HPF1 and nucleosomes mediate a dramatic switch in activity of PARP1 from polymerase to hydrolase
Figures
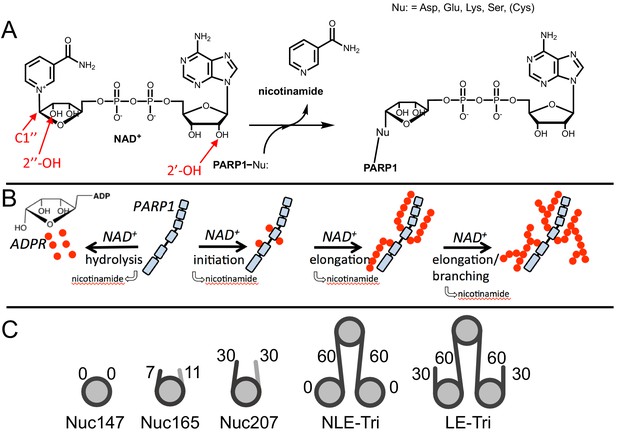
Chemical mechanism of poly(ADP-ribose) polymerase 1 (PARP1) and structures of nucleosomes used here.
(A) Chemical mechanism of PARP1 highlighting in red important groups of NAD+ as related to initiation (C-1′′), elongation (2′-OH), and branching (2′′-OH). (B) Cartoon representation of the four reactions catalyzed by PARP1 (in light blue): initiation, elongation, branching, and hydrolysis. ADP-ribose (ADPR) monomers (red circles) form PAR chains or free ADPR. (C) Cartoon representation of the different nucleosome constructs utilized in this study. The number in the name indicates the length of DNA (e.g., Nuc147 has 147 bp of DNA), and the small numbers next to the linkers or ends indicate the length of these regions.
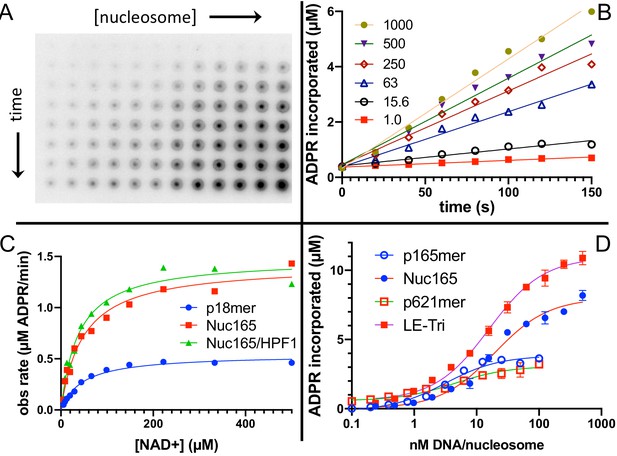
Nucleosomes are better activators of poly(ADP-ribose) polymerase 1 (PARP1) than free DNA.
(A) Representative image from the filter binding assay monitoring the incorporation of 32P-NAD into PARP1 (autoPARylation) in the presence of Nuc165. Different time points (0, 20, 40, 60, 80, 100, 120, and 150 s) are represented in the vertical direction and different concentrations of Nuc165 (0.5–1000 nM by factors of 2 in concentration) are represented in the horizontal direction. (B) Representative data from monitoring the incorporation of 32P-NAD into PARP1 (autoPARylation) in the presence of Nuc165 (different concentrations indicated in nM). Good linearity of rates is observed at all concentrations of nucleosome up to 150 s. (C) Representative curve determining the Km for NAD+ in the presence of p18mer, Nuc165, or Nuc165 in the presence of HPF1. The Km values for p18mer, Nuc165, and Nuc165 in the presence of HPF1 are 38 ± 9 µM (n = 8), 39 ± 10 µM (n = 3), and 54 ± 8 µM (n = 3), respectively. The kcat values for p18mer, Nuc165, and Nuc165 in the presence of HPF1 are 4.4 ± 1.9 s−1, 2.4 ± 0.8 s−1, and 4.6 ± 1.0 s−1, respectively. (D) Representative activation curves for p165mer, Nuc165, p621mer, and LE-Tri demonstrating that nucleosomes lead to greater maximal autoPARylation activity. Indicated error bars are from triplicate assay points. Derived apparent values for kcat for these and other activators of PARP1 and their replicates are shown in Table 1.
