Disintegration promotes protospacer integration by the Cas1-Cas2 complex
Figures
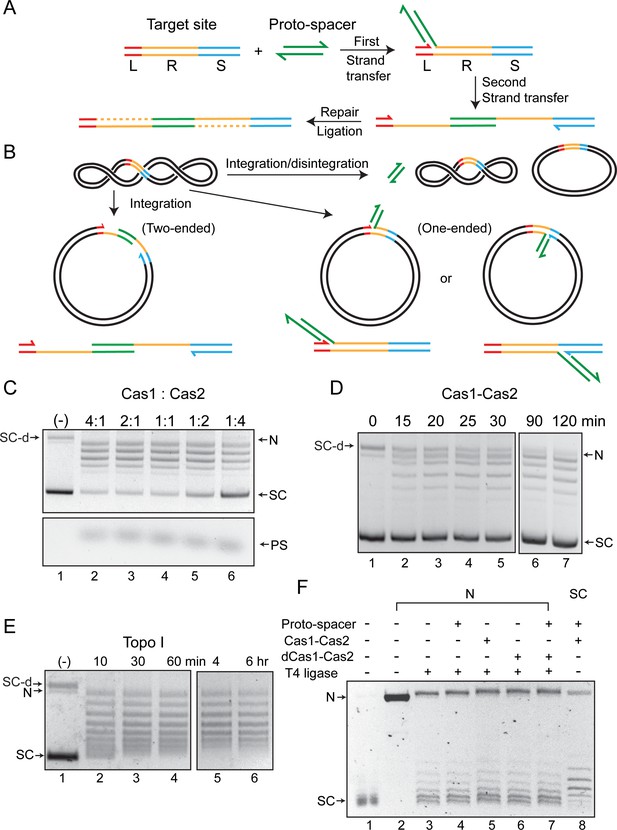
Cas1-Cas2 integrates and disintegrates protospacers at the CRISPR locus in a supercoiled plasmid.
(A) Illustration of protospacer (PS, green) integration into a minimal CRISPR locus comprised of the leader (L, red), repeat (R, yellow), and spacer (S, blue). The reaction follows the cut-and-paste DNA transposition mechanism with a duplication of the repeat sequence. The 3′-hydroxyls that perform nucleophilic attacks at the L-R junction (top strand) and the R-S junction (bottom strand) are indicated by the split arrowheads. (B) In vitro reactions were performed with a supercoiled acceptor plasmid containing the minimal CRISPR locus (L=11 bp; R=36 bp; S=27 bp). Each strand of the protospacer was 26 nt long (DNA sequence in Figure 2A), with four single-stranded 3′-proximal nucleotides. The products of two-ended (complete) and one-ended (partial) protospacer integration and of protospacer integration-disintegration are diagrammed. Plasmid supercoiling will be reduced if the free DNA end undergoes rotation between integration and disintegration. (C) The target plasmid and the protospacer were reacted with Cas1-Cas2 mixtures in the indicated molar ratios. After 1 hr incubation, reactions were analyzed by agarose gel electrophoresis and ethidium bromide staining. A split view of the top and bottom portions of the gel is presented to show the plasmid and protospacer bands. (D) Reactions similar to those shown in C utilized a Cas1 to Cas2 molar ratio of 2:1 and were analyzed over a time course. (E) The substrate plasmid used for the reactions in (C) and (D) was treated with a sub-optimal amount of Escherichia coli topoisomerase I to follow the pattern of DNA relaxation over time. (F) The T4 ligase reactions (lanes 3–7) were performed on the plasmid containing the L-R-S target site nicked with Nb.BtsI (lane 2). The Cas1-Cas2 integration-disintegration reaction (lane 8) utilized the same plasmid in its supercoiled form (lane 1). Reactions were analyzed by gel electrophoresis in the presence of chloroquine (0.4 μg/ml). The DNA bands from the T4 ligase (lanes 3–7) and Cas1-Cas2 (lane 8) reactions are offset by one-half twist/writhe, as indicated by their staggered patterns. N = nicked plasmid; PS = protospacer; SC = supercoiled plasmid; SC-d = supercoiled plasmid dimer .
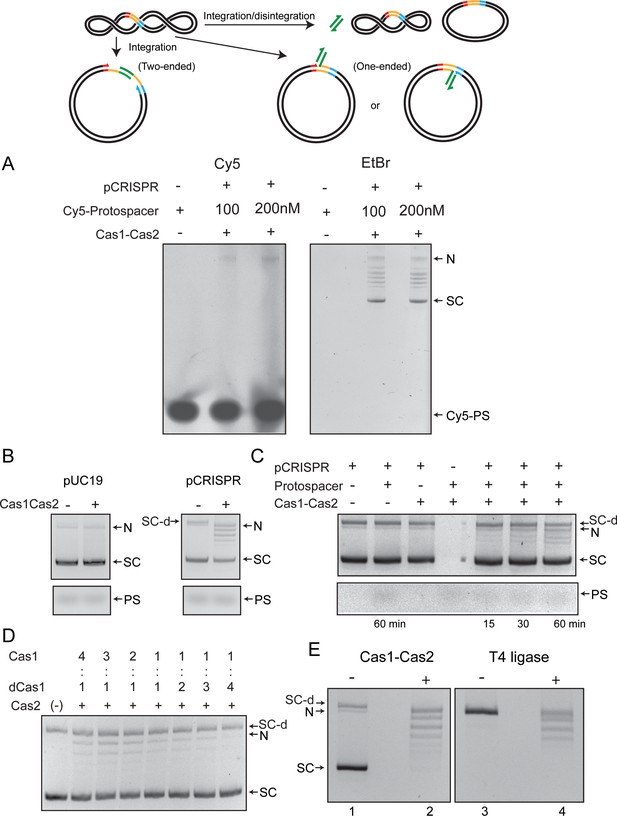
Integration-disintegration by Streptococcus pyogenes (Spy) Cas1-Cas2 is associated with plasmid relaxation.
