External signals regulate continuous transcriptional states in hematopoietic stem cells
Figures
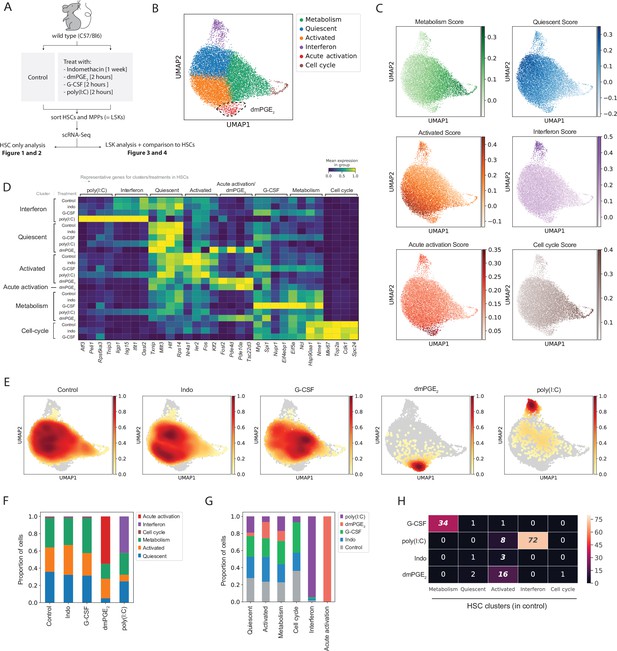
Hematopoietic stem cells (HSCs) are transcriptionally heterogeneous and niche perturbations rapidly shift cells into different states.
(A) Schematic of stimulant treatment before HSC and multipotent progenitor (MPP) isolation, see also Figure 1—figure supplement 1. (B) Uniform manifold approximation and projection (UMAP) plot of HSC clusters (n = 15,355 cells), with 16,16-dimethyl prostaglandin E2 (dmPGE2)-induced cluster (red) traced with a dashed line, see also Figure 1—figure supplement 2A-G. (C) UMAP plot with transcriptional scores for each cluster. (D) Heatmap of selected enriched genes for each HSC cluster and treatment (columns, scaled expression) averaged gene expression for all cells within a cluster and treatment (rows, only clusters shown with >20 cells), see also Figure 1—figure supplement 2I and Figure 1—figure supplement 4. (E) UMAP density graphs of HSC distribution for each external stimulant. (F) Proportion of HSCs within clusters for each perturbation. (G) Proportion of HSCs of each perturbation within a cluster normalized for total cell number per treatment. (H) Heatmap with number of common genes between the 100 top induced genes per HSC treatment (rows) and HSC clusters (columns), false discovery rate (FDR)-corrected hypergeometric p-values < 0.01 are italicized, exact p-values in Figure 1—source data 1. For separate analysis of male and female HSCs, see Figure 1—figure supplement 3.
-
Figure 1—source data 1
Excel spreadsheet containing quantitative data for Figure 1.
- https://cdn.elifesciences.org/articles/66512/elife-66512-fig1-data1-v1.xlsx
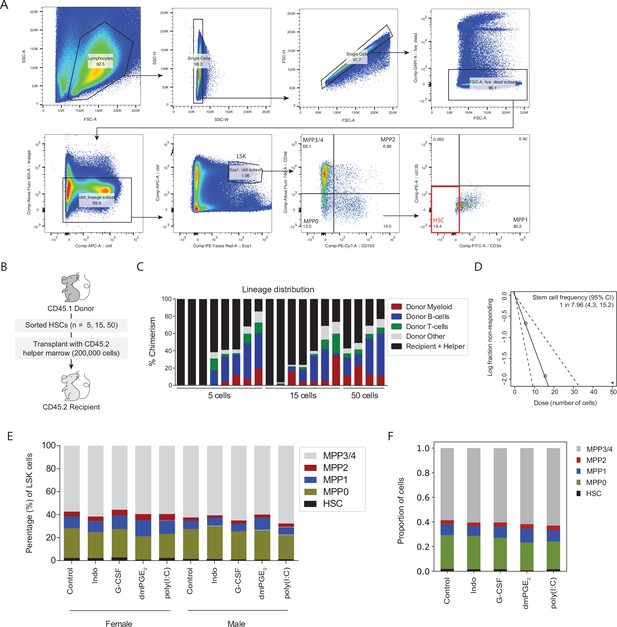
Functional characterization of hematopoietic stem cells (HSCs) confirms high regenerative capacity.
(A) Sorting scheme of multipotent progenitors (MPPs) and HSCs. Cells were lineage depleted prior to sort. (B) Schematic of limiting dilution transplantation assay (LDTA) experiment. (C) Chimerism and lineage distribution per mouse 4 months post-transplant. (D) Extreme limiting dilution assay (ELDA) for LDTA experiment. (E) Percentage of MPPs and HSCs within Lin-, c-Kit+, Sca1+ (LSK) cells at baseline and after niche stimulation (pooled cells from five mice for each condition). (F) Proportion of each surface phenotype within all five experimental conditions after computational reassembly of the LSK compartment.
