negative group, and the + symbols indicate a significant difference between sero-low and sero-high groups (q < 0.1, Mann–Whitney test). (b) Scatter plots for indicated immune factors significantly correlated with seroconversion indices among COVID19 patients. Points are colored by density; lines represent linear model fit with 95% confidence interval. (c) Sina plots showing values for immune factors correlated with seroconversion comparing controls (Neg.) to COVID19 patients divided into seroconversion low and high status. Data are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers above brackets are q-values for Mann–Whitney tests. (d) Scatter plots showing correlations between circulating levels of IFNA2 measured by MSD and the indicated cell types measured by mass cytometry. Values for immune cells correspond to frequency among all live cells. Points are colored by density; lines represent linear model fit with 95% confidence interval. See also Figure 3—figure supplement 1.
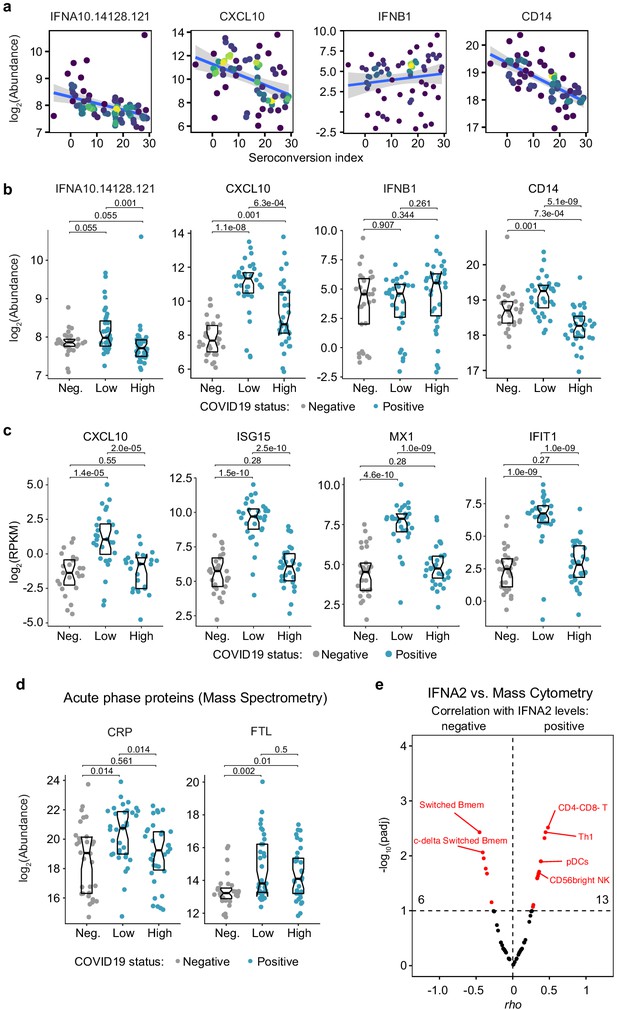
Seroconversion associates with differential abundance of circulating immune factors.
(a and b) XY scatter plots (a) and Sina plots (b) for select circulating immune factors. Points in (a) are colored by density; lines represent linear model fit with 95% confidence interval. Data in (b) are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers above brackets are q-values for Mann–Whitney tests. (c) Sina plots showing expression of select IFN-inducible mRNAs in a whole blood transcriptome analysis comparing controls (Neg., negative) to COVID19 patients divided into seroconversion low (Low) and high (High) status. Data are presented as in b. (d) Sina plots for the acute phase proteins CRP and FTL (Ferritin Light Chain) detected by MS comparing controls (Neg., negative) to COVID19 patients divided into seroconversion low (Low) and high (High) status. Data are presented as in b. (e) Volcano plot showing associations between circulating levels of IFNA2 measured by MSD versus immune cell subsets among all live peripheral blood mononuclear cells. X axes show Spearman rho values. Y axes show −log10 p-values adjusted with Benjamini–Hochberg method. Dashed vertical line indicates rho = 0. Dashed horizontal line indicates a false discovery rate (FDR) threshold of 10% (q < 0.1).
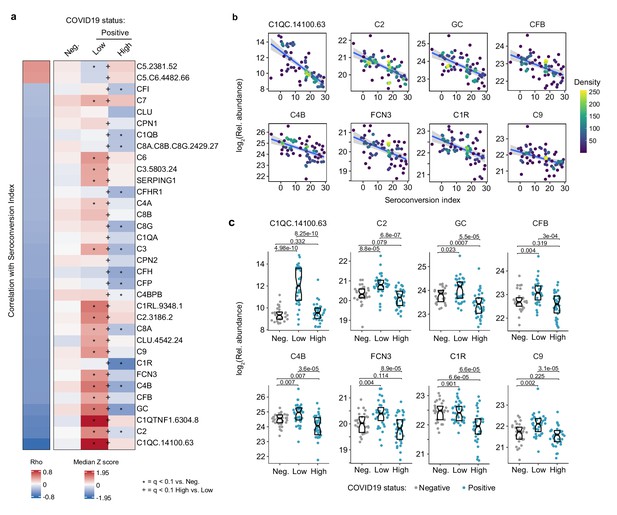
Seroconversion correlates with decreased markers of systemic complement activation.