(A) The reactions were performed using the same target site-containing supercoiled plasmid substrate as that used for the assays shown in Figure 1. The protospacer was the same as well, except that it contained the Cy5 fluorophore at the 5′-end on both strands. The agarose gel in which the reactions were fractionated was analyzed by fluorography (left panel) and was subsequently stained with ethidium bromide to visualize all DNA bands. Note that there was no fluorescence associated with the supercoiled plasmid or the relaxed plasmid bands. Faint fluorescence was detected in a band that migrates at the position of the nicked plasmid. (B) Reactions utilized the same plasmid as in A (pCRISPR) or one lacking the integration target site (pUC19). (C) Individual components were omitted from the reactions with the pCRISPR plasmid, as indicated. (D) These reactions contained Cas1 and dCas1 (catalytically inactive due to the H205A mutation) in the indicated molar ratios. The molar ratio of (Cas1+dCas1) to Cas2 was maintained at 2:1 in all reactions. (E) Integration-disintegration reactions were performed with Cas1-Cas2 (left panel). The nicked plasmid was ligated in the absence of Cas1-Cas2 using T4 DNA ligase (right panel). The gel was run in the absence of an intercalator and was then stained with ethidium bromide. N = Nicked plasmid; PS = protospacer; SC = Supercoiled plasmid; SC-d = Supercoiled plasmid dimer. Supplementary text 1. Nearly all of the protospacer integration events in the target plasmid appear to be one-ended (transfer of only one strand), with integration being rapidly reversed by disintegration to give covalently closed plasmid molecules. Integration-disintegration requires the target site, the protospacer, and the functional Cas1-Cas2 complex. The strand transfer reaction is performed by the Cas1 active site. The nicked strand resulting from protospacer integration is not impeded from rotation before nick closure. As a result, a relaxed distribution of plasmid circles is formed by disintegration.
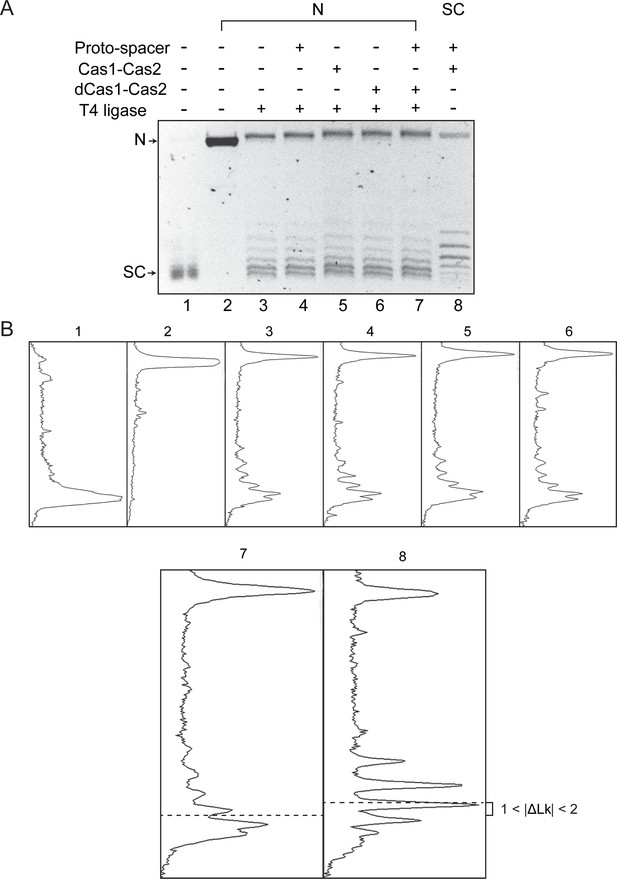
Cas1-Cas2 unwinds DNA within the target site during protospacer integration-disintegration.
(A) The image of the gel panel shown in Figure 1F (electrophoresis done in the presence of 0.4 μg/ml chloroquine) is reproduced here. (B) The densitometric tracings of individual DNA bands in each lane are shown in the panels below. The tracings for lanes 7 and 8 are enlarged, and the centers of their distributions are marked by the dashed lines.
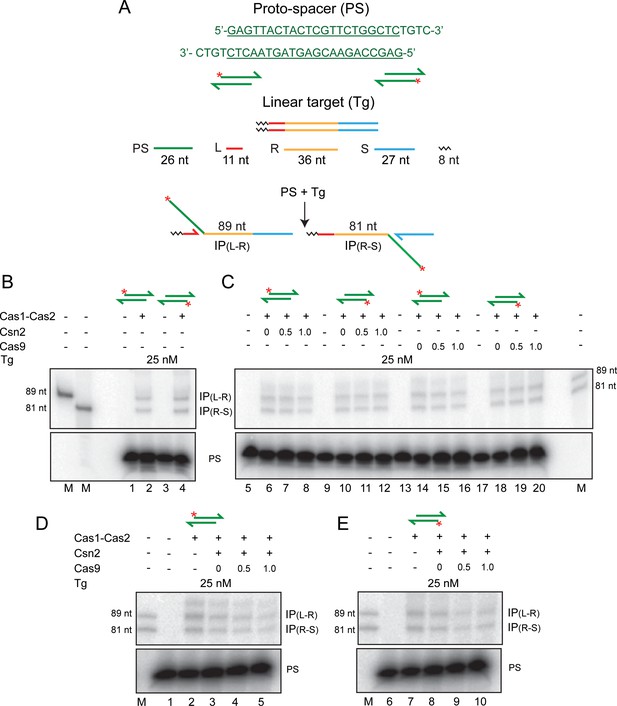
Cas1-Cas2 integrates a protospacer into a linear DNA target.