-
Figure 2—source data 1
Nucleosomes are better activators of PARP1 than free DNA.
- https://cdn.elifesciences.org/articles/65773/elife-65773-fig2-data1-v2.xlsx
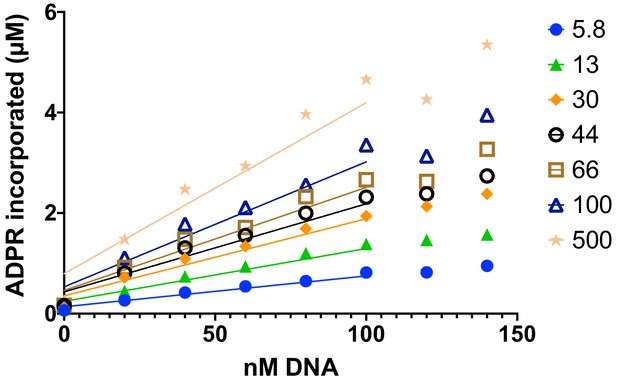
Addition of Histone PARylation Factor 1 reduces the linearity of poly(ADP-ribose) polymerase 1 (PARP1 )in a plate-based assay monitoring incorporation of ADP-ribose at short times of reaction.
Representative data from monitoring the incorporation of 32P-NAD into PARP1 (autoPARylation) and Nuc165 (transPARylation, different concentrations indicated in nM). Reasonable linearity of rates is observed at all concentrations of nucleosome up to 100 s, with a significant decline thereafter.
-
Figure 2—figure supplement 1—source data 1
Addition of HPF1 reduces the linearity of PARP1 in a plate-based assay monitoring incorporation of ADPR at short times of reaction.
- https://cdn.elifesciences.org/articles/65773/elife-65773-fig2-figsupp1-data1-v2.xlsx
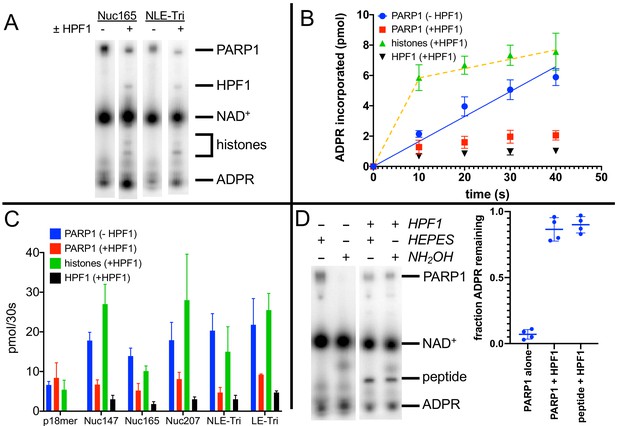
Histone PARylation Factor 1 (HPF1) mediates transPARylation onto nucleosomes and serines.
