Developing a multivariate prediction model of antibody features associated with protection of malaria-infected pregnant women from placental malaria
Figures
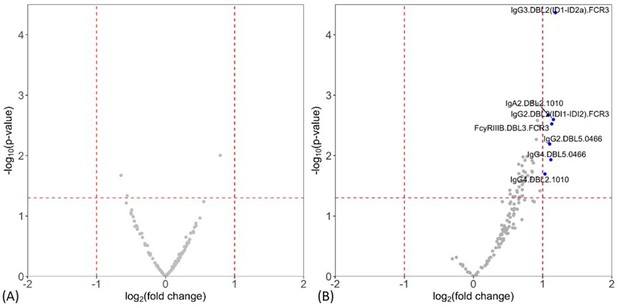
Individual antibody features to recombinant VAR2CSA Duffy binding-like (DBL) domain proteins measured by multiplex comparing women with placental malaria at delivery to (A) non-infected women and (B) women with non-placental infection.
Fold-change (log2 transformed), characterizing the magnitude of difference between the antibody levels of two groups (x-axis), is plotted against the -log10 p-value, characterizing the statistical significance of the difference (y-axis). The vertical dotted lines (log2(2) and log2(0.5)) mark a threshold for a twofold change, and the horizontal dotted lines (log10(0.05)) mark the statistical significance threshold (p≤0.05, Welch’s t-test). Antibody features beyond the thresholds are shown in blue and labeled.
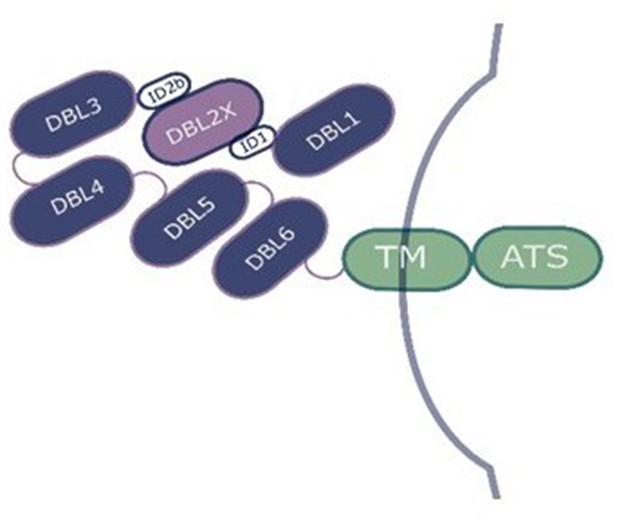
Diagram of the Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) VAR2CSA on the surface of the infected erythrocyte.
DBL2, which is involved in binding to chondroitin sulfate A, is highlighted in light purple. DBL: Duffy binding-like domain; ID: interdomain region; TM: transmembrane segment; ATS: acid terminal segment.
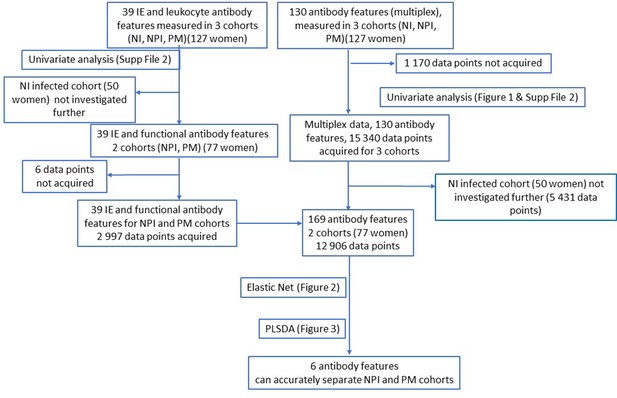
Flow chart of data acquisition and analysis.
NPI: non-placental infection; PM: placental malaria; NI: non-infected; IE: infected erythrocyte; PLSDA: partial least squares discriminant analysis.