(a) Heatmap displaying changes in circulating levels of components of the various complement pathways that are significantly correlated, either positively or negatively, with seroconversion status. The left column represents Spearman rho values, while the right columns display median Z-scores for each complement factor for controls (negative, Neg.) versus COVID19 patients (positive) divided into seroconversion low (Low) and high (High) status. Factors are ranked from most positively correlated (top, high rho values) to most anti-correlated (bottom, low rho values) with seroconversion status. Z-scores were calculated from the adjusted concentration values for each analyte in each sample, based on the mean and standard deviation of COVID19-negative samples. Asterisks indicate a significant difference relative to the control COVID19-negative group, and the + symbols indicate a significant difference between sero-low and sero-high groups (q < 0.1, Mann–Whitney test). (b) Scatter plots for indicated complement factors significantly correlated with seroconversion indices among COVID19 patients. Points are colored by density; lines represent linear model fit with 95% confidence interval. (c) Sina plots showing values for complement factors correlated with seroconversion comparing controls (Negative, Neg.) to COVID19 patients divided into seroconversion low (Low) and high (high) status. Data are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers above brackets are q-values for Mann–Whitney tests. See also Figure 4—figure supplement 1.
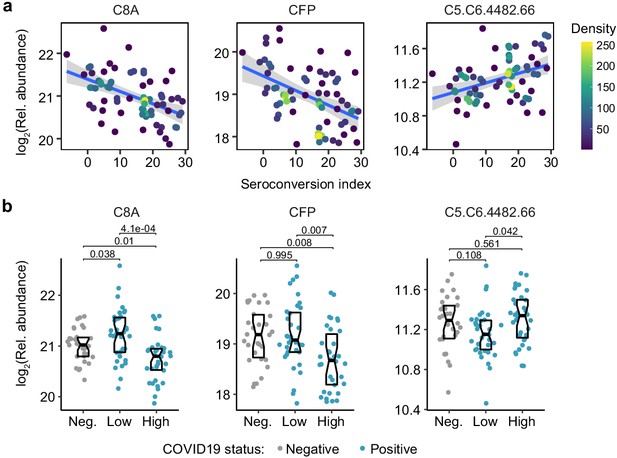
Seroconversion associates with decreased markers of systemic complement activation.
(a and b) XY scatter plots (a) and Sina plots (b) for select complement factors significantly associated, either positively or negatively, with seroconversion indices among COVID19 patients. Points in (a) are colored by density; lines represent linear model fit with 95% confidence interval. Data in (b) are presented as modified Sina plots with boxes indicating median and interquartile range comparing controls (Neg., negative) to COVID19 patients divided into seroconversion low (Low) and high (High) status. Numbers above brackets are q-values for Mann–Whitney tests.
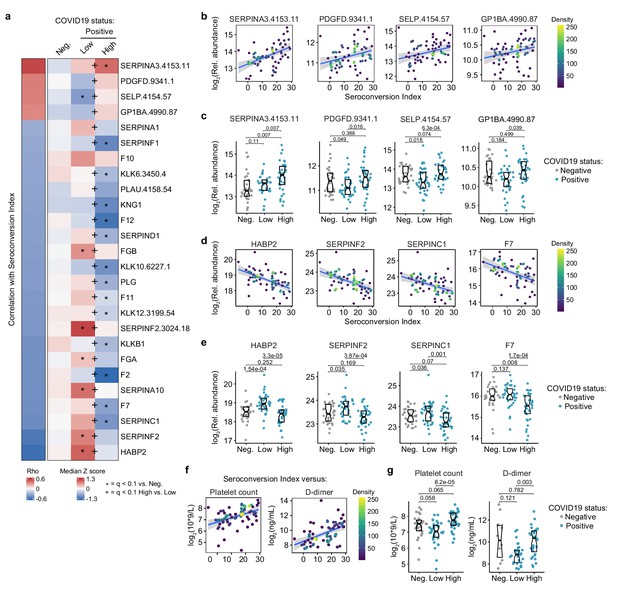
Seroconversion associates with remodeling of the hemostasis control network.