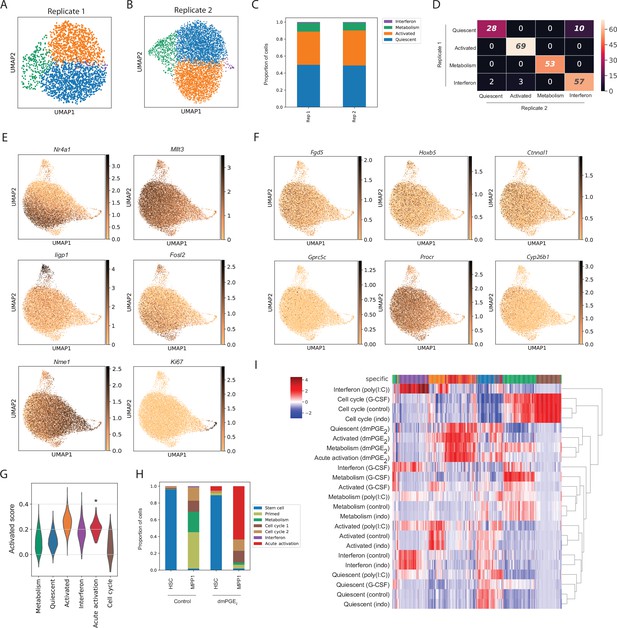
Evaluation of single-cell RNA sequencing (scRNA-Seq) clustering with independent replicates, candidate genes, and transcriptional scores.
(A–D) Comparison of clustering of control hematopoietic stem cells (HSCs) in two biological replicates. Uniform manifold approximation and projection (UMAP) plots for replicates 1 (A, n = 2382 cells) and 2 (B, n = 5334 cells) and summarized cell proportions in each cluster (C). (D) Heatmap with number of common genes between the 100 top enriched genes for Replicate 1 (rows) and Replicate 2 (columns) clusters, false discovery rate (FDR)-corrected hypergeometric p-values < 0.01 are italicized, exact p-values listed in Figure 1—source data 1. (E) UMAP plot with expression of representative genes for each HSC cluster. (F) UMAP plots with expression of previously described HSC markers. (G) Violin plot of transcriptional scores in HSC clusters computed from HSC ‘activation’ cluster with highest mean ‘Activated’ score (besides the ‘Activated’ cluster) indicated with an asterisk, for exact p-values and confidence intervals, see Figure 1—source data 1. (H) Proportion of 16,16-dimethyl prostaglandin E2 (dmPGE2) and control cells within clusters split by surface phenotype for HSCs and multipotent progenitors 1 (MPP1s). (I) Unified heatmap with top 100 genes per cluster and treatment. Single-cell expression is averaged within a single-cell cluster, scaled to z-scores and similar genes (columns) and clusters (rows) are aggregated by hierarchical clustering. Top row (‘specific’) indicates the cluster or treatment each gene is assigned to, the color scheme is the same as in Figure 1F (clusters) and Figure 1G (treatments).
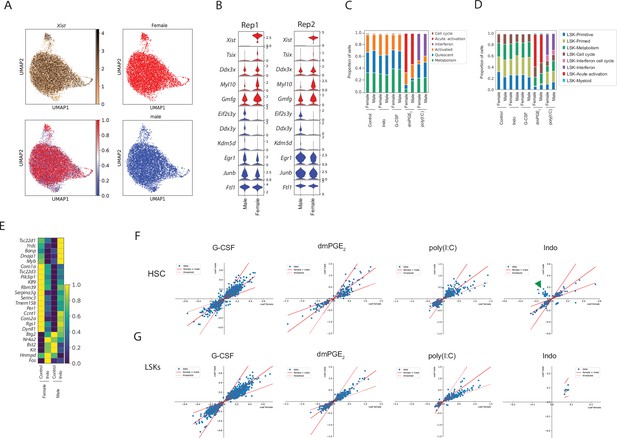
Minimal sexual dimorphism in hematopoietic stem cells (HSCs) and Lin-, c-Kit+, Sca1+ (LSKs) in steady state and upon stimulation.
(A) Xist expression, classification of male and female cells and male and female cells plotted separately. (B) Stacked violin plots of all consistent sexually dimorphic genes (red; female, blue; male) for two independent biological replicates. (C–D) Proportions of male and female HSCs (C) and LSK cells (D) within clusters for each drug treatment (p-value(DPA) > 0.05 for all male vs. female comparisons). (E) Heatmap of average expression for ‘opposite directionality’ genes in HSCs for indomethacin (‘Indo’) and control. (F) Scatter plot of differential expression coefficient (converted to log2 scale) induced by stimulants in HSCs (F) and LSKs (G) between male (y-axis) and female (x-axis). Solid red line indicates equal expression coefficients (coef female = coef male) and dashed line indicates a twofold deviation (2*coef female = coef male or vice versa). Green arrowhead indicates ‘opposite directionality’ genes in indomethacin (shown also in E).