(A) The protospacer sequence, with the double-stranded region underlined, is shown at the top. In the schematics, the protospacer (32P-labeled at the 5′-end; asterisk) and the linear target site are color-coded as in Figure 1. The labeled products of integration at the L-R and R-S junctions are predicted to be 89 nt and 81 nt, respectively. (B–E) Reactions were performed using the indicated proteins, the protospacer labeled on the top or bottom strand and the unlabeled target. For all reactions, Cas1:Cas2 molar ratio was held at 2:1. The numbers above individual lanes against Csn2 or Cas9 indicate their molar ratios to Cas1. In (D and E), the molar ratio of Csn2:Cas1 was 1:1. The reactions were analyzed by electrophoresis in 12% denaturing polyacrylamide gels, and bands were visualized by phosphorimaging. Only the portions of the gel containing the relevant products and the unreacted protospacer are shown here. The markers in lane M are synthetic oligonucleotides identical in sequence to the predicted integration products. IP = integration product; PS = protospacer.
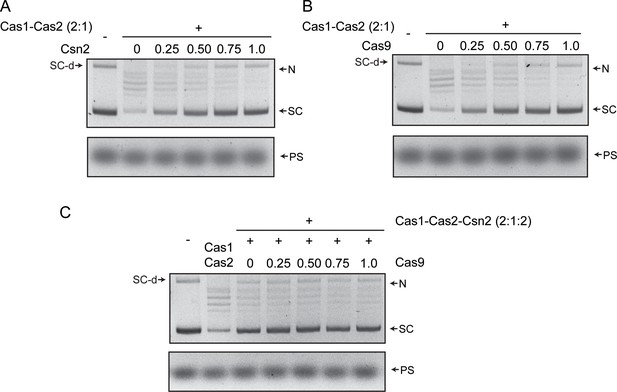
Protospacer integration by Cas1-Cas2 into a supercoiled plasmid target is not stabilized by Csn2, Cas9, or both.
(A–C) Integration reactions containing the indicated proteins were carried out and analyzed as described in Figure 1 and Figure S1. In all reactions, Cas1:Cas2 molar ratio was kept at 2:1. The numbers above the lanes are the molar ratios of Csn2 or Cas9 relative to Cas1. The reactions in (C) contained a 1:1 molar ratio of Cas1:Csn2. N = nicked plasmid; PS = protospacer; SC = supercoiled plasmid; SC-d = supercoiled plasmid dimer.
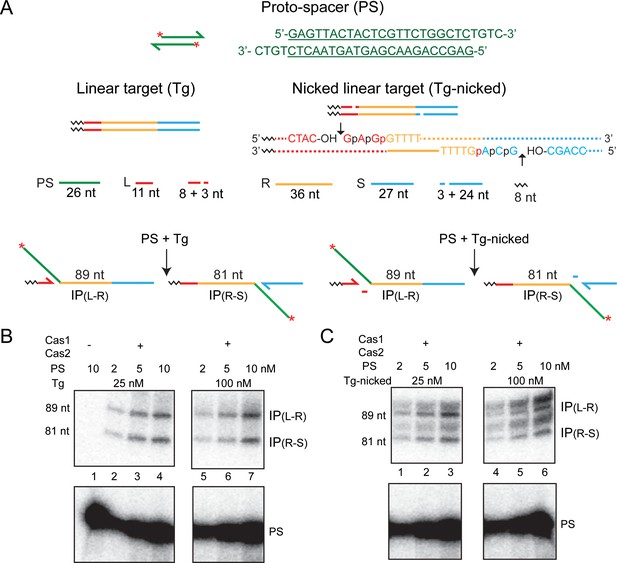
A nicked linear target is active in protospacer integration by Cas1-Cas2.
(A) The schematics of the protospacer, the linear target, and the predicted products of integration are similar to those in Figure 2. In the nicked target, the nicks were placed three nucleotides from the repeat, within the leader and the spacer on the top and bottom strands, respectively. The leader-repeat and repeat-spacer sequences spanning the nicks (short vertical arrows) are spelled out. The leader, repeat, and spacer bases are in red, yellow, and blue fonts, respectively. The normal target phosphodiester bonds for integration are shown as ‘p’. The two phosphodiester bonds neighboring each on the 5′ side are indicated by ‘p’. (B, C) Cas1-Cas2 reactions with the linear (B) and the nicked linear (C) target sites were analyzed as described in Figure 2. Supplementary Text S2. The uncharacterized doublet bands migrating above the 81 nt and 89 nt products could indicate integrations at phosphodiester positions marked as ‘p’ in the nicked target. The expected product lengths for the upper doublet are 90 nt and 91 nt, and those for the lower doublet are 82 nt and 83 nt. The slightly uneven spacing of the 82 nt and 83 nt bands from the 81 nt band is likely due to the size difference in the additional base(s) they harbor. The 82 nt and 81 nt products differ by an extra ‘A’ gained from the spacer. The second additional base present in the 83 nt product is the smaller ‘C’. IP = integration product; PS = protospacer.
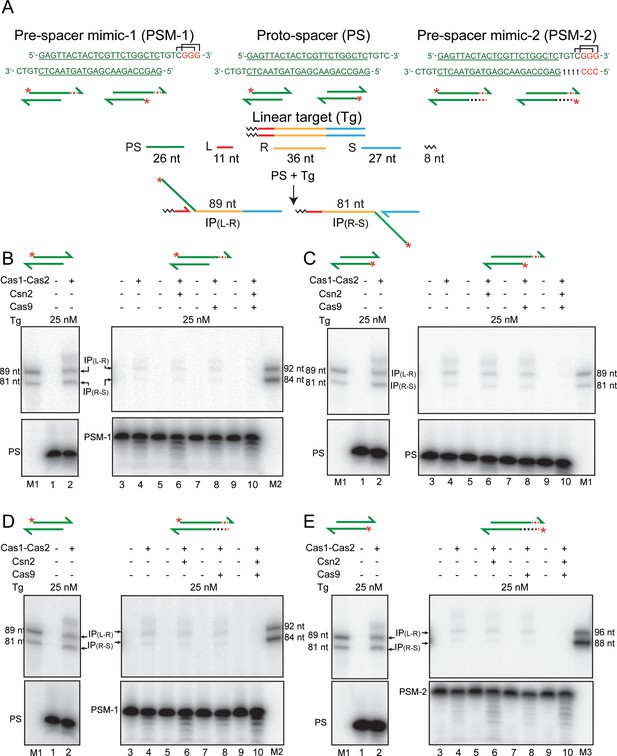
PAM-containing pre-spacer mimics do not confer strand selectivity on integration by Cas1-Cas2.