(a) Heatmap displaying changes in circulating levels of known modulators of hemostasis that are significantly correlated, either positively or negatively, with seroconversion status. The left column represents Spearman rho values, while the right columns display row-wise Z-scores for each factor for controls (negative, Neg.) versus COVID19 patients divided into seroconversion low (Low) and high (High) status. Factors are ranked from most positively correlated (top, high rho values) to most anti-correlated (bottom, low rho values) with seroconversion status. Z-scores were calculated from the adjusted concentration values for each analyte in each sample, based on the mean and standard deviation of COVID19-negative samples. Asterisks indicate a significant difference relative to the control COVID19-negative group, and the + symbols indicate a significant difference between sero-low and sero-high groups (q < 0.1, Mann–Whitney test). (b and c) Scatter plots (b) and Sina plots (c) for factors positively correlated with seroconversion indices. (d and e) Scatter plots (d) and Sina plots (e) for factors negatively correlated with seroconversion indices. (f and g) Scatter plot (f) and Sina plots (g) displaying the correlations between seroconversion index and platelet counts and D-dimer values obtained from clinical laboratory testing. Points in (b), (d), and (f) are colored by density; lines represent linear model fit with 95% confidence interval. Data in (c), (e), and (g) are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers above brackets are q-values for Mann–Whitney tests. See also Figure 5—figure supplement 1.
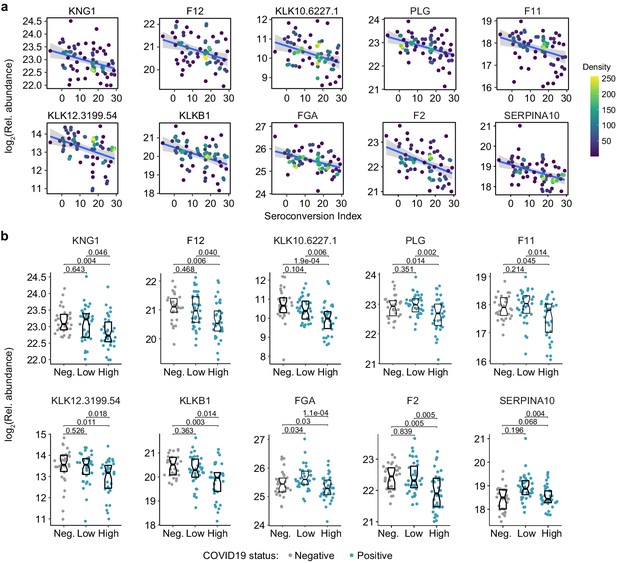
Seroconversion associates with remodeling of the hemostasis network.
(a and b) XY scatter plots (a) and Sina plots (b) for select factors involved in control of hemostasis significantly associated, either positively or negatively, with seroconversion indices among COVID19 patients. Points in (b) are colored by density; lines represent linear model fit with 95% confidence interval. Data in (b) are presented as modified Sina plots with boxes indicating median and interquartile range comparing controls (Neg., negative) to COVID19 patients divided into seroconversion low (Low) and high (High) status. Numbers above brackets are q-values for Mann–Whitney tests.
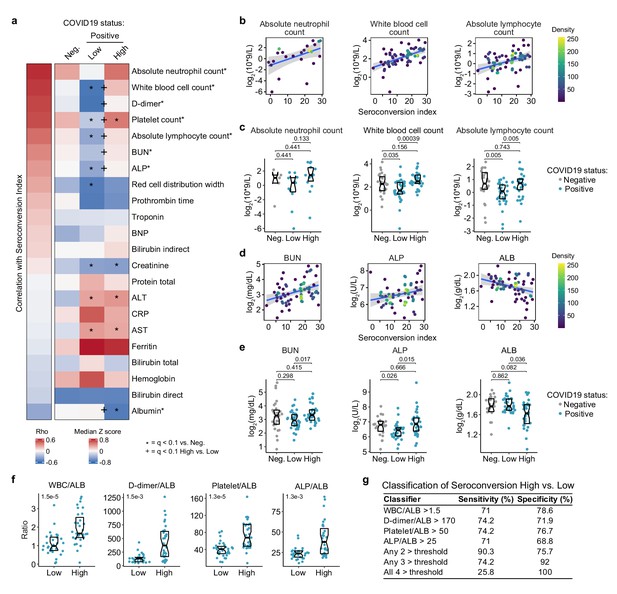
Seroconversion associates with recovery in blood cell numbers and hypoalbuminemia.