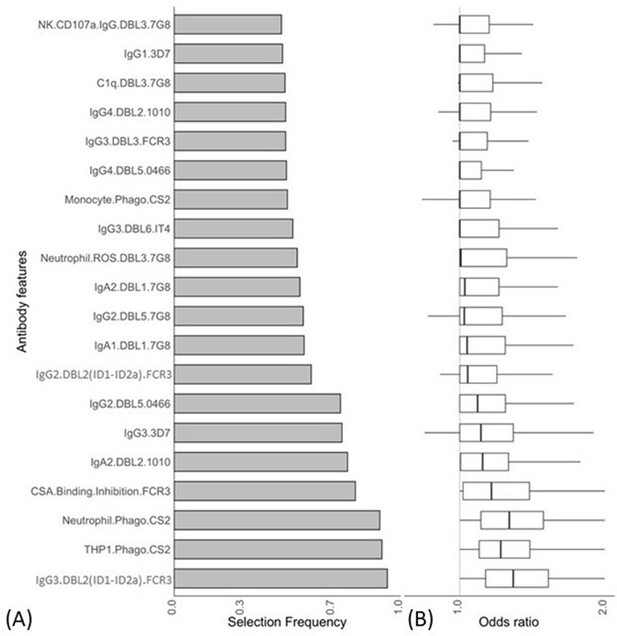
Antibody features that are influential in distinguishing between malaria-infected women with and without placental infection identified by the elastic net-regularized logistic regression model.
Resampling (5000 repeats of 10-fold cross-validation) was used to obtain the selection frequencies and the odds ratios. (A) The top 20 antibody features are ranked in ascending order of selection frequency. (B) Boxplots of the estimated odds ratios, an odds ratio >1 indicates the antibody feature is positively associated with non-placental infection at delivery. Boxplots are median, IQR, whiskers (the lowest data point that falls between Q1 and 1.5 * Q1 IQR, the highest data point that falls between Q3 and 1.5 * Q3 IQR). IQR: interquartile range.
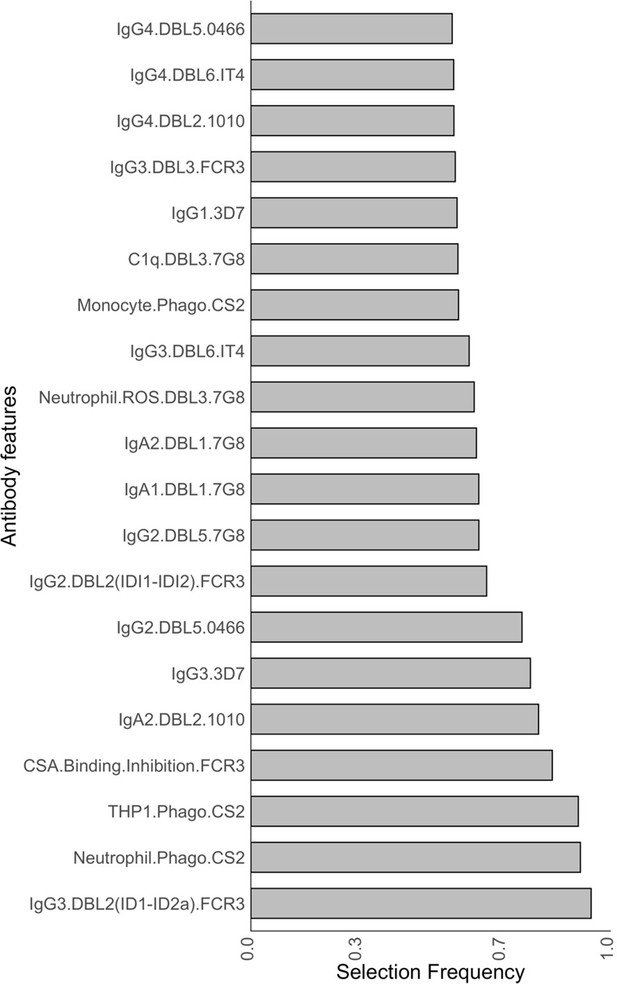
Selection frequencies estimated across all values {0, 0.25, 0.5, 0.75 1} using the resampling of elastic net-regularized logistic regression.
See section Identification of key antibody features.