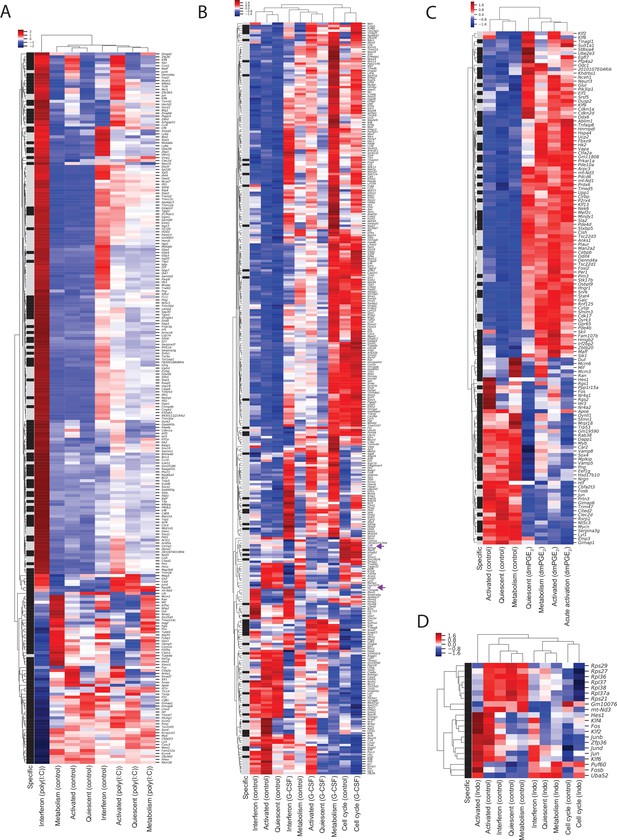
Heatmaps of differentially expressed genes in hematopoietic stem cells (HSCs) enables identification of genes and single-cell clusters with similar expression patterns.
(A–D) Heatmap of differentially expressed genes between external stimulants and control in HSCs. Single-cell expression is averaged within a single-cell RNA sequencing (scRNA-Seq) cluster, scaled to z-scores and similar genes (rows) and clusters (columns) are aggregated by hierarchical clustering. Row label (‘specific’) in black indicates HSC-specific genes, gray label marks genes differentially expressed in both HSCs and Lin-, c-Kit+, Sca1+ (LSKs). (A) Poly (I:C) at 1.5-fold cutoff. (B) Granulocyte colony-stimulating factor (G-CSF) at 1.2-fold cutoff, (C) 16,16-Dimethyl prostaglandin E2 (dmPGE2) at 1.5-fold cutoff, (D) indomethacin at 1.2-fold cutoff.
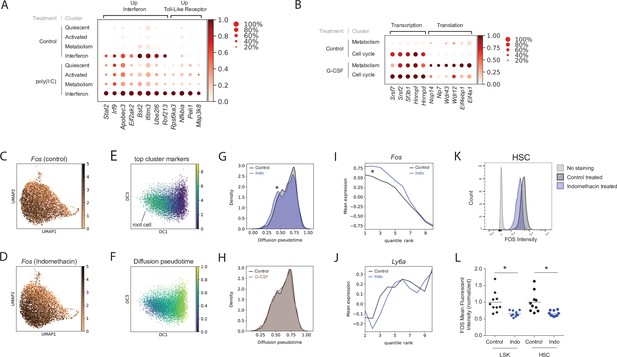
Poly(I:C), granulocyte colony-stimulating factor (G-CSF), and indomethacin induce cluster-specific transcriptional changes in hematopoietic stem cells (HSCs).
(A) Dot plot of representative genes from poly(I:C) treated and control HSC clusters (scaled expression across columns). (B) Dot plot of representative genes from the G-CSF-treated and control HSC clusters (scaled expression across columns). (C–J) Diffusion pseudotime analysis. Uniform manifold approximation and projection (UMAP) plot of Fos expression in control (C) and upon indomethacin (D) treatment, see also Figure 2—figure supplement 1A-B. Diffusion map embedding with combined expression of top ‘Activated’ genes to select root cell (E) and cells colored by pseudotime (F). Kernel density of pseudotime distribution comparing indomethacin and control (G, asterisk: p-value [Mann–Whitney U-test] = 5.8*10–12) and G-CSF and control (H). Average expression of Fos (I) and Ly6a (J) across cells ranked by pseudotime (cells split into 10 bins to decrease noise), change in transcript levels indicated by asterisk in I, see also Figure 2—figure supplement 1C-D. (K) Histogram of FOS levels via intracellular fluorescence-activated cell sorting (FACS) of HSCs, ‘no stain’ is FACS-negative control, ‘control’ is FOS in untreated mice. (L) Normalized mean fluorescent intensity (MFI) for FOS in control and indomethacin-treated HSCs (p-value = 6.2 * 10–3, Welch-corrected t-test, asterisk) and LSK cells (p-value = 6.6 * 10–3, Welch-corrected t-test, asterisk) across two independent biological replicate experiments, n(mice) = 20.
-
Figure 2—source data 1
Excel spreadsheet containing quantitative data for Figure 2.
- https://cdn.elifesciences.org/articles/66512/elife-66512-fig2-data1-v1.xlsx
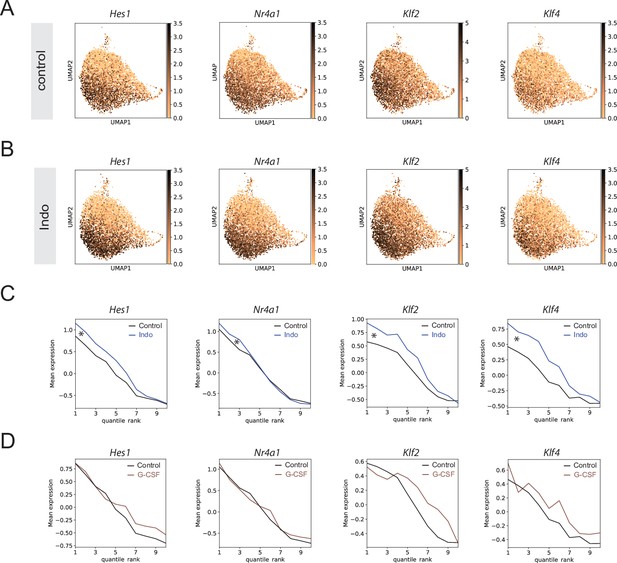
Indomethacin affects transcriptional state of immediate early genes (IEGs).