(A) The sequences of the native protospacer and the PAM-containing pre-spacer mimics derived from it are shown in the top row. In their schematic representations below, the 5′-end label on the top or bottom strand is shown by the asterisk. The linear target and the integration products formed from the native protospacer are schematically diagrammed. In the sequence of the native protospacer (green font), the double-stranded region is underlined. The additional sequences of the pre-spacer mimics are shown in lower case or in red font. Two overlapping PAM sequences present on the top strand are bracketed. (B–E) Integration assays were performed using the indicated proteins and the protospacer or pre-spacer mimics containing the 5′-end label on the top or bottom strand. Supplementary Text S3. In vitro, Cas1-Cas2 reactions lack PAM-specific strand cleavage and orientation-specific strand transfer. In principle, the orientation problem of integration may be circumvented if processing occurs only when two appropriately spaced PAM sequences are recognized on the two strands of the pre-spacer. In this case, insertion in either orientation would give immunity. For a PAM of the type 5′NGG3′, a phage or plasmid genome large enough to be potentially harmful to the host bacterium is likely to contain at least one, and probably several, functional protospacers. IP = integration product. M = marker lanes; PAM = protospacer adjacent motif; PS = protospacer; PSM = pre-spacer mimic.
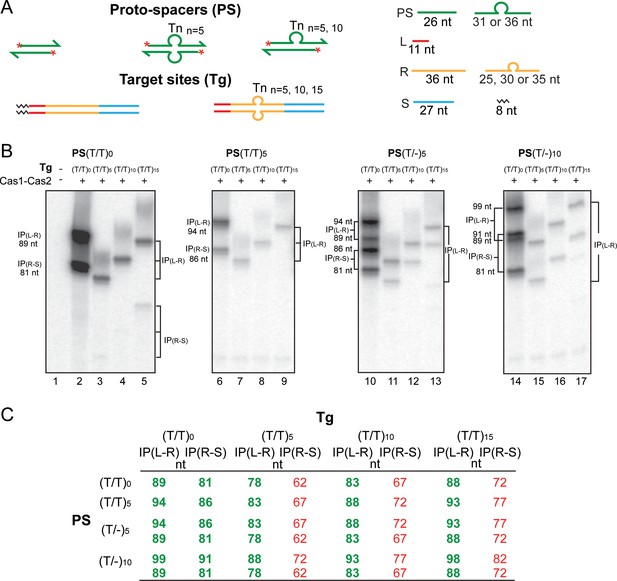
Cas1-Cas2 integrates modified protospacers into modified target sites.
(A) Schematics of the native and modified versions of the protospacers (labeled at the 5′-ends on both strands; asterisks) and target sites used for integration assays are shown. (B) The reactions were performed and analyzed as described in Figure 2 (see Figure 3—figure supplement 1 for uncropped gel images). (C) The expected integration products from the various protospacer and target site pairings are summarized. The (T/T)0 designation refers to the unmodified protospacer and target. The observed products from the reactions shown in (B) are in bold green font; those not detected or detected in extremely small amounts are in normal red font. IP = integration product.
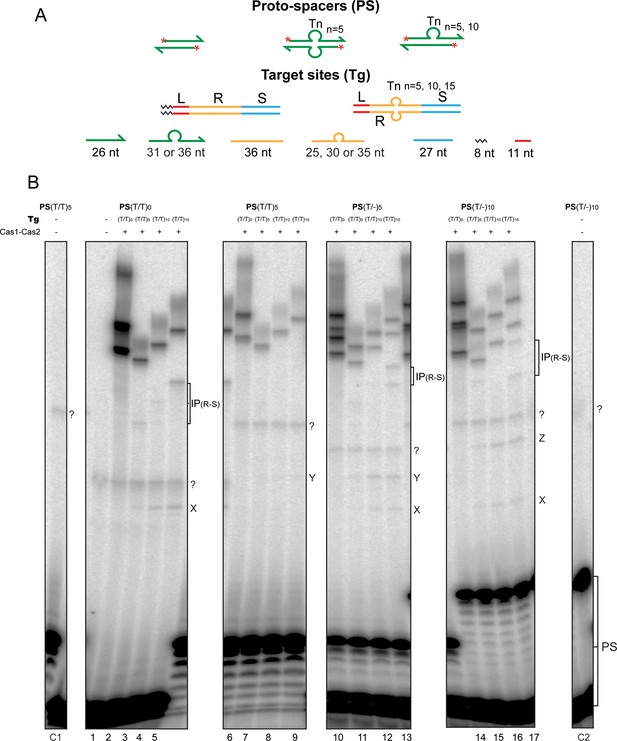
Cas1-Cas2 integrates protospacers predominantly at the L-R junction in modified target sites.