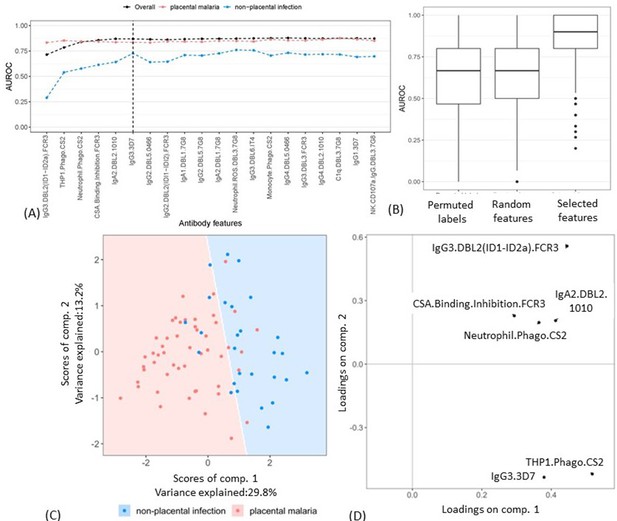
Selecting a minimal set of antibody features by partial least squares discriminant analysis (PLSDA).
(A) Performance of PLSDA at classifying women as having placental malaria (PM) or non-placental infection (NPI) when the features (ranked by selection frequency using the elastic net; Figure 2) are added one by one to the model from highest to lowest rank (500 repeats of 10-fold cross-validation were performed to estimate accuracy for each model). The three lines represent the accuracy of classification of all women in the cohort (black), those with NPI (blue), and those with PM (red). The vertical dashed line denotes the cutoff beyond which the accuracy does not change significantly by adding more antibody features (used for selecting the minimal set of features). (B) Comparing the performance of the elastic net-regularized logistic regression (ENLR) + PLSDA applied on the original data of the top six variables (rightmost boxplot) with two permutation tests (null cases): (1) the PLSDA model was fitted to six randomly selected antibody features and the performance was computed for 500 repeats of 10-fold cross-validation resampling; (2) 100 datasets were generated by randomly permuting the group labels (PM and NPI) and the same analysis performed for the original dataset (i.e., building PLSDA models using the top six frequently selected antibody features found by resampling of elastic net) was repeated for each dataset. (C) Segregation of women with NPI (blue) and PM (red) using the scores of the two components of the PLSDA model with data from the selected six antibody features. The background colors show the predicted classification of the women for all the possible score values in the depicted range. (D) Feature loadings on the components of the PLSDA of the six selected antibody features (see Figure 3—source data 1 for more details about the factor loadings and group prediction using the PLSDA). Boxplots are median, IQR, whiskers (the lowest data point that falls between Q1 and 1.5 * Q1 IQR, the highest data point that falls between Q3 and 1.5 * Q3 IQR). IQR: interquartile range; AUROC: area under the receiver operating characteristic curve.
-
Figure 3—source data 1
Partial least squares discriminant analysis (PLSDA) prediction model.
A PLSDA model with two components and using the six selected antibody features can be formed for prediction of the group of the pregnant women. The estimated loading factors of the model for the two components (shown in Figure 3) are listed here. The softmax technique was used to normalize the scores for each class (placental malaria and non-placental infection) that works as the probability of an observation belonging to a certain class (Kuhn, 2008). The predicted class is the one with the largest model prediction or, equivalently, the largest class probability.
- https://cdn.elifesciences.org/articles/65776/elife-65776-fig3-data1-v3.docx
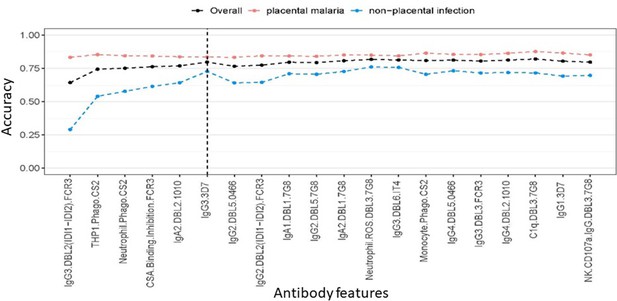
Accuracy of partial least squares discriminant analysis (PLSDA) at classifying women as having placental malaria or non-placental infection when the features (ranked by selection frequency using the elastic net; Figure 2) are added one by one to the model from highest to lowest rank (500 repeats of 10-fold cross-validation were performed to estimate accuracy for each model).