(A) Uniform manifold approximation and projection (UMAP) plot with expression of selected ‘Activated’ genes in control (A) and indomethacin (B). Average expression of the same genes across cells ranked by pseudotime (cells split into 10 bins to decrease noise) comparing indomethacin and control (C, difference indicated by asterisk) or granulocyte colony-stimulating factor (G-CSF) and control (D).
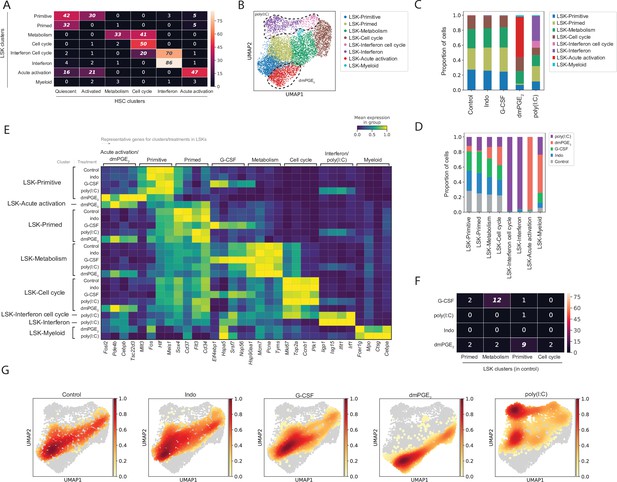
Comparative analysis of Lin-, c-Kit+, Sca1+ (LSK) response to external stimulants.
(A) Heatmap with number of common genes between the 100 top enriched genes for LSK (rows) and hematopoietic stem cell (HSC) (columns) clusters, false discovery rate (FDR)-corrected hypergeometric p-values < 0.01 are italicized, exact p-values listed in Figure 3—source data 1. (B) Uniform manifold approximation and projection (UMAP) plot of LSK clustering (n = 8191 cells), with induced clusters by 16,16-dimethyl prostaglandin E2 (dmPGE2) (red) and poly(I:C) (pink and purple) traced with dashed line, see also Figure 3—figure supplement 1. (C) Proportion of LSK cells within clusters for each perturbation. (D) Proportion of LSK cells of each perturbation within a cluster normalized for total cell number per treatment. (E) Heatmap of selected enriched genes for each LSK cluster and treatment (columns, scaled expression) averaged gene expression for all cells within a cluster and treatment (rows, only clusters shown with >20 cells), see also Figure 3—figure supplement 2. (F) Heatmap with number of common genes between the 100 top induced genes per LSK treatment (rows) and LSK clusters (columns), FDR-corrected hypergeometric p-values < 0.01 are italicized, exact p-values listed in Figure 3—source data 1. (G) UMAP density graphs of LSK distribution for each external stimulant.
-
Figure 3—source data 1
Excel spreadsheet containing quantitative data for Figure 3.
- https://cdn.elifesciences.org/articles/66512/elife-66512-fig3-data1-v1.xlsx
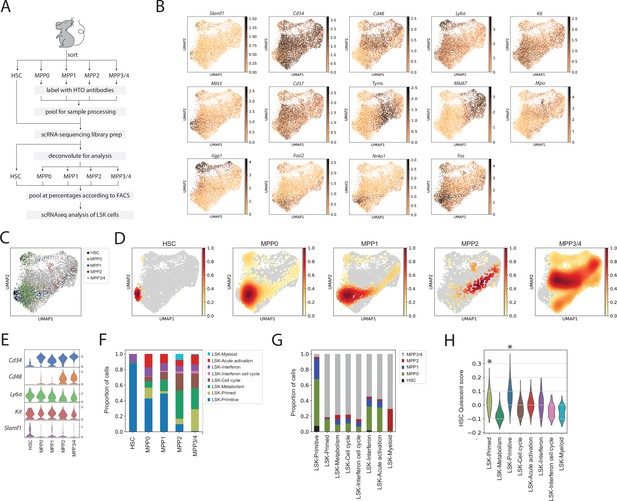
Multipotent progenitor (MPP) surface marker expression validates Lin-, c-Kit+, Sca1+ (LSK) cluster definitions.
(A) Schematic of LSK pooling and cellular indexing of transcriptomes and epitopes by sequencing (CITE-Seq) surface hashtag (hashtag oligonucleotide [HTO]) methodology. (B) Uniform manifold approximation and projection (UMAP) plots with expression of selected genes in LSK cells. (C) UMAP plot of surface receptor phenotypes in LSK cells. (D) UMAP density graphs visualizing the distribution of cells by surface phenotype. (E) Stacked violin plots of gene expression for surface markers within MPPs and hematopoietic stem cells (HSCs). (F) Proportion of LSK cells belonging to different clusters for each surface phenotype. (G) Proportion of surface phenotypes within each LSK cluster. (H) Violin plot of transcriptional scores computed from HSC ‘quiescent’ cluster in LSKs with the two highest mean ‘quiescent’ scores indicated with asterisks, for exact p-values and confidence intervals, see Figure 3—source data 1.