(A) Representative autoradiogram from an assay gel demonstrating the reduction of autoPARylation in the presence of HPF1 with the concomitant appearance of PARylated histones and HPF1. The remaining substrate 32P-NAD+ is by far the most prominent band on the image, indicating that we are monitoring the early time points of the reaction. (B) Time dependence of ADP-ribose (ADPR) incorporation onto poly(ADP-ribose) polymerase 1 (PARP1), histones, and HPF1. Note the linearity seen for autoPARylation in the absence of HPF1 and the non-linearity seen for PARylation in the presence of HPF1, as emphasized by the dotted line. (C) Bar graph of ADPR incorporation onto PARP1, histones, and HPF1 at 30 s demonstrating that in the presence of HPF1 histones become the primary target of PARylation to the detriment of PARP1. Levels of HPF1 PARylation are low despite the high concentration of HPF1 (2 µM) compared to nucleosomes (300–700 nM) in the reaction mixture. For the reaction indicated with p18mer, activation is by free DNA and the PARylated product is the H3 peptide. Each experiment was performed four separate times, and the data shown are mean values with standard deviations. (D) Representative gel demonstrating that PARylation in the absence of HPF1 is directed towards hydroxylamine labile Asp/Glu residues and in the presence of HPF1 becomes stable to this treatment, consistent with PARylation of Ser residues. Quantitation in the bar graph is the summary of 8–12 replicates.
-
Figure 3—source data 1
HPF1 mediates transPARylation onto nucleosomes and serines.
- https://cdn.elifesciences.org/articles/65773/elife-65773-fig3-data1-v2.xlsx
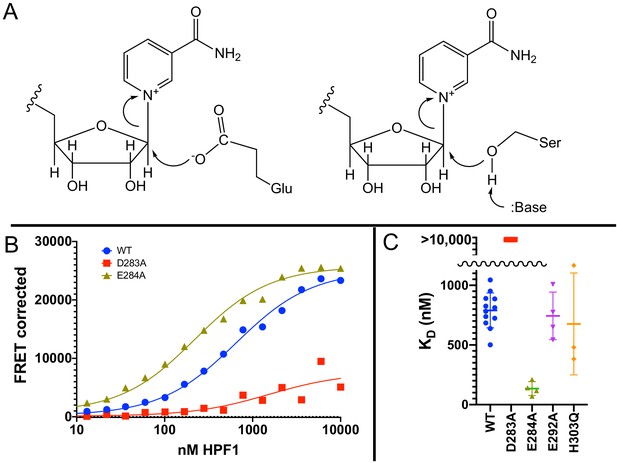
Glu284 of Histone PARylation Factor 1 (HPF1) is the catalytic base required for transPARylation.
(A) Chemical mechanism of PARylation of glutamate (Glu, on left) does not require a catalytic base, whereas PARylation of serine (Ser, on right) requires deprotonation of serine by a catalytic base. (B) Representative curves demonstrating the binding of HPF1 (WT, D283A, and E284A) to the poly(ADP-ribose) polymerase 1 (PARP1)–Nuc165 complex using FRET between labeled HPF1 and labeled PARP1. (C) Bar graph for binding of HPF1 to PARP1–Nuc165 complex as determined by FRET assay demonstrating that the E284A mutant of HPF1 binds more tightly than WT, and that the D283 mutant does not bind with measurable affinity. The E292A and H303Q mutants of HPF1 are shown to bind with similar affinity as WT HPF1. Data for these findings with standard deviations and number of replicates can be found in Table 2.
-
Figure 4—source data 1
Glu284 of HPF1 is the catalytic base required for transPARylation.
- https://cdn.elifesciences.org/articles/65773/elife-65773-fig4-data1-v2.xlsx
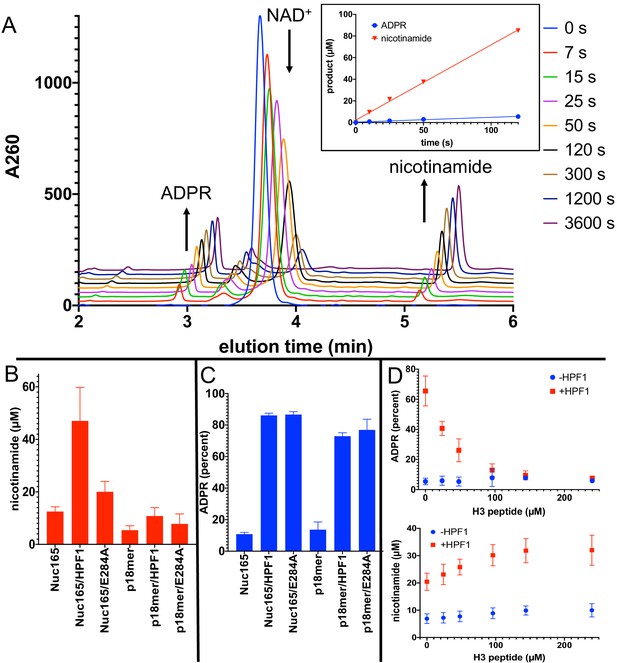
Addition of Histone PARylation Factor 1 (HPF1) converts poly(ADP-ribose) polymerase 1 (PARP1) predominantly into an NAD+ hydrolyase.