(a) Heatmap displaying correlations between clinical laboratory values and seroconversion status. The left column represents Spearman rho values, while the right columns display row-wise Z-scores for each variable for controls (negative, Neg.) versus COVID19 patients divided into seroconversion low (Low) and high (High) status. Measures are ranked from most positively correlated (top, high rho values) to most anti-correlated (bottom, low rho values) with seroconversion status. Asterisks after the clinical parameter name indicate a significant correlation. Z-scores were calculated from the adjusted concentration values for each analyte in each sample, based on the mean and standard deviation of COVID19-negative samples. Asterisks indicate a significant difference relative to the control COVID19-negative group, and the + symbols indicate a significant difference between sero-low and sero-high groups (q < 0.1, Mann–Whitney test). (b–e) Scatter plots (b and c) and Sina plots (c and e) for indicated clinical laboratory values significantly correlated with seroconversion indices among COVID19 patients. In b and c, points are colored by density; lines represent linear model fit with 95% confidence interval. In c and e, Sina plots show values for clinical laboratory tests correlated with seroconversion comparing controls (Negative, Neg.) to COVID19 patients divided into seroconversion low (Low) and high (High) status. Data are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers above brackets are q values for Mann–Whitney tests. (f) Differences in the indicated ratios of clinical laboratory values between sero-low and sero-high COVID19 patients. Data are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers at upper left of each plot are p-values for Mann–Whitney tests. (g) Table showing how the indicated ratios of the specified clinical values could be potentially used to gauge the seroconversion status of a hospitalized patient with moderate pathology. The units employed for calculating these ratios are 103/mcL for white blood cells (WBC) and platelets; g/dL for albumin (ALB); ng/mL for D-dimer; and U/L for alkaline phosphatase (ALP). ALT: alanine aminotransferase, AST: aspartate aminotransferase; BUN: blood urea nitrogen; BNP: brain natriuretic peptide; CRP: C-reactive protein. See also Figure 6—figure supplement 1.
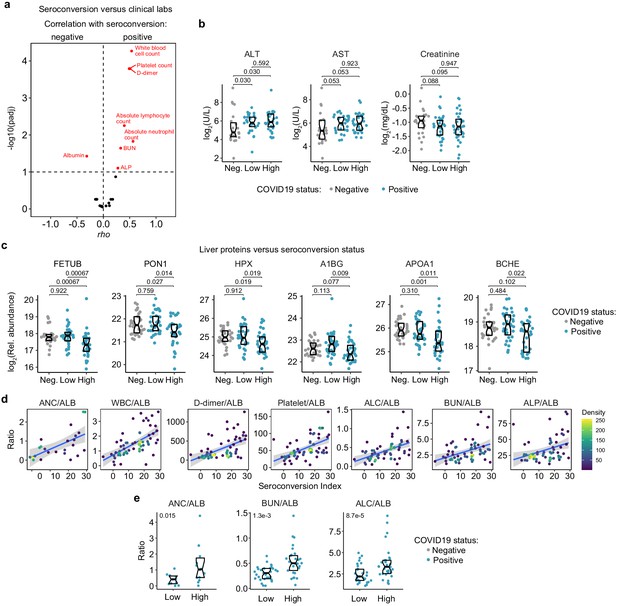
Seroconversion associates with recovery of blood cell counts and hypoalbuminemia.
(a) Volcano plot displaying the correlations between seroconversion indices and clinical laboratory values. X axes show Spearman rho values. Y axes show −log10 p-values adjusted with Benjamini–Hochberg method. Dashed vertical line indicates rho = 0. Dashed horizontal line indicates a false discovery rate (FDR) threshold of 10% (q < 0.1). (b) Sina plots for select clinical laboratory values. Data are presented as modified Sina plots with boxes indicating median and interquartile range comparing controls (Neg., negative) to COVID19 patients divided into seroconversion low (Low) and high (High) status. Numbers above brackets are q-values for Mann–Whitney tests. (c) Sina plots showing values for liver proteins detected by MS that are significantly correlated with seroconversion comparing controls (Negative, Neg.) to COVID19 patients divided into seroconversion low (Low) and high (High) status. Data are presented as modified Sina plots with boxes indicating median and interquartile range. Numbers at upper left of each plot are p-values for Mann–Whitney tests. (d) Scatter plots showing the correlation between the indicated ratios of clinical values and the seroconversion index. ALB: albumin; ALC: absolute lymphocyte count; ANC: absolute neutrophil count; ALT: alanine aminotransferase; AST: aspartate aminotransferase; BUN: blood urea nitrogen, WBC: white blood cell count. (e) Sina plots for indicated clinical laboratory ratios. Data are presented as modified Sina plots with boxes indicating median and interquartile range comparing controls (Neg., negative) to COVID19 patients divided into seroconversion low (Low) and high (High) status. Numbers at upper right of each plot are q-values for Mann–Whitney tests.