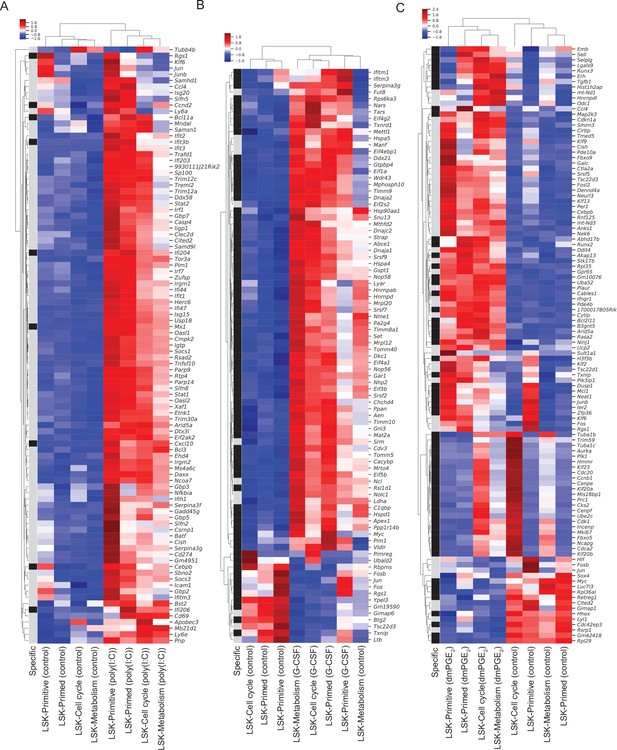
Heatmaps of differentially expressed genes in Lin-, c-Kit+, Sca1+ (LSKs) enable identification of genes and single-cell clusters with similar expression patterns.
(A–D) Heatmap of differentially expressed genes between external stimulants and control in LSKs. Single-cell expression is averaged within a single-cell RNA sequencing (scRNA-Seq) cluster, scaled to z-scores and similar genes (rows) and clusters (columns) are aggregated by hierarchical clustering. Row label (‘specific’) in black indicates LSK-specific genes, gray label marks genes differentially expressed in both hematopoietic stem cells (HSCs) and LSKs. (A) Poly (I:C) at 1.5-fold cutoff. (B) Granulocyte colony-stimulating factor (G-CSF) at 1.5-fold cutoff, (C) 16,16-dimethyl prostaglandin E2 (dmPGE2) at 1.5-fold cutoff.
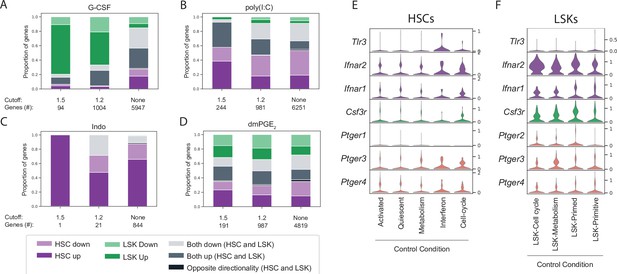
Lin-, c-Kit+, Sca1+ (LSK) and hematopoietic stem cell (HSC) cluster-specific differential gene expression cannot be explained by receptor expression.
(A–D) Stacked bar graphs with proportion of differentially expressed genes that are unique for HSCs (purple), LSKs (green) or common (gray) upon granulocyte colony-stimulating factor (G-CSF) (A), poly(I:C) (B), Indo (C), or 16,16-dimethyl prostaglandin E2 (dmPGE2) (D) treatment. Below each bar graph the total number of differentially expressed genes (‘genes #’) for each fold-change (‘cutoff’) is listed. (E–F) Violin plots of receptor expression in control HSCs (E) and LSKs (F) split by cluster (only clusters with >20 cells displayed).
-
Figure 4—source data 1
Excel spreadsheet containing quantitative data for Figure 4.
- https://cdn.elifesciences.org/articles/66512/elife-66512-fig4-data1-v1.xlsx
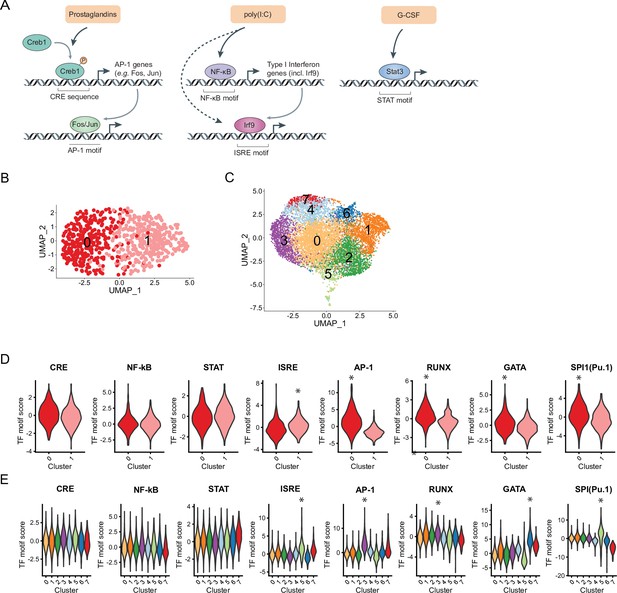
Heterogeneous distribution of interferon signaling response element (ISRE) and AP-1 motif in hematopoietic stem cells (HSCs) and Lin-, c-Kit+, Sca1+ (LSKs) and specific motif co-occurrences in HSCs.