(A) Representative HPLC traces from a reaction of PARP1 using p18 as an activator that simultaneously monitors depletion of NAD+ (200 µM initial) and formation of both ADP-ribose (ADPR) and nicotinamide. Each successive time point trace is offset in both the x- and y-axis to allow for better visualization. The inset shows that the assay is linear with respect to formation of nicotinamide and ADPR for at least 90 s. (B) Comparison of activity of PARP1 (100 nM) as measured by formation of nicotinamide using either Nuc165 (300 nM) or p18mer (100 nM) as activators in the presence or absence of HPF1 (2 µM, wild-type vs. E284A mutant) after 1 min of reaction time. (C) Comparison of percent of turnover of PARP1 that leads to free ADPR under the same conditions as in (B). Error bars in (C) and (D) are derived from three experiments, each performed using four replicates. (D) The hydrolase activity of PARP1 (100 nM) is suppressed (top panel) with a modest increase in overall activity as measured by nicotinamide formation (bottom panel) by high concentrations of H3 histone tail peptide (varied as indicated), but only in the presence of HPF1 (2 µM). Reactions were performed at 200 µM NAD+ for 1 min, and the data and standard deviations shown are derived from four separate experiments.
-
Figure 5—source data 1
Addition of HPF1 converts PARP1 predominantly into an NAD+ hydrolyase.
- https://cdn.elifesciences.org/articles/65773/elife-65773-fig5-data1-v2.xlsx
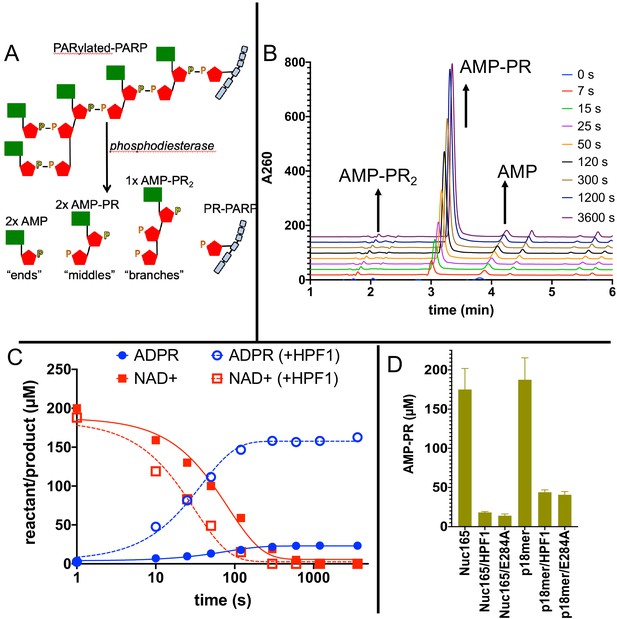
Addition of Histone PARylation Factor 1 (HPF1) accelerates consumption of NAD+ and leads to significantly shorter PAR chains.