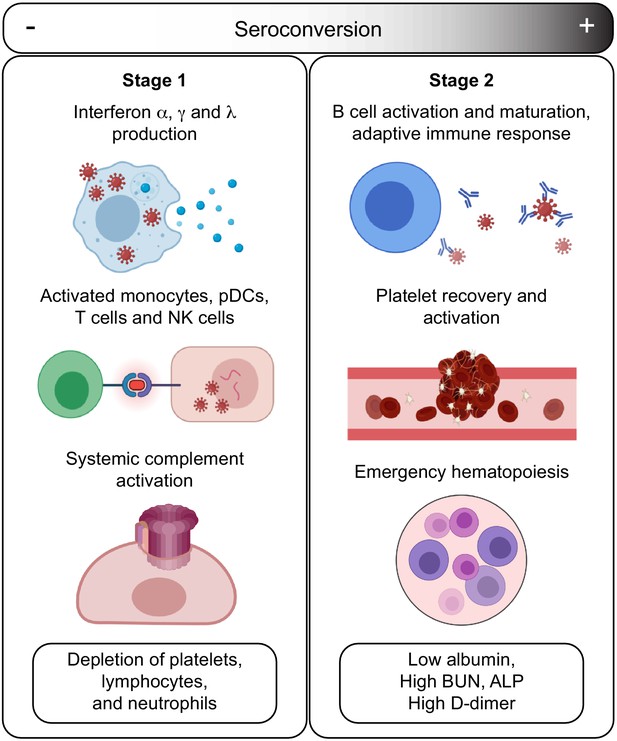
Model for staging COVID19 pathophysiology based on seroconversion status.
Stage 1 applies to COVID19 patients with low degree of seroconversion and involves high levels of circulating IFNs, signs of strong systemic complement activation, hyperactive T cells, activated monocytes, and cytokine-producing NK cells, as well as depletion of key blood cell types. Stage 2 applies to COVID19 patients with high degree of seroconversion and is characterized by increased blood cell numbers, increased levels of markers of platelet degranulation, elevated D-dimer, and markers of increased liver dysfunction and/or interstitial leakage.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-Human CD45 | Fluidigm | Cat# 3089003B, RRID:AB_2661851 | Monoclonal-Clone: HI30 Dilution: 1/200 |
| Antibody | Anti-Human CD57 | Biolegend | Cat# 322302, RRID:AB_2661815 | Mouse-Monoclonal-Clone: HCD57 Dilution: 1/100 |
| Antibody | Anti-Human CD11c | BD bioscience | Cat# 555390, RRID:AB_395791 | Mouse-Monoclonal-Clone: B-ly6 Dilution: 1/100 |
| Antibody | Anti-Human CD16 | eBioscience | Cat# 16-0167-85, RRID:AB_11040983 | Mouse-Monoclonal-Clone: B73.1 Dilution: 1/50 |
| Antibody | Anti-Human CD196 (CCR6) | Biolegend | Cat# 353402, RRID:AB_10918625 | Mouse-Monoclonal-Clone: 11a9 Dilution: 1/50 Stain live |
| Antibody | Anti-Human CD19 | Fluidigm | Cat# 3142001B, RRID:AB_2651155 | Monoclonal-Clone: HIB19 Dilution: 1/100 |
| Antibody | Anti-Human CD123 | Fluidigm | Cat# 3143014B, RRID:AB_2811081 | Mouse-Monoclonal-Clone: 6H6 Dilution: 1/123 |
| Antibody | Anti-Human CCR5 | Fluidigm | Cat# 3144007A | Monoclonal-Clone: NP6G4 Dilution: 1/25 Stain live |
| Antibody | Anti-Human IgD | Fluidigm | Cat# 3146005B, RRID:AB_2811082 | Mouse-Monoclonal-Clone: IA6-2 Dilution: 1/100 |
| Antibody | Anti-Human CD1c | Miltenyi | Cat# 130-108-032, RRID: AB_2661165 | Mouse-Monoclonal-Clone: AD5-8E7 Dilution: 1/30 |
| Antibody | Anti-Human CD38 | Biolegend | Cat# 303502, RRID:AB_314354 | Mouse-Monoclonal-Clone: HIT2 Dilution: 1/50 |
| Antibody | Anti-Human CD127 | Fluidigm | Cat# 3149011B, RRID:AB_2661792 | Monoclonal-Clone: A019D5 Dilution: 1/100 Stain live |
| Antibody | Anti-Human CD86 | Fluidigm | Cat# 3150020B, RRID:AB_2687852 | Monoclonal-Clone: IT2.2 Dilution: 1/100 |
| Antibody | Anti-Human ICOS | Biolegend | Cat# 313502, RRID:AB_416326 | Armenian hamster -Monoclonal-Clone: DX29 Dilution: 1/50 |
| Antibody | Anti-Human CD141 | Biolegend | Cat# 344102, RRID:AB_2201808 | Mouse-Monoclonal-Clone: M80 Dilution: 1/50 |
| Antibody | Anti-Human Tim3 | Fluidigm | Cat# 3153008B, RRID:AB_2687644 | Monoclonal-Clone: MBSA43 Dilution: 1/100 |
| Antibody | Anti-Human TIGIT | Fluidigm | Cat# 3154016B, RRID:AB_2888926 | Mouse-Monoclonal-Clone: F38-2E2 Dilution: 1/50 Stain live |
| Antibody | Anti-Human CD27 | Fluidigm | Cat# 3155001B, RRID:AB_2687645 | Mouse-Monoclonal-Clone: L128 Dilution: 1/100 |
| Antibody | Anti-Human CXCR3 | Fluidigm | Cat# 3156004B, RRID:AB_2687646 | Monoclonal-Clone: G025H7 Dilution: 1/100 Stain live |
| Antibody | Anti-Human CD45RA | Biolegend | Cat# 304102, RRID:AB_314406 | Mouse-Monoclonal-Clone: HI100 Dilution: 1/50 |
| Antibody | Anti-Human PD-1 | Biolegend | Cat# 329941, RRID:AB_2563734 | Mouse-Monoclonal-Clone: EH12.2H7 Dilution: 1/50 |
| Antibody | Anti-Human PDL1 | Fluidigm | Cat# 3159029B, RRID:AB_2861413 | Mouse-Monoclonal-Clone: 29E.2A3 Dilution: 1/100 |