The three lines represent the accuracy of classification of all women in the cohort (black), those with non-placental infection (blue), and those with placental malaria (red). The vertical dashed line denotes the cutoff beyond which the accuracy does not change significantly by adding more antibody features (used for selecting the minimal set of features).
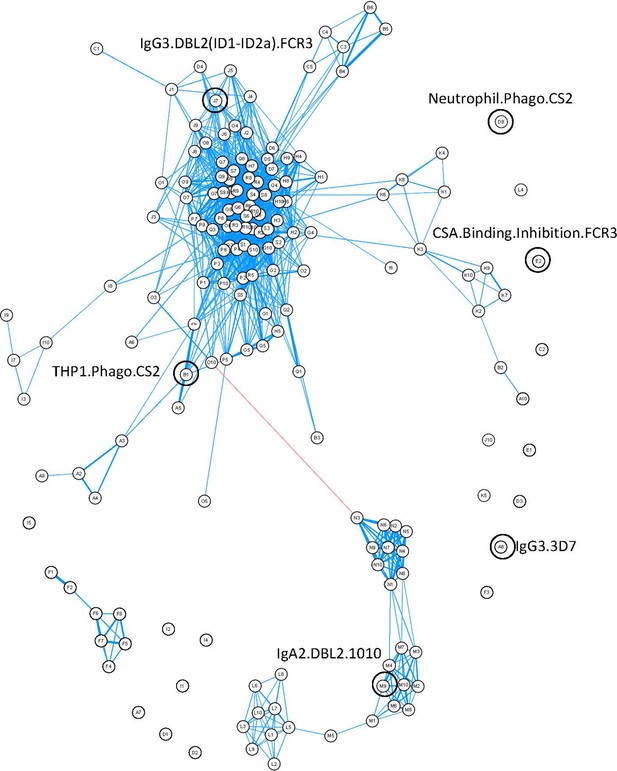
Correlation network of antibody features.
Correlation network of all antibody features in both women with non-placental infection and those with placental malaria. The six selected antibody features do not cluster together. Antibody features with similar functions (denoted by the same letter) tend to correlate with each other. Blue: positive correlation; red: negative correlation; line width and closeness of variables increase with increasing correlation coefficients; only significant correlations (after Bonferroni correction for multiple comparisons) are shown. Selected antibody features identified by elastic net are highlighted and labeled. See supplementary file 3 for a full list of feature names.
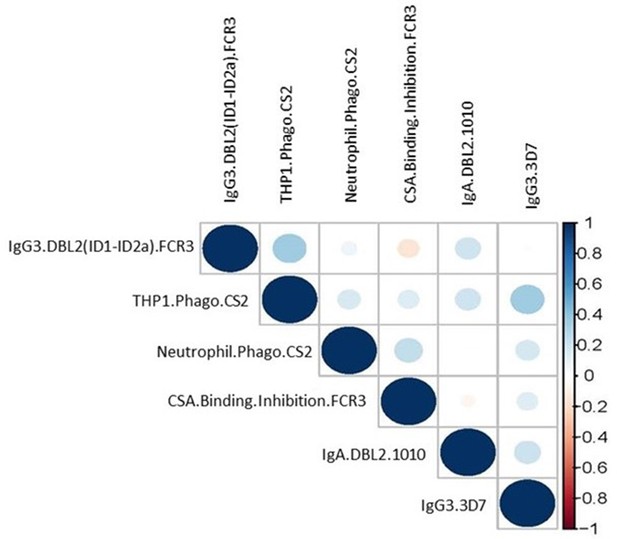
Correlation matrix of the six selected antibody features from women with non-placental infection at delivery and women with placental malaria.
Color and size of dots represent estimates of the correlation coefficient (r) (the further the correlation coefficient from 0 the larger the dot).