(A) Schematic of downstream transcriptional signaling pathways for externalstimulants. (B–C) Uniform manifold approximation and projection (UMAP) plot of HSC (B) single-cell chromatin accessibility sequencing (scATAC-Seq) clusters (n = 730 cells) or LSK (C) scATAC-Seq clusters (n = 10,750 cells), see also Figure 5—figure supplement 1A-B. (D–E) Violin plots of transcription factor (TF) motif scores enriched in HSCs (D) and LSKs (E) with selected significant p-values (logistic regression) indicated by asterisks, see also Supplementary file 11 and Figure 5—figure supplement 1C-D.
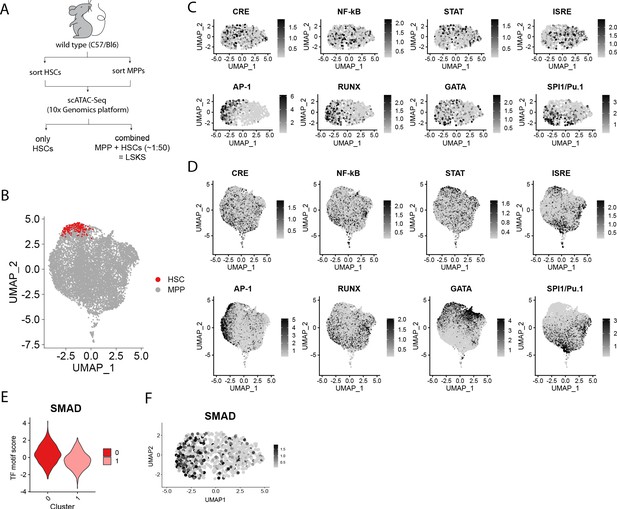
Uniform distribution of motif activity immediately downstream of external stimulants and differential enrichment for secondary signals in hematopoietic stem cells (HSCs) and Lin-, c-Kit+, Sca1+ (LSKs).
(A) Schematic of fluorescence-activated cell sorting (FACS) and analysis of the single-cell chromatin accessibility sequencing (scATAC-Seq) experiment. (B) Uniform manifold approximation and projection (UMAP) plot of combined LSKs (n = 10,750 cells) colored by sorted HSCs or multipotent progenitors (MPPs). (C–D) UMAP plots of various transcription factor (TF) motif scores in HSCs (C) or LSKs (D). (E–F) Violin plot (E) and UMAP plot (F) of SMAD TF motif score in HSCs.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus)Male and female | Replicate 1 | Jackson Laboratory | RRID:IMSR_JAX:016617 | |
| Genetic reagent (Mus musculus)Male and female | Replicate 2, CD45.2 (transplant recipients) | Jackson Laboratory | RRID:IMSR_JAX:000664 | Used for pharmacological perturbations |
| Genetic reagent (Mus musculus)Female only | Transplant donors | Jackson Laboratory | RRID:IMSR_JAX:002014 | |
| Antibody | Anti-CD117 (c-Kit), ACK2, APC (rat monoclonal) | Thermo Fisher Scientific(17-1172-83) | RRID:AB_469434 | FACS (1:100) |
| Antibody | Anti-CD11b/Mac1, M1/70, eFluor 450 (rat monoclonal) | Thermo Fisher Scientific(48-0112-80) | RRID:AB_1582237 | FACS (1:100) |
| Antibody | Anti-CD11b/Mac1, M1/70, PE-Cyanine5 (rat monoclonal) | Thermo Fisher Scientific(15-0112-83) | RRID:AB_468715 | FACS (1:100) |
| Antibody | Anti-CD11b/Mac1, M1/70, Alexa Fluor 700 (rat monoclonal) | BD Pharmingen(557960) | RRID:AB_396960 | FACS (1:300) |
| Antibody | Anti-CD135 (Flt3), A2F10, PE (rat monoclonal) | Thermo Fisher Scientific(12-1351-81) | RRID:AB_465858 | FACS (1:100) |
| Antibody | Anti-CD150, TC15-12F12.2, PE/Cy7 (rat monoclonal) | Biolegend(115914) | RRID:AB_439797 | FACS (1:100) |
| Antibody | Anti-CD3, 17A2, APC (rat monoclonal) | Thermo Fisher Scientific(17-0032-82) | RRID:AB_10597589 | FACS (1:100) |
| Antibody | Anti-CD34, RAM34, eFluor 450 (rat monoclonal) | Thermo Fisher Scientific(48-0341-80) | RRID:AB_2043838 | FACS (1:33) |
| Antibody | Anti-CD34, RAM34, FITC (rat monoclonal) | Thermo Fisher Scientific(11-0341-85) | RRID:AB_465022 | FACS (1:33) |
| Antibody | Anti-CD3e, 145–2C11, eFluor 450 (armenian hamster monoclonal) | Thermo Fisher Scientific(48-0031-80) | RRID:AB_10733280 | FACS (1:100) |
| Antibody | Anti-CD3e, 145–2C11, PE-Cyanine5 (armenian hamster monoclonal) | Thermo Fisher Scientific(15-0031-83) | RRID:AB_468691 | FACS (1:100) |
| Antibody | Anti-CD45.1, A20, FITC (mouse monoclonal) | BD Pharmingen(553775) | RRID:AB_395043 | FACS (1:100) |
| Antibody | Anti-CD45.2, 104, PE (mouse monoclonal) | BD Pharmingen(560695) | RRID:AB_1727493 | FACS (1:100) |
| Antibody | Anti-CD45R (B220), RA3-6B2, eFluor 450 (rat monoclonal) | Thermo Fisher Scientific(48-0452-80) | RRID:AB_1548763 | FACS (1:100) |
| Antibody | Anti-CD45R (B220), RA3-6B2, PE-Cyanine5 (rat monoclonal) | Thermo Fisher Scientific(15-0452-83) | RRID:AB_468756 | FACS (1:100) |
| Antibody | Anti-CD45R/(B220), RA3-6B2, pacific Blue (rat monoclonal) | Biolegend(103227) | RRID:AB_492876 | FACS (1:100) |
| Antibody | Anti-CD48, HM48-1, Alexa Fluor 700 (armenian hamster monoclonal) | Biolegend(103425) | RRID:AB_10612754 | FACS (1:100) |
| Antibody | Anti-CD5, 53–7.3, eFluor 450 (rat monoclonal) | Thermo Fisher Scientific(48-0051-80) | RRID:AB_1603252 | FACS (1:100) |
| Antibody | Anti-CD8a, 53–6.7, eFluor 450 (rat monoclonal) | Thermo Fisher Scientific(48-0081-80) | RRID:AB_1272235 | FACS (1:100) |