| Antibody | Anti-Human CD14 | Fluidigm | Cat# 3160001B, RRID:AB_2687634 | Monoclonal-Clone: M5E2 Dilution: 1/100 |
| Antibody | Anti-Human Tbet | Fluidigm | Cat# 3161014B, RRID:AB_2858233 | Monoclonal-Clone: 4b10 Dilution: 1/100 |
| Antibody | Anti-Human Ki67 | Fluidigm | Cat# 3162012B, RRID:AB_2888928 | Mouse-Monoclonal-Clone: B56 Dilution: 1/100 |
| Antibody | Anti-Human CD33 | Fluidigm | Cat# 3163023B, RRID:AB_2687857 | Monoclonal-Clone: WM53 Dilution: 1/100 |
| Antibody | Anti-Human CD95 | Fluidigm | Cat# 3164008B, RRID:AB_2858235 | Monoclonal-Clone: DX2 Dilution: |
| Antibody | Anti-Human Foxp3 | Biolegend | Cat# 14-4774-82, RRID:AB_467552 | Mouse-Monoclonal-Clone: 150D/E4 Dilution: 1/50 |
| Antibody | Anti-Human Eomes | Biolegend | Cat# 14-4877-82, RRID:AB_2572882 | Mouse-Monoclonal-Clone: WD1928 Dilution: 1/100 |
| Antibody | Anti-Human CCR7 | Fluidigm | Cat# 3167009A, RRID:AB_2858236 | Monoclonal-Clone: G043H7 Dilution: 1/100Stain live |
| Antibody | Anti-Human CD8a | Fluidigm | Cat# 3168002B | Monoclonal-Clone: SK1 Dilution: 1/100 |
| Antibody | Anti-Human CD25 | Fluidigm | Cat# 3169003B, RRID:AB_2661806 | Monoclonal-Clone: 2A3 Dilution: 1/100 Stain live |
| Antibody | Anti-Human CD3 | Fluidigm | Cat# 3170001B, RRID:AB_2811085 | Mouse-Monoclonal-Clone: UCHT1 Dilution: 1/100 |
| Antibody | Anti-Human CXCR5 | Fluidigm | Cat# 3171014B, RRID:AB_2858239 | Monoclonal-Clone: 51505 Dilution: 1/100 Stain live |
| Antibody | Anti-Human IgM | Fluidigm | Cat# 3172004B, RRID:AB_2810858 | Mouse-Monoclonal-Clone: MHM-88 Dilution: 1/100 Stain live |
| Antibody | Anti-Human HLA-DR | Fluidigm | Cat# 3173005B, RRID:AB_2810248 | Monoclonal-Clone: L243 Dilution: 1/100 |
| Antibody | Anti-Human CD4 | Fluidigm | Cat# 3174004B, RRID:AB_2687862 | Monoclonal-Clone: SK3 Dilution: 1/100 |
| Antibody | Anti-Human CCR4 | R and D | Cat# MAB1567-500 | Mouse-Monoclonal-Clone: 205410 Dilution: 1/50 Stain live |
| Antibody | Anti-Human CD56 | Miltenyi | Cat# 130-113-312, RRID:AB_2726090 | Monoclonal-Clone: HCD56 Dilution: 1/200 |
| Antibody | Anti-Human CD11b | Fluidigm | Cat# 3209003B, RRID:AB_2687654 | Monoclonal-Clone: ICRF44 Dilution: 1/200 |
| Commercial assay or kit | U-PLEX Biomarker Group 1 (hu) 71-Plex | Meso Scale Discovery (MSD) | Cat# K15081K | |
| Commercial assay or kit | V-PLEX Vascular Injury Panel 2 Human Kit | Meso Scale Discovery (MSD) | Cat# K15198D | |
| Commercial assay or kit | V-PLEX Angiogenesis Panel 1 Human Kit | Meso Scale Discovery (MSD) | Cat# K15190D | |
| Commercial assay or kit | PAXgene Blood RNA Tubes | PreAnalytiX/Qiagen | Cat# 762165 | |
| Commercial assay or kit | PAXgene Blood RNA Kit | Qiagen | Cat# 762164 | |
| Commercial assay or kit | Universal Plus mRNA-Seq with NuQuant, Human Globin AnyDeplete | Tecan | Cat# 0521-A01 | |
| Software, algorithm | R | R Foundation for Statistical Computing | v4.0.1 RRID:SCR_001905 | https://www.R-project.org/ |
| Software, algorithm | RStudio | RStudio, Inc | v1.3.959 RRID:SCR_000432 | http://www.rstudio.com/ |
| Software, algorithm | Bioconductor | N/A | v3.11 RRID:SCR_006442 | https://bioconductor.org/ |
| Software, algorithm | Tidyverse collection of packages for R | N/A | N/A RRID:SCR_019186 | https://www.tidyverse.org/ |