(A) The schematic diagrams of the 5′-end-labeled protospacers and target sites are arranged as in Figure 3A. (B) A full view of the source gel for the data for Figure 3B is shown. Phosphorimaging time was extended relative to that in Figure 3B to reveal weak reaction products at the R-S junction, IP(R-S), in the modified targets. For the unmodified protospacer (T/T)0, IP(R-S) was now readily detectable in the (T/T)15 target (72 nt; lane 5). Even weaker R-S reaction was seen in the (T/T)10-target (67 nt; lane 4) and the (T/T)5 target (62 nt; lane 3). Bands for IP(R-S) in the (T/T)15 target were also visible for the (T/-)5-protospacer (77 nt and 72 nt; lane 13) and the (T/-)10-protospacer (82 nt and 72 nt; lane 17). For the (T/-)5-protospacer and the (T/T)5 or (T/T)10-target combinations, IP (R-S) was barely above background (67 nt and 62 nt; lane 11, and 72 nt and 67 nt; lane 12). In all reactions with the modified target sites, the L-R junction far outmatched the R-S junction for insertion efficiency. The bands labeled as X, Y, and Z are explained in Supplementary Text S4 (see below). Unexpected co-migrating sets of bands from reactions of individual protospacers with the modified and unmodified target sites, as well as bands that were independent of Cas1-Cas2, are marked with a ‘?’ symbol. These were potential contaminants from the oligonucleotides used for protospacer assembly. The incubation mixtures run in lanes C1, 1, and C2 contained the labeled protospacer (PS(T/T)5 in C1; PS(T/T)0 in 1; and PS(T/-)10 in C2) together with the unmodified target (T/T)0 but no Cas1-Cas2. The C1 and C2 control lanes were not included in the gel panels of Figure 3B. Lanes 1–17 here correspond to lanes 1–17 of Figure 3B. Supplementary Text S4. The bands X, Y, and Z were formed only in reactions containing the modified (T/T)n targets and were dependent on Cas1-Cas2. Their relative spacing (depending on the sizes of the protospacer strands) suggests a unique site of insertion common to all the targets. For example, the smallest band X was formed from the native (T/T)oprotospacer (with 26 nt on either strand) in all three of the modified targets, (T/T)5, (T/T)10, and (T/T)15 (lanes 3–5). The mobility of the fainter band Y formed from the (T/T)5protospacer (with 31 nt strands) was consistent with a 5 nt increase in length compared to X (lanes 7–9). With the (T/-)5protospacer (strand lengths of 31 nt and 26 nt), both X and Y were formed (lanes 11–13). And with the (T/-)10protospacer (strand lengths of 36 nt and 26 nt), X and Z were formed (lanes 15–17), the migration of Z suggesting it to be longer than X and Y by 10 nt and 5 nt, respectively. Taken together, X, Y, and Z appear to be formed by insertions of the 26 nt, 31 nt, and 36 nt protospacer strands, respectively, at the leader-proximal junction between the Tn-insertion and the repeat sequence on the bottom strand. The sizes of the X, Y, and Z bands would then be 47 nt, 52 nt, and 57 nt, respectively, and match their observed gel mobilities. The double-strand/single-strand junction may mimic the DNA conformation organized by Cas1-Cas2 at the L-R or R-S junction to promote protospacer insertion during the native reaction. IP = integration product; PS = protospacer.
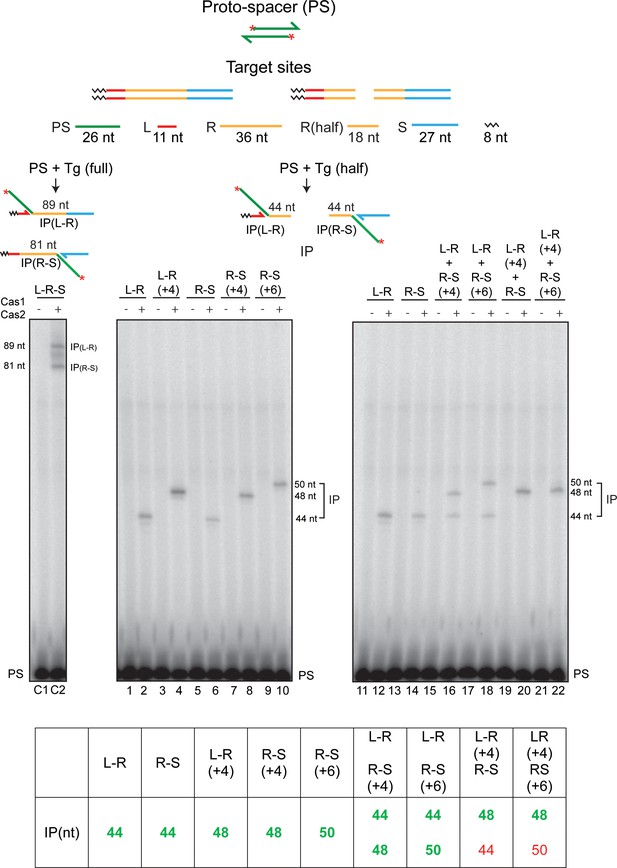
Cas1-Cas2 transfers the protospacer into linear half-target sites.
The protospacer labeled with 32P at its 5′-ends and the linear full- and half-target sites are diagrammed at the top. The minimal L-R or R-S half-site contained 18 bp of the L-proximal or R-proximal repeat sequence, respectively (Figure 4—figure supplement 1). Each half-site is the product of splitting a full-site into two exactly at the center of the 36 bp repeat. Reactions contained the labeled protospacer together with the indicated minimal L-R or R-S half-sites, or half-sites containing longer repeat sequences (+4 or +6 bp) (Figure 4—figure supplement 1), as well as combinations thereof. Reactions were analyzed as described in Figures 2 and 3. The lanes C1 and C2 denote the control reaction with a normal target site containing the full-length repeat. The predicted integration products from the individual and mixed half-target site reactions are listed at the bottom. The experimentally observed products are distinguished from the undetected ones by the bold green and normal red fonts, respectively. IP = integration product; PS = protospacer.

Half-target sites used for protospacer integration assays.
The complete sequences of the full- and half-target sites used for the assays shown in Figure 4 are listed. The leader (L), repeat (R), and spacer (S) sequences are shown in red, yellow, and blue fonts, respectively. The sequences present to the left of L are not relevant to the reaction.