| Antibody | Anti-c-Fos, H15-S, FITC (rabbit monoclonal) | Abcam(ab175647) | RRID:AB_2893164 | FACS (10 µl for 1 MIO cells) |
| Antibody | Anti-Ly-6A/E (Sca-1), D7, PE-eFluor 610 (rat monoclonal) | Thermo Fisher Scientific(61-5981-80) | RRID:AB_2574647 | FACS (1:100) |
| Antibody | Anti-Ly-6A/E (Sca-1), D7, APC/Cy7 (rat monoclonal) | Biolegend(108125) | RRID:AB_10639725 | FACS (1:100) |
| Antibody | Anti-Ly-6G (Gr-1), RB6-8C5, eFluor 450 (rat monoclonal) | Thermo Fisher Scientific(48-5931-80) | RRID:AB_1548797 | FACS (1:100) |
| Antibody | Anti-Ly-6G (Gr-1), RB6-8C5, PE-Cyanine5 (rat monoclonal) | Thermo Fisher Scientific(15-5931-83) | RRID:AB_468814 | FACS (1:100) |
| Antibody | Anti-Ly-6G (Gr-1), RB6-8C5, PE-Cyanine7 (rat monoclonal) | Thermo Fisher Scientific(25-5931-82) | RRID:AB_469663 | FACS (1:100) |
| Antibody | Anti-TER-119/Erythroid Cells, TER-119, eFluor 450 (rat monoclonal) | Thermo Fisher Scientific(48-5921-80) | RRID:AB_1518809 | FACS (1:100) |
| Antibody | Anti-TER-119/Erythroid Cells, TER-119, PE-Cyanine5 (rat monoclonal) | Thermo Fisher Scientific(15-5921-83) | RRID:AB_468811 | FACS (1:100) |
| Antibody | Anti-TER-119/Erythroid Cells, TER-119, APC/Cy7 (rat monoclonal) | Biolegend(116223) | RRID:AB_2137788 | FACS (1:100) |
| Antibody | TotalSeq-A0301 anti-mouse Hashtag 1 Antibody, M1/42; 30-F11 (rat monoclonal) | Biolegend(155801) | RRID:AB_2750032 | Cell hashing (1 µg per reaction) |
| Antibody | TotalSeq-A0302 anti-mouse Hashtag 2 Antibody, M1/42; 30-F12 (rat monoclonal) | Biolegend(155803) | RRID:AB_2750033 | Cell hashing (1 µg per reaction) |
| Antibody | TotalSeq-A0303 anti-mouse Hashtag 3 Antibody, M1/42; 30-F13 (rat monoclonal) | Biolegend(155805) | RRID:AB_2750034 | Cell hashing (1 µg per reaction) |
| Antibody | TotalSeq-A0304 anti-mouse Hashtag 4 Antibody, M1/42; 30-F14 (rat monoclonal) | Biolegend(155807) | RRID:AB_2750035 | Cell hashing (1 µg per reaction) |
| Reagent, commercial | Streptavidin, -, PE-Cyanine5 | Thermo Fisher(15-4317-82) | RRID:AB_10116415 | FACS (1:100) |
| Reagent, commercial | Streptavidin, -, eFluor 450 | Thermo Fisher Scientific(48-4317-82) | RRID:AB_10359737 | FACS (1:100) |
| Commercial assay or kit | scRNA-Seq kit V2 – replicate 1 | 10× Genomics | PN-120267 | |
| Commercial assay or kit | scRNA-Seq kit V3– replicate 2 | 10× Genomics | PN-1000075 | |
| Commercial assay or kit | scATAC-Seq kit | 10× Genomics | PN-1000111 | |
| Commercial assay or kit | Lineage depletion kit | Miltenyi Biotech | 130-090-858 | |
| Chemical compound, drug | Poly(I:C) HMW | Invivogen | tlrl-pic-5 | |
| Chemical compound, drug | DmPGE2 | Cayman | 14750 | |
| Chemical compound, drug | G-CSF | Thermo Fisher | PHC2031 | |
| Chemical compound, drug | Indomethacin | Sigma | PHR1247-500MG | |
| Software, algorithm | GraphPad Prism | GraphPad(Version 6.05) | RRID:SCR_002798 | https://www.graphpad.com/ |
| Software, algorithm | FlowJo (Tree Star) | FlowJo(Version 10.5.3) | RRID:SCR_008520 | https://www.flowjo.com/ |
| Software, algorithm | Cellranger | 10× Genomics | v3.0.1v2.1.0 (Replicate 1) v1.2.0 (scATAC-Seq) | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| Software, algorithm | CITE-Seq count | https://hoohm.github.io/CITE-seq-Count/(version 1.4.3) | RRID:SCR_019239 | https://github.com/Hoohm/CITE-seq-Count, Roelli, 2021 |
| Software, algorithm | Scanpy | (Wolf et al., 2018)Various versions, see jupyter notebooks + dockerhub for documentation | RRID:SCR_018139 | https://scanpy.readthedocs.io/en/stable/ |
| Software, algorithm | pegasuspy | Gaublomme et al., 2019 | Version 0.17.1 | https://github.com/klarman-cell-observatory/pegasus/tree/0.17.1, Yang, 2021 |
| Software, algorithm | Signac | (Stuart et al., 2019)Version 0.2.5 | RRID:SCR_021158 | https://satijalab.org/signac/ |
| Software, algorithm | GitHub | This paper | https://github.com/evafast/scrnaseq_paper, copy archived at swh:1:rev:231286dc1447516f938bed8191839edb554a4fd3 (Fast, 2021) | Code for all analyses + description |
| Software, algorithm | Dockerhub | This paper | https://hub.docker.com/u/evafast1 | Docker images for analysis |
| Software, algorithm | UCSC cell browser | Speir et al., 2021 | https://cells.ucsc.edu/ | Interactive app |
Additional files
-
Supplementary file 1
Table of sequencing metrics.