| Software, algorithm | limma package for R | N/A | v3.44.3 RRID:SCR_010943 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| Software, algorithm | CellEngine | Primity Bio Inc | N/A | https://primitybio.com/cellengine.html |
| Software, algorithm | bcl2fastq | Illumina, Inc | v2.20.0.422 RRID:SCR_015058 | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| Software, algorithm | FASTQC | N/A | v0.11.5 RRID:SCR_014583 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Software, algorithm | FastQ Screen | N/A | v0.11.0 RRID:SCR_000141 | https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/ |
| Software, algorithm | bbduk/BBTools | N/A | v37.99 RRID:SCR_016968 | https://jgi.doe.gov/data-and-tools/bbtools/ |
| Software, algorithm | fastq-mcf/ea-utils | N/A | v1.05 RRID:SCR_005553 | https://expressionanalysis.github.io/ea-utils/ |
| Software, algorithm | HISAT2 | N/A | v2.1.0 RRID:SCR_015530 | http://daehwankimlab.github.io/hisat2/ |
| Other | Human genome reference fasta | N/A | GRCh38 RRID:SCR_014966 | ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_33/GRCh38.primary_assembly.genome.fa.gz |
| Other | Human genome annotation GTF file | Gencode | v33 RRID:SCR_014966 | ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_33/gencode.v33.basic.annotation.gtf.gz |
| Software, algorithm | Samtools | N/A | v1.5 | http://www.htslib.org/ |
| Software, algorithm | HTSeq-count | N/A | v0.6.1 RRID:SCR_005514 | https://htseq.readthedocs.io/en/master/ |
| Software, algorithm | DESeq2 package for R | N/A | v1.28.1 RRID:SCR_015687 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Software, algorithm | Hmisc package for R | N/A | v4.4–0 | https://cran.r-project.org/web/packages/Hmisc/index.html |
| Software, algorithm | ggplot2 package for R | N/A | v3.3.1 RRID:SCR_014601 | https://ggplot2.tidyverse.org/ |
| Software, algorithm | rstatix package for R | N/A | v0.6.0 | https://cran.r-project.org/web/packages/rstatix/index.html |
| Software, algorithm | ComplexHeatmap package for R | N/A | v2.4.2 RRID:SCR_017270 | https://www.bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| Software, algorithm | ggforce package for R | N/A | v0.3.1 | https://ggforce.data-imaginist.com/reference/index.html |
Additional files
-
Supplementary file 1
Cohort characteristics.
Table summarizing cohort characteristics. Information pertaining to less than 10 participants is indicated as <10 to prevent potential reidentification. For clinical labs, values represent the mean ± standard deviation. Acronyms for clinical laboratory measurements are as follows: BUN: blood urea nitrogen; CRP: C-reactive protein; ALT: alanine aminotransferase; ALP: alkaline phosphatase; AST: aspartate aminotransferase; BNP: brain natriuretic peptide. Comorbidities affecting each group are listed based on two different annotations: Carlson and Elixhauser. Acronyms for comorbidities are as follows: CHF: chronic heart failure; DM: diabetes mellitus; DMCX: diabetes with complications; METS: metastatic cancer; MI: myocardial infarction; PUD: peptic ulcer disease; PVD: peripheral vascular disease; HTN: hypertension; PHTN: pulmonary hypertension. Fisher’s exact test was used to calculate p-values for differences in % among groups, and the Mann–Whitney test was used to calculate p-values for differences in clinical lab values.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp1-v1.xlsx
-
Supplementary file 2
Seroconversion versus mass spectrometry (MS) proteomics.