(A) Cartoon representation of the reaction products generated by phosphodiesterase treatment of PARylated poly(ADP-ribose) polymerase 1 (PARP1). (B) Representative HPLC traces from the analysis of PARylated PARP1 that used p18 as an activator, simultaneously monitoring AMP (‘ends’), AMP-PR (‘middles’), and AMPR-PR2 (‘branches’). Each successive time point trace is offset in both the x- and y-axis to allow for better visualization. (C) Comparison of a time course for accumulation of ADP-ribose (ADPR) and depletion of NAD+ for PARP1 with Nuc165 in the absence (solid symbols and lines) or presence (open symbols, dotted lines) of HPF1 (wild-type). Note the faster consumption of NAD+ in the presence of HPF1 and the much higher levels of ADPR observed. The appearance of nicotinamide (not shown for clarity) was also monitored in these assays and mirrored the depletion of NAD+. (D) Quantitation of AMP-PR (chain middles) released from PARylated PARP1 and histones after complete consumption of 200 µM NAD+. Note that in the absence of HPF1 ~90% of the ADPR is attached to protein, indicating long PAR chains. In contrast, the addition of HPF1 (either wild-type or E284A mutant) led to much shorter PAR chains, consistent with the high amount of free ADPR formed (Figures 5D, C). Error bars in (D) are derived from three experiments.
-
Figure 6—source data 1
Addition of HPF1 accelerates consumption of NAD+ and leads to significantly shorter PAR chains.
- https://cdn.elifesciences.org/articles/65773/elife-65773-fig6-data1-v2.xlsx
Tables
Comparison of the binding and activity of PARP1 using nucleosome and free DNA as activators, in the presence or absence of HPF1.
| PARylation product | PARP1 | Histones | |||
|---|---|---|---|---|---|
| ± HPF1 | No HPF1 | No HPF1 | No HPF1 | With HPF1 | With HPF1 |
| DNA/nucleosome | CC50 (nM) | kcat (s−1) | ADPR (pmol/30 s) | ADPR (pmol/30 s) | ADPR (pmol/30 s) |
| p18mer | 11.5 ± 3.2 (n = 6) | 2.4 ± 0.8 (n = 14) | 6.6 ± 0.9 | 8.4 ± 3.8 | 5.4 ± 2.4 (peptide) |
| Nuc147 | 13.6 ± 6.4 (n = 4) | 4.0 ± 0.9 (n = 3) | 17.8 ± 2.1 | 6.7 ± 1.2 | 27 ± 5 |
| p165mer | 10.1 ± 4.3 (n = 5) | 2.7 ± 0.8 (n = 6) | n.d. | n.d. | n.d. |
| Nuc165 | 6.1 ± 2.7 (n = 8) | 4.4 ± 1.0 (n = 15) | 13.9 ± 2.0 | 5.2 ± 1.8 | 10.1 ± 1.3 |
| Nuc207 | n.d. | n.d. | 17.9 ± 4.5 | 8.1 ± 1.7 | 28 ± 12 |
| NLE-Tri | 1.6 ± 0.4 (n = 5) | 4.5 ± 1.2 (n = 4) | 20.3 ± 4.3 | 4.7 ± 1.3 | 15 ± 6 |
| p621mer | 4.2 ± 1.7 (n = 4) | 1.6 ± 0.4 (n = 2) | n.d. | n.d. | n.d. |
| LE-Tri | 1.9 ± 0.7 (n = 6) | 5.1 ± 1.3 (n = 3) | 18.5 ± 0.5 | 9.2 ± 0.2 | 29 ± 1 |
-
Column 1: The apparent competitive concentration for 50% binding (CC50) values as determined by titrating nucleosomes or free DNA into a solution containing pre-bound PARP1 bound to fluorescent p18mer (p18mer*) and monitoring the release of p18mer* by fluorescence polarization. Column 2: The apparent kcat values (determined at NAD+=40 µM = Km for NAD+) were determined from fitting the measurements of activity for incorporation of ADPR as shown in Figure 2B in the plate-based assay at varying concentrations of DNA/nucleosome (Figure 2D). Data reflect at least three different replicates wherein each time point was collected in triplicate. Columns 3–6: incorporation of ADPR into PARP1, HPF1, and histones was determined using the gel-based assay using 10 µM NAD+ after 30 s of reaction. Data shown are from 3 to 4 replicates for each assay condition. Activation by p18mer monitored addition of ADPR onto H3-tail peptide as there are no histones present in this reaction. All indicated errors are standard deviations from the mean. ADPR: ADP-ribose; PARP1: poly(ADP-ribose) polymerase 1; HPF1: Histone PARylation Factor 1.