Sequencing metric output from cellranger.
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp1-v1.xlsx
-
Supplementary file 2
Table with overlap of differentially regulated genes in male and female hematopoietic stem cells (HSCs) and Lin-, c-Kit+, Sca1+ (LSKs).
Table summarizing number of differentially regulated genes within male and female HSCs and LSKs. Over-representation analysis odds ratio and p-value were calculated using a Fisher’s exact test.
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp2-v1.csv
-
Supplementary file 3
Table of differential expression result (model-based analysis of single-cell transcriptomics [MAST]) by sex.
Each tab contains a treatment vs. control comparison (16,16-dimethyl prostaglandin E2 [dmPGE2], Indo, poly(I:C), granulocyte colony-stimulating factor [G-CSF]). Each cluster was compared to its respective control cluster separated by sex. Log fold change and adjusted p-value from the Hurdle model are listed for genes with p-values < 0.01.
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp3-v1.xlsx
-
Supplementary file 4
Table with marker gene enrichments in single-cell RNA sequencing (scRNA-Seq) clusters.
Marker gene enrichment was calculated using a Wilcoxon rank-sum test. Score (suffix ‘_s’) indicates the z-score of each gene on which p-value computation is based. Other fields are log fold change = suffix ‘_l’ and false discovery adjusted p-value – suffix ‘_p’.
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp4-v1.xlsx
-
Supplementary file 5
Table with curated pathways used for over-representation analysis.
Gene lists curated from literature search and MsigDB.
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp5-v1.xlsx
-
Supplementary file 6
Table of pathway enrichment for hematopoietic stem cell (HSC) and Lin-, c-Kit+, Sca1+ (LSK) clusters and treatments.
Over-representation analysis for genes induced by external stimulants or enriched in single-cell RNA sequencing (scRNA-Seq) clusters (top 3–5 pathways shown for each database or curated pathway set with adjusted p-value < 0.05). Granulocyte colony-stimulating factor (G-CSF) has an additional tab listing all enrichments (including adjusted p-value > 0.05) of curated pathways.
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp6-v1.xlsx
-
Supplementary file 7
Table of differential gene expression result (model-based analysis of single-cell transcriptomics [MAST]).
Each tab contains a treatment vs. control comparison (16,16-dimethyl prostaglandin E2 [dmPGE2], Indo, poly(I:C), granulocyte colony-stimulating factor [G-CSF]). Each cluster was compared to its respective control cluster. Log fold change and adjusted p-value from the Hurdle model are listed for genes with p-values < 0.01.
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp7-v1.xlsx
-
Supplementary file 8
Table of average expression per cluster of differentially regulated genes in hematopoietic stem cells (HSCs).
Count normalized and log transformed UMI counts were averaged across cells in HSC clusters for differentially regulated genes from model-based analysis of single-cell transcriptomics (MAST).
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp8-v1.xlsx
-
Supplementary file 9
Table of coocurrence of top 100 genes across Lin-, c-Kit+, Sca1+ (LSK) and hematopoietic stem cell (HSC) treatments and clusters.
Comparison of top 100 genes between LSK or HSC treatments and clusters (no duplicates) and comparison of top 100 genes between LSK and HSC clusters (with duplicates) and in HSC clusters Replicate 1 vs. Replicate 2.
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp9-v1.xlsx
-
Supplementary file 10
Table of average expression per cluster of differentially regulated genes in Lin-, c-Kit+, Sca1+ (LSKs).
Count normalized and log transformed UMI counts were averaged across cells in LSK clusters for differentially regulated genes from model-based analysis of single-cell transcriptomics (MAST).
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp10-v1.xlsx
-
Supplementary file 11
Table of ChromVar transcription factor (TF) motif activity score enrichment in Lin-, c-Kit+, Sca1+ (LSK) and hematopoietic stem cell (HSC) single-cell chromatin accessibility sequencing (scATAC) clusters.
ChromVar motif activity score enrichment for HSC and LSK scATAC clusters.
- https://cdn.elifesciences.org/articles/66512/elife-66512-supp11-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66512/elife-66512-transrepform1-v1.docx