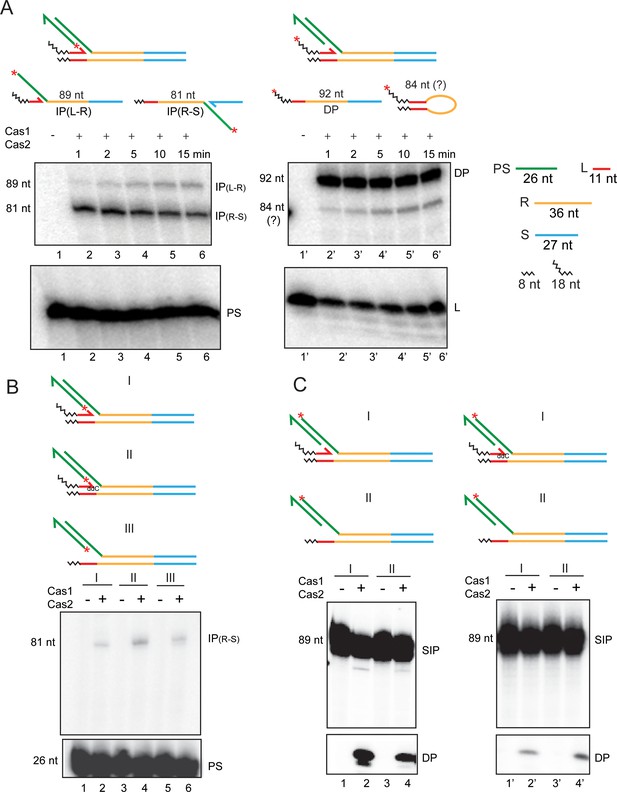
Second integration and disintegration occur in substrates mimicking a semi-integrated protospacer.
(A) Schematics of semi-integrated protospacers at the L-R junction (top row) and the possible products formed from them by Cas1-Cas2 action (bottom row) are shown. The 32P-label was placed at the 5′-end of the unintegrated protospacer strand or the leader strand to follow the second integration and disintegration events, respectively. (B) Integration of the 32P-labeled protospacer strand at the R-S junction was monitored in the indicated substrates. (C) Disintegration was assayed in the same substrates as in (B), except that the 32P-label was placed at the 5′-end of the integrated protospacer strand. ddC = dideoxy C; DP = disintegration product; IP = integration product; PS = protospacer; SIP = semi-integrated protospacer.
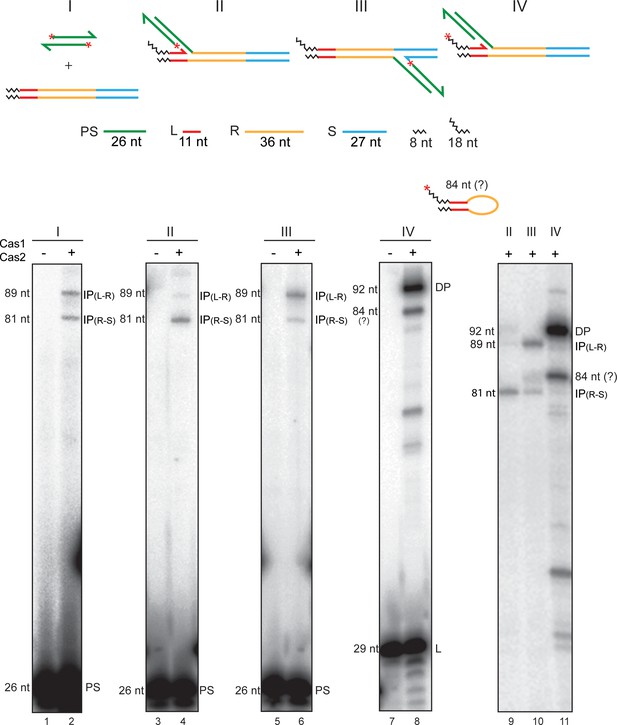
Cas1-Cas2 mediates integration and disintegration of a semi-integrated protospacer.
The substrates for the standard protospacer integration reaction and those for the second strand transfer or the disintegration reaction from a semi-integrated protospacer are schematically illustrated at the top. The 5′-end label (32P) placed on the protospacer strands (I), the unintegrated strand of a semi-integrated protospacer (II and III), or on the leader strand (IV) is signified by the asterisks. The integration products from a protospacer in a linear target site at the L-R junction (lane 2; 89 nt) and the R-S junction (lane 2; 81 nt) provide the reference frame for interpreting the product bands from the ‘semi-integrated’ substrates. The primary products from the L-R and R-S semi-integrated substrates were integrations of the second protospacer strand at the R-S junction (lane 4; 81 nt) and the L-R junction (lane 6; 89 nt), respectively. The weaker 89 nt (lane 4) and 81 nt (lane 6) bands are consistent with the reintegration of a protospacer (following its release by disintegration) at the L-R and R-S junctions, respectively. These junctions would be re-formed during disintegration mediated by the 3′-hydroxyls of the leader and spacer strands, respectively. When the 32P-label was present at the 5′-end of the leader strand, the prominent product was the resealed L-R junction (via disintegration) (92 nt; lane 8). The migration of the weaker band below in lane 8 (see also Figure 5A; lanes 2′−6′) is consistent with a size of 84 nt (Supplementary Text S5 below). Its mobility difference from the 81 nt band (IP[R-S]) was more conspicuous in a longer run geI in which the labeled protospacer and leader strands were run off the gel bottom (lanes 9–11). Supplementary Text S5. An 84 nt product is expected if the leader strand were to be transferred to the R-S junction on the opposite strand in an aberrant integration-like reaction. This reaction is formally similar to ‘hairpinning’ carried out by the Rag1-Rag2 recombinase/transposase and the transposases of the cut-and-paste transposons Tn5 and Tn10 (Bhasin et al., 1999; Kennedy et al., 1998; McBlane et al., 1995). Bringing the leader 3′-hydroxyl in proximity to the R-S junction would require considerable distortion of the repeat DNA or its reconfiguration into a cruciform-like structure (Nuñez et al., 2015a). DP = disintegration product; IP = integration product; L = leader; PS = protospacer.