Results of Spearman correlation analysis between seroconversion indices and proteins identified by MS. Column A indicates the protein name, column B indicates the SwissProt ID, column C indicates the Spearman rho value, column D indicates the p-value, and column E indicates the adjusted p-value using the Benjamini–Hochberg method.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp2-v1.xlsx
-
Supplementary file 3
Seroconversion versus SOMAcan proteomics.
Results of Spearman correlation analysis between seroconversion indices and proteins identified by SOMAscan technology. Column A indicates the SOMAmer identification number, column B indicates the target protein recognized by the SOMAmer, column C indicates the SwissProt ID, column D indicates the gene symsol, column E indicates the Spearman rho value, column F indicates the p-value, and column G indicates the adjusted p-value using the Benjamini–Hochberg method.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp3-v1.xlsx
-
Supplementary file 4
Seroconversion versus MSD cytokine profiling.
Results of Spearman correlation analysis between seroconversion indices and cytokines, chemokines, and other immune factors measured by multiplex immunoassays using Meso Scale Discovery (MSD) technology. Column A indicates the MSD analyte name, column B indicates the Spearman rho value, column C indicates the p-value, and column D indicates the adjusted p-value using the Benjamini–Hochberg method.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp4-v1.xlsx
-
Supplementary file 5
Seroconversion versus mass cytometry (MC).
Results of Spearman correlation analysis between seroconversion indices and immune cell subsets measured by MC. Separate tabs are used for the results obtained from different parent lineages: all live cells, all CD45+ live cells, B cells, T cells, CD4+ T cells, CD8+ T cells, myeloid dendritic cells (mDCs), and monocytes. In each tab, column A indicates the population measured, column B indicates parent lineage, column C indicates the Spearman rho value, column D indicates the p-value, and column E indicates the adjusted p-value using the Benjamini–Hochberg method.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp5-v1.xlsx
-
Supplementary file 6
Seroconversion versus immune factors.
Results of Spearman correlation analysis between seroconversion indices and circulating immune factors measured by mass spectrometry, SOMAscan assays, or multiplex immunoassays with MSD technology. Column A indicates the analyte name in the platform, column B indicates the name used for display in the corresponding figure, column C indicates the platform used to measure the indicated analyte, column D indicates the Spearman rho value, column E indicates the p-value, and column F indicates the adjusted p-value using the Benjamini–Hochberg method.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp6-v1.xlsx
-
Supplementary file 7
IFNA2 versus mass cytometry (MC).
Results of Spearman correlation analysis between levels of IFNA2 measured by multiplex immunoassays using MSD technology versus immune cell subsets measured by MC. Column A indicates the immune cell population among all live cells, column B indicates the lineage used to calculate cell frequencies, column C indicates the Spearman rho value, column D indicates the p-value, and column E indicates the adjusted p-value using the Benjamini–Hochberg method.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp7-v1.xlsx
-
Supplementary file 8
Seroconversion versus complement.
Results of Spearman correlation analysis between seroconversion indices and components of the complement pathways measured by mass spectrometry or SOMAscan assays. Column A indicates the unique identifier within the platform, column B indicates the name use for display in the corresponding figure, column C indicates the platform used to measure the indicated analyte, column D indicates the Spearman rho value, column E indicates the p-value, and column E indicates the adjusted p-value using the Benjamini–Hochberg method.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp8-v1.xlsx
-
Supplementary file 9
Seroconversion versus hemostasis network.
Results of Spearman correlation analysis between seroconversion indices and factors involved in control of hemostasis measured by mass spectrometry or SOMAscan assays. Column A indicates the unique identifier within the platform, column B indicates the name used for display in the corresponding figure, column C indicates the platform used to measure the indicated analyte, column D indicates the Spearman rho value, column E indicates the p-value, and column E indicates the adjusted p-value using the Benjamini–Hochberg method.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp9-v1.xlsx
-
Supplementary file 10
Seroconversion versus clinical laboratory values.
Results of Spearman correlation analysis between seroconversion indices and clinical laboratory values closest to the time of the research blood draw. Column A indicates the clinical laboratory parameter, column B indicates the Spearman rho value, column C indicates the p-value, and column D indicates the adjusted p-value using the Benjamini–Hochberg method.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp10-v1.xlsx
-
Supplementary file 11
Mass cytometry (MC) antibody table.
List of antibodies used in MC. Column A indicates the antibody target, column B indicates the element conjugated to the antibody, column C indicates the mass of the element, column D indicates the manufacturer, column E indicates the catalog number, column F indicates the clone number, and column G indicates the type of stain protocol used (fixed, live, or fixed with permeabilization).
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp11-v1.xlsx
-
Supplementary file 12
Immune cell type definition.
List of immune cell subsets defined by mass cytometry. Column A indicates the population identified, column B indicates definition based on gating strategy employed, and column C indicates the parent lineage.
- https://cdn.elifesciences.org/articles/65508/elife-65508-supp12-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65508/elife-65508-transrepform-v1.pdf