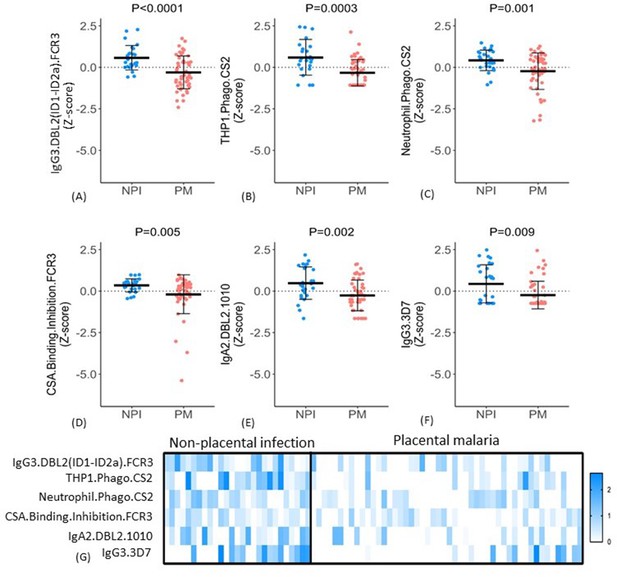
Distribution of antibody features in women with non-placental infection (NPI) and placental malaria (PM).
(A–F) Levels of each of the selected antibody features in individual women in the two groups (G) No single antibody feature was present in all individuals with NPI (or was absent in all those with PM). Errors bars are mean (SD), p-values derived from Welch’s t-test. IE: infected erythrocyte; Z-score: distribution of the features was centered and scaled to have zero mean and a standard deviation of 1.
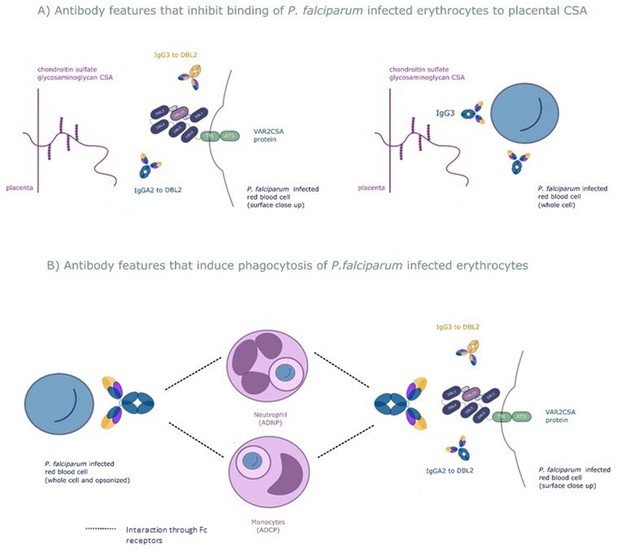
The six selected antibody features may protect women from placental malaria by (A) inhibiting infected red blood cells from binding chondroitin sulfate A (CSA) and sequestering in the placenta and/or (B) promoting phagocytosis of infected red blood cells by monocytes and/or neutrophils.
Selected features that may inhibit placental sequestration include IgG3 to the whole infected red blood cell and IgG3 and IgA2 to VAR2CSA's CSA binding domain DBL2. Selected features that may promote parasite clearance by antibody dependent phagocytosis include IgG3 to the whole infected red blood cell and to DBL2, IgA2 to DBL2, and neutrophil and monocyte phagocytosis of whole infected red blood cells. ADNP: antibody-dependent neutrophil phagocytosis; ADCP: antibody-dependent cellular phagocytosis; VAR2CSA: a parasite protein expressed on the surface of the infected red blood cell, made up of Duffy binding-like domains (DBL).
Tables
Clinical characteristics of the three groups of pregnant women at the time of antibody feature measurement at enrollment (14–26 weeks’ gestation) and also at delivery.