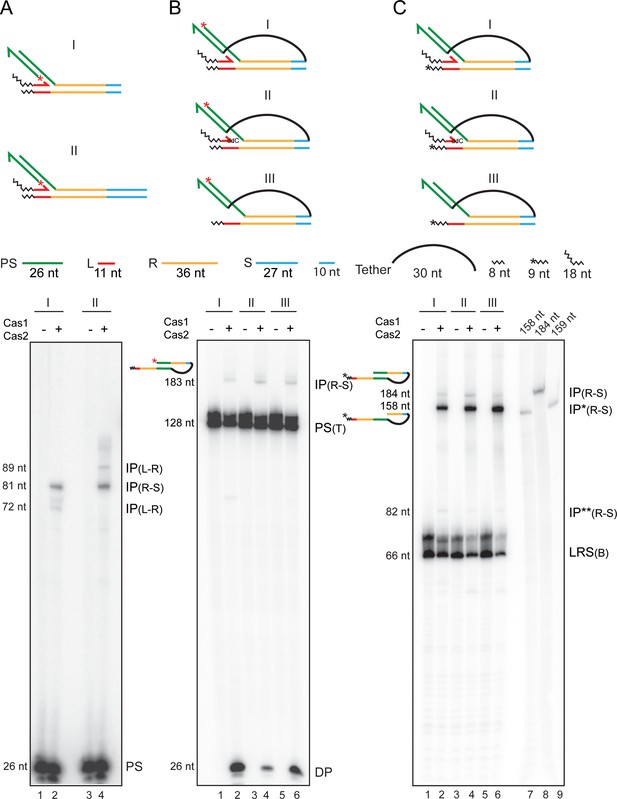
Second strand integration by Cas1-Cas2 occurs in concert with first-strand disintegration.
Schematics of the substrates used for the individual sets of assays (A–C) are placed above the respective gel panels. The layout of the trombone substrates (B, C) from a perspective of the Cas1-Cas2-protospacer structure is shown in Figure 6—figure supplement 1. (A) Integration reactions were performed in the two substrates that differed only in their spacer lengths. The truncated spacer (10 bp long) lacked the 17 bp distal to the R-S junction. (B, C) The substrate configurations were similar for the two reaction sets, except for the position of the 32P label. In (B), the integrated protospacer strand carried the label at the 5′-end (red asterisk). In (C), one labeled nucleotide (shown by the asterisk) was added to the 3′-end of the bottom strand. DP = disintegration product; IP = integration product; IP* = integration product coupled to disintegration; IP** = integration product uncoupled from (but preceded by) disintegration; PS = protospacer; PS (T) = protospacer (tethered); LRS (B) = Substrate bottom strand. Lanes 7–9 in (C), oligonucleotide length markers.
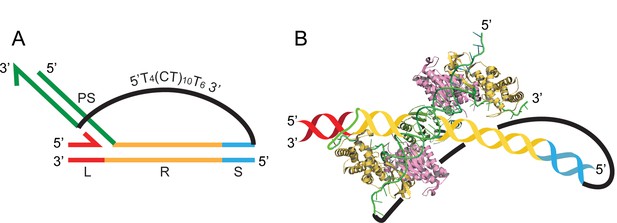
Structural perspective of a trombone substrate in which an integrated protospacer strand is linked to the unintegrated strand by a single-stranded tether.
(A) The schematic representation of a typical trombone substrate (see Figure 6B,C) is reproduced here (L = leader; R = repeat; S = spacer; PS = protospacer). The truncated spacer of the target DNA (containing 10 bp adjacent to the repeat) is linked to the 5′-end of the unintegrated protospacer strand by the 5′T4(CT)10T63′ tether. (B) This idealized structural perspective of (A) shows the Escherichia coli Cas1-Cas2 complex with a bound protospacer DNA (Wang et al., 2015) (PDB 5DLJ), one strand of which is integrated at the L-R junction. The target DNA (color-coded as in A) is an add-on, and its placement with respect to the protospacer is arbitrary. The covalent linkage of the protospacer strand to the repeat DNA at L-R was manipulated using Adobe Illustrator. The single-strand tether between the spacer and the 5′-end of unintegrated protospacer strand in effect covalently connects the two protospacer strands.
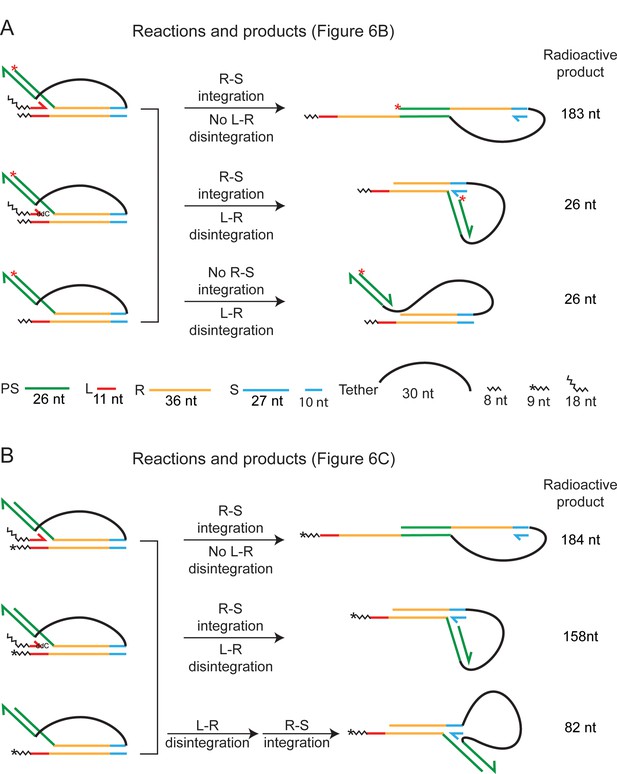
Distinct products formed by Cas1-Cas2 action on trombone substrates.
The observed reaction products (Figure 6B and C) are schematically illustrated, and the steps involved in their formation are indicated. The unlabeled leader strand is omitted from the figures of the products. The 32P-label (5′-end; red asterisk) in the substrates in (A) is positioned to identify protospacer integration at the R-S junction without its disintegration from the L-R junction. In addition, disintegration of the protospacer from the L-R junction (which may or may not be coupled to integration at the R-S junction) is also reported by these substrates. The labeling position in (B) (one 32P-labeled nucleotide at the 3′-end; asterisk) distinguishes integration events at R-S occurring with or without disintegration at L-R. In principle, disintegration and integration may occur in a concerted or sequential manner. The sequential reaction is reported by the 82 nt labeled product, signifying the initial release of the protospacer strand inserted at L-R and its subsequent insertion at R-S.