HPF1 mutant analysis.
| HPF1 | Melting temperature (°C) | Histone PARylation (pmol ADPR/30 s) | KD (nM) |
|---|---|---|---|
| WT | 50.6 ± 0.10 n = 5 | 10.1 ± 1.3 n = 4 | 790 ± 147 n = 12 |
| D283A | 50.8 ± 0.11 n = 5 | 0 | >10,000,000 n = 5 |
| E284A | 50.3 ± 0.86 n = 5 | 0 | 135 ± 58 n = 4 |
| E292A | 49.0 ± 0.08 n = 5 | 17.5 ± 2.1 n = 4 | 743 ± 200 n = 4 |
| H303Q | 50.2 ± 0.10 n = 5 | 21.1 ± 7.6 n = 4 | 676 ± 427 n = 3 |
| Column 1: protein stability was measured by Thermo Fisher Protein Thermal Shift kit. Column 2: incorporation of ADPR onto histones was determined using the gel-based assay using 10 µM NAD+ after 30 s of reaction. Data shown are from 3 to 4 replicates for each assay condition. Column 3: binding constant of HPF1 to the PARP1–Nuc165 complex was determined using the FRET assay shown in Figure 4B. Data shown are the mean and standard deviation of the indicated number of replicates. HPF1: Histone PARylation Factor 1; PARP1: poly(ADP-ribose) polymerase 1; ADPR: ADP-ribose. | |||
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Peptide, recombinant protein | PARP1 | UniProt | P09874 | As described in Rudolph et al., 2018 |
| Peptide, recombinant protein | HPF1 | UniProt | Q9NWY4 | As described in manuscript |
| Sequenced-based reagent | p18mer DNA | IDT | 5′-phosphate-GGGTTGCGGCCGCTTGGG-3′; double-stranded | |
| Strain, strain background (Escherichia coli) | Rosetta DE3 pLys | EMD Millipore | 70956 | Chemically competent cells |
| Other | 384-well plates | Corning | 3575 | |
| Other | 96-well plates | Corning | 3898 | |
| Other | Whatman GF/C glass paper | Whatman | 28497-619-PK | |
| Chemical compound, drug | Olaparib | SelleckChem | S1060 | |
| Commercial assay or kit | QuikChange II Mutagenesis kit | Agilent Technologies | 200523 | |
| Other | Synergi Fusion-RP column | Phenomenex | 00F-4424-EO | |
| Peptide, recombinant protein | Nuc147, Nuc165, Nuc207, NLE-Tri, LE-Tri | Prepared in-house | See Muthurajan et al., 2016 | |
| Chemical compound, drug | ADP-ribose | Sigma-Aldrich | A0752 | |
| Chemical compound, drug | Nicotinamide | Sigma-Aldrich | N3376 | |
| Chemical compound, drug | NAD+ | Sigma-Aldrich | N0632 | |
| Peptide, recombinant protein | H3 peptide | Anaspec | AS-61701 | |
| Chemical compound, drug | 32P-NAD+ | PerkinElmer | NEG023X | |
| Chemical compound, drug | Snake venom phosphodiesterase | Worthington | LS003926 | |
| Software, algorithm | GraphPad Prism 9.0 | GraphPad Prism | Version 9.0.1 (128) |