| Non-infected at delivery | Placental malaria at delivery | Non-placental infection at delivery | ||||
|---|---|---|---|---|---|---|
| N = 50 | N = 50 | N = 27 | ||||
| Enrollment | ||||||
| Mean age (years), SD | 24.5 | 5.3 | 24.0 | 5.0 | 23.1 | 4.4 |
| Residence, N (%) | ||||||
| Rural | 37 | (74.0) | 38 | (76.0) | 18 | (66.7) |
| Non-rural | 13 | (26.0) | 12 | (24.0) | 9 | (33.3) |
| Ethnicity, N (%) | ||||||
| Sepik | 6 | (12.0) | 11 | (22.0) | 3 | (11.1) |
| Madang/Morobe | 39 | (78.0) | 30 | (60.0) | 22 | (81.5) |
| Highlander | 3 | (6.0) | 5 | (10.0) | 1 | (3.7) |
| Other | 2 | (4.0) | 4 | (8.0) | 1 | (3.7) |
| Formal schooling, N (%) | 46 | (92.0) | 46 | (92.0) | 25 | (92.6) |
| Smoking, N (%) | 9 | (18.0) | 11 | (22.0) | 6 | (22.2) |
| Betel nut user, N (%)† | 41 | (82.0) | 41 | (82.0) | 24 | (88.9) |
| Alcohol, N (%) | 2 | (4.0) | 2 | (4.0) | 2 | (7.4) |
| Gravidity, N (%) | ||||||
| Primigravidae | 26 | (52.0) | 29 | (58.0) | 14 | (51.9) |
| Secundigravidae | 8 | (16.0) | 7 | (14.0) | 8 | (29.6) |
| Multigravidae | 16 | (32.0) | 14 | (28.0) | 5 | (18.5) |
| IPTp regime, N (%) | ||||||
| SPCQ | 27 | (54.0) | 30 | (60.0) | 18 | (66.7) |
| SPAZ | 23 | (46.0) | 20 | (40.0) | 9 | (33.3) |
| Mean gestational age (days), SD | 145.9 | 31.4 | 147.5 | 31.3 | 152.2 | 19.8 |
| Mean maternal weight (kg), SD* | 54.7 | 13.1 | 53.5 | 8.2 | 54.1 | 7.4 |
| Mean maternal height (cm), SD† | 154.3 | 5.9 | 154.6 | 6.9 | 154.4 | 6.0 |
| Bed net use, N (%) | 34 | (68.0) | 40 | (80.0) | 21 | (77.8) |
| Hb (g/dL), mean SD‡ | 9.7 | 1.2 | 9.3 | 2.0 | 9.4 | 1.2 |
| Light microscopy positive for Pf, N (%) | 2 | (4.0) | 4 | (8.0) | 4 | (14.8) |
| PCR positive for Pf, N (%) | 4 | (8.0) | 5 | (10.0) | 5 | (18.5) |
| Delivery | ||||||
| Placenta light microscopy positive for Pf, N (%) | 0 | (0) | 50 | (100) | 0 | (0) |
| Peripheral blood light microscopy positive for Pf, N (%) | 0 | (0) | 10 | (20) | 13 | (48.2) |
| Peripheral blood PCR positive for Pf, N (%) | 0 | (0) | 13 | (26) | 21 | (77.8) |
| Placental blood PCR positive for Pf, N (%)§ | 1 | (2.0) | 10 | (20) | 10 | (37.04) |
| Birthweight (g), SD | 3062 | 546 | 2827 | 501 | 2840 | 416 |
| Gestation at delivery (days), SD¶ | 278 | 18 | 279 | 16 | 280 | 13 |
| Mean Hb (g/dL), SD | 10.1 | 2 | 9.5 | 1.9 | 10.2 | 1.4 |
-
* One participant with missing data on betel nut use in placental malaria.
†One participant with missing data on weight in the non-infected group.
-
‡ Missing Hb data, five in non-infected group, four in placental malaria group, and two in the non-placental infection group.
§ Missing placental PCR data in five non-infected, eight placental malaria, and four non-placental infection women.
-
¶One participant with missing data on gestation length at delivery in the non-infected group.