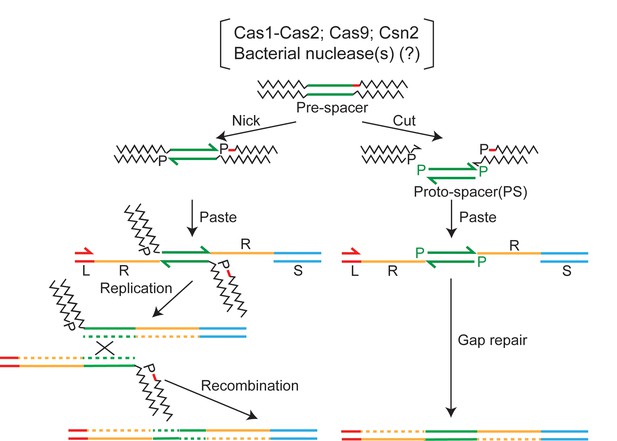
Two-ended integration of the protospacer by the spacer acquisition complex.
Two possible schemes for protospacer integration are outlined. The pathway shown at the left is equivalent to replicative DNA transposition. The pre-spacer is nicked at the protospacer 3′-ends, at least one cleavage being adjacent to a PAM sequence (shown by the red dash). The 3′-hydroxyls are then transferred to the leader-repeat (L–R) and repeat-spacer (R–S) junctions of the target site. Replication across L and PS (protospacer) followed by recombination within the duplicated PS completes integration. An alternative mode of processing the strand transfer intermediate involves removal of the pre-spacer DNA flanking PS, and gap repair by limited DNA synthesis and ligation (Figure 7—figure supplement 1). The integration pathway shown at the right follows cut-and-paste DNA transposition. The steps involve excision of PS from the pre-spacer, strand transfer to L-R and R-S junctions, gap repair, and ligation. In either pathway (replicative or cut-and-paste), directing the PAM-adjacent 3′-hydroxyl to the L-R junction will ensure the functional orientation of the inserted spacer. PAM, protospacer adjacent motif.
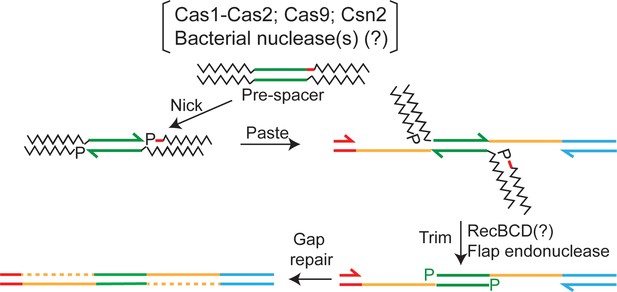
Spacer acquisition by a mechanism that combines aspects of replicative and cut-and-paste transposition mechanisms.
An integration event may be initiated using the single-stranded ends generated by Cas1 (or Cas9) activity without the protospacer being excised from the pre-spacer DNA (as in replicative transposition; see Figure 7). The flanking DNA is then trimmed by a nuclease such as RecBCD, perhaps with assistance from a flap endonuclease, to generate a structure that resembles the intermediate of cut-and-paste transposition. In effect, the mechanism is ‘nick-paste-and-trim’. The final integration product is formed by gap-filling replication and ligation. The process would be analogous to the removal of host DNA attached to the ends of the infecting phage Mu genome after it has been partially integrated into the Escherichia coli chromosome (Jang and Harshey, 2015). The red dash within the pre-space denotes the PAM sequence. PAM, protospacer adjacent motif.
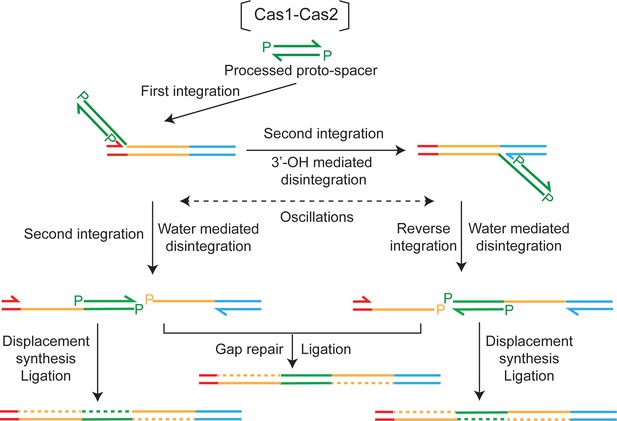
A model for disintegration-assisted integration.
Integration of the protospacer by Cas1-Cas2 is predominantly one-ended. Following the initial strand transfer to the L-R junction, transfer of the second strand to the R-S junction is associated with disintegration at L-R. Oscillations of the semi-integrated protospacer become possible when disintegration mediated by the 3′-hydroxyl re-forms the L-R or R-S junction at the disintegration site. Disintegration mediated by water during a second strand transfer step will block oscillations. However, the nick between L and R or R and S will be preserved, and can be utilized by DNA repair machineries to complete protospacer insertion. Formation of the repair-suitable intermediate shown at the left is dependent only on water-mediated disintegration, the 3′-hydroxyl-mediated disintegration being dispensable. However, both types of disintegration are required to generate the intermediate shown at the right.
Additional files
-
Source data 1
Set 1 of source data files.
- https://cdn.elifesciences.org/articles/65763/elife-65763-data1-v1.zip
-
Source data 2
Set 2 of source data files.
- https://cdn.elifesciences.org/articles/65763/elife-65763-data2-v1.zip
-
Supplementary file 1
List of oligonucleotides.
- https://cdn.elifesciences.org/articles/65763/elife-65763-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65763/elife-65763-transrepform-v1.pdf