SD: standard deviation; Hb: hemoglobin; PCR: polymerase chain reaction; IPTp: intermittent preventive treatment in pregnancy; SPAZ: sulfadoxine pyrimethamine-azithromycin; SPCQ: sulfadoxine pyrimethamine-chloroquine.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Plasmodium falciparum, CS2) | CS2 | Chandrasiri et al., 2014 | ||
| Strain, strain background (Plasmodium falciparum, 3D7) | 3D7 | Chandrasiri et al., 2014 | Selected for chondroitin sulfate A (CSA) adhesion | |
| Strain, strain background (Plasmodium falciparum, FCR3) | FCR3 | Nielsen and Salanti, 2015 | Selected for CSA adhesion (parent of CS2) | |
| Strain, strain background (Plasmodium falciparum, NF54) | NF54 | Nielsen and Salanti, 2015 | Selected for CSA adhesion (parent of 3D7) | |
| Biological sample (Homo sapiens) | Plasma | Unger et al., 2015 | ||
| Biological sample (Homo sapiens) | Primary monocytes | This paper | Freshly isolated cells (see Materials and methods – Primary leukocytes) | |
| Biological sample (Homo sapiens) | Primary neutrophils | This paper | Freshly isolated cells (see Materials and methods – Primary leukocytes) | |
| Biological sample (Homo sapiens) | Primary NK cells | This paper | Freshly isolated cells (see Materials and methods – Primary leukocytes) | |
| Cell line (Homo sapiens) | THP1 | Ataíde et al., 2010 | RRID:CVCL_0006 | Monocytic cell line |
| Peptide, recombinant protein | DBL1-7G8 | Avril et al., 2011 | MV-1398 | Parasite line 7G8 |
| Peptide, recombinant protein | DBL2(ID1-ID2a)-FCR3 | Doritchamou et al., 2016 | MV1942 | Parasite line FCR3 |
| Peptide, recombinant protein | DBL2(ID1-ID2a)-FCR3 | Mordmüller et al., 2019 | Parasite line FCR3 | |
| Peptide, recombinant protein | DBL2-isolate | Doritchamou et al., 2016 | MV 1940 | Parasite isolate 1010 |
| Peptide, recombinant protein | DBL3- FCR3 | Nielsen et al., 2009 | MP1028 | Parasite line FCR3 |
| Peptide, recombinant protein | DBL3- 7G8 | Avril et al., 2011 | MV-1914 | Parasite line 7G8 |
| Peptide, recombinant protein | DB4-FCR3 | Fried et al., 2018 | MP2369 | Parasite line FCR3 |
| Peptide, recombinant protein | DBL4-isolate | Doritchamou et al., 2016 | MV1700 | Parasite isolate I 0711 |
| Peptide, recombinant protein | DBL5-3D7 | Avril et al., 2011 | 1218 | Parasite line 3D7 |
| Peptide, recombinant protein | DBL5-7G8 | Avril et al., 2011 | 1269 | Parasite line 7G8 |
| Peptide, recombinant protein | DBL5-isolate | Doritchamou et al., 2016 | MV 1749 | Parasite isolate I 0466 |
| Peptide, recombinant protein | DBL6-IT4 | Avril et al., 2011 | MV-1137 | Parasite line IT4 |
| Peptide, recombinant protein | MSP-1 | Barua et al., 2019 | ||
| Biological sample (Plasmodium falciparum) | Schizont extract | Barua et al., 2019 | ||
| Antibody | Goat anti-human IgG (polyclonal) | Mabtech | 3820-4-250 | (1:2000) |
| Antibody | Mouse anti-human IgG-PE (polyclonal) | SouthernBiotech | 9040-09 RRID:AB_2796601 | (1:77) |
| Antibody | Mouse anti-human IgG1-PE (monoclonal) | SouthernBiotech | 9052-09 RRID:AB_2796621 | (1:77) |
| Antibody | Mouse anti-human IgG2-PE (monoclonal) | SouthernBiotech | 9070-09 RRID:AB_2796639 | (1:77) |
| Antibody | Mouse anti-human IgG3-PE (monoclonal) | SouthernBiotech | 9210-09 RRID:AB_2796701 | (1:77) |
| Antibody | Mouse anti-human IgG4-PE (monoclonal) | SouthernBiotech | 9200-09 RRID:AB_2796693 | (1:77) |
| Antibody | Mouse anti-human IgA1-PE (monoclonal) | SouthernBiotech | 9130-09 RRID:AB_2796656 | (1:77) |
| Antibody | Mouse anti-human IgA2-PE (monoclonal) | SouthernBiotech | 9140-09 RRID:AB_2796664 | (1:77) |
| Antibody | Mouse anti-human IgM-PE (monoclonal) | SouthernBiotech | 9020-09 RRID:AB_2796577 | (1:77) |
| Antibody | Rabbit anti-human IgG (polyclonal) | DAKO | A0425 | (1:100) |
| Antibody | Mouse anti-human IgG1 HP6069 (monoclonal) | Merck Millipore | 411451 | (1:50) |
| Antibody | Mouse anti-human IgG2 HP6002 (monoclonal) | Merck Millipore | MAB1308 | (1:50) |
| Antibody | Mouse anti-human IgG3 HP6050 (monoclonal) | Sigma | I7260.2ml | (1:50) |
| Antibody | Mouse anti-human IgG4 HP6023 (monoclonal) | Merck Millipore | MAB1312-K | (1:50) |
| Antibody | Goat anti-mouse IgG AlexaFluor 647 (polyclonal) | Life Technologies | A-21235 RRID:AB_2535804 | (1:500) |
| Antibody | Donkey anti-rabbit 647 (polyclonal) | Life Technologies | A-31573 RRID:AB_2536183 | (1:500) |
| Peptide, recombinant protein | C1q | MP Biomedicals | 80295-33-6 | 1.3 µg/ml |
| Peptide, recombinant protein | FcγRI | R&D Systems | 1257-FC | |
| Peptide, recombinant protein | FcγRIIa | Wines et al., 2016 | ||
| Peptide, recombinant protein | FcγRIIIa | Wines et al., 2016 | ||
| Peptide, recombinant protein | FcγRIIIb | R&D Systems | 1875 CD | |
| Antibody | Mouse anti-human CD16-Brilliantviolet 605 (monoclonal) | BD | 563172 | (1:50) |
| Antibody | Mouse anti-human CD56-Brilliantultraviolet 737 (monoclonal) | BD | 347344 | (1:800) |
| Antibody | Mouse anti-human CD3-peridinin-chlorophyll-protein (monoclonal) | BD | 552127 | (1:200) |
| Antibody | Mouse anti-human IFNγ-PE (monoclonal) | BD | 554701 | (1:200) |
| Antibody | Mouse anti-human TNFα-BV-785 (monoclonal) | BioLegend | 502947 RRID:AB_2565857 | (1:200) |
| Software, algorithm | R software, caret package | R software Kuhn, 2008 | ||
| Software, algorithm | R software, glmnet package | R software Zou and Hastie, 2005 | ||
| Software, algorithm | R software, PLS package | R software Mevik and Wehrens, 2007 | ||
| Software, algorithm | R software, mixOmics package | R software Rohart et al., 2017 | ||
| Software, algorithm | R software, qgraph package | R software Epskamp et al., 2012 | ||
| Commercial assay or kit | EasySep Direct Human Neutrophil Isolation Kit | STEMCELL Technologies | 19666 | |
| Commercial assay or kit | RosetteSep Human Monocyte Enrichment Cocktail | STEMCELL Technologies | 15068 | |
| Commercial assay or kit | RosetteSep Human NK Enrichment Cocktail | STEMCELL Technologies | 15065 | |
| Commercial assay or kit | Melon Gel purification kits | Thermo Fisher Scientific | 45212 | |
| Commercial assay or kit | EZ-link Sulfo NHS-LC-Biotin kit | Thermo Fisher Scientific | 24520 | |
| Other | Bio-Plex magnetic carboxylated microspheres | Bio-Rad | #MC100xx-01 | |
| Other | Streptavidin-PE | SouthernBiotech | 7105-09 |
Additional files
-
Supplementary file 1
Recombinant VAR2CSA DBL domain proteins used to measure antibody features.
- https://cdn.elifesciences.org/articles/65776/elife-65776-supp1-v3.docx
-
Supplementary file 2
Univariate analysis non-infected vs. placental malaria and non-placental infection vs. placental malaria.
- https://cdn.elifesciences.org/articles/65776/elife-65776-supp2-v3.docx
-
Supplementary file 3
Antibody feature code, name, and description.
- https://cdn.elifesciences.org/articles/65776/elife-65776-supp3-v3.docx
-
Supplementary file 4
Table 1: Association between selected antibody features and graviditya in 77 women (women with placental malaria or non-placental infection at delivery) and Table 2: univariate analysis of selected antibody features placental malaria v non-placental infection, in women uninfected at enrollment.
- https://cdn.elifesciences.org/articles/65776/elife-65776-supp4-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65776/elife-65776-transrepform-v3.docx
-
Reporting standard 1
Tripod checklist.
- https://cdn.elifesciences.org/articles/65776/elife-65776-repstand1-v3.docx





