Mosquito community composition shapes virus prevalence patterns along anthropogenic disturbance gradients
Figures
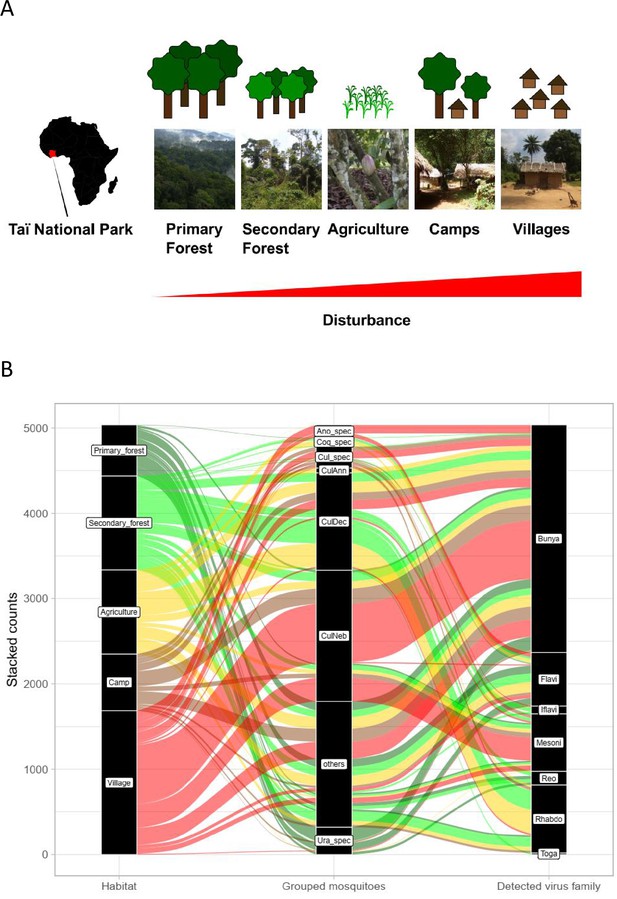
Study sites and mosquito distribution.
(A) Study site location and overview over the five habitat types. The different habitat types along the anthropogenic disturbance gradient are depicted by photos and drawings. (B) Alluvial plot showing the distribution of the main mosquito species groups and main virus families to the five respective habitats. CulAnn: Culex annulioris; CulDec: Culex decens; CulNeb: Culex nebulosus; Cul_spec: other Culex species; Ano_spec: Anopheles species; Ura_spec: Uranotaenia species; Coq_spec: Coquillettidia species; others: all other grouped species (see main text). Bunya: order Bunyavirales containing Phenuiviridae, Peribunyaviridae, and Phasmaviridae; Flavi: Flaviviridae; Toga: Togaviridae; Rhabdo: Rhabdoviridae; Reo: Reoviridae; Iflavi: Iflaviridae; Meso: Mesoniviridae.

Biodiversity analyses.
Mosquito species richness per habitat. Rarefaction curves (with 95% confidence intervals) show the observed (solid lines) and interpolated (dotted line) mosquito diversity in the five habitats.
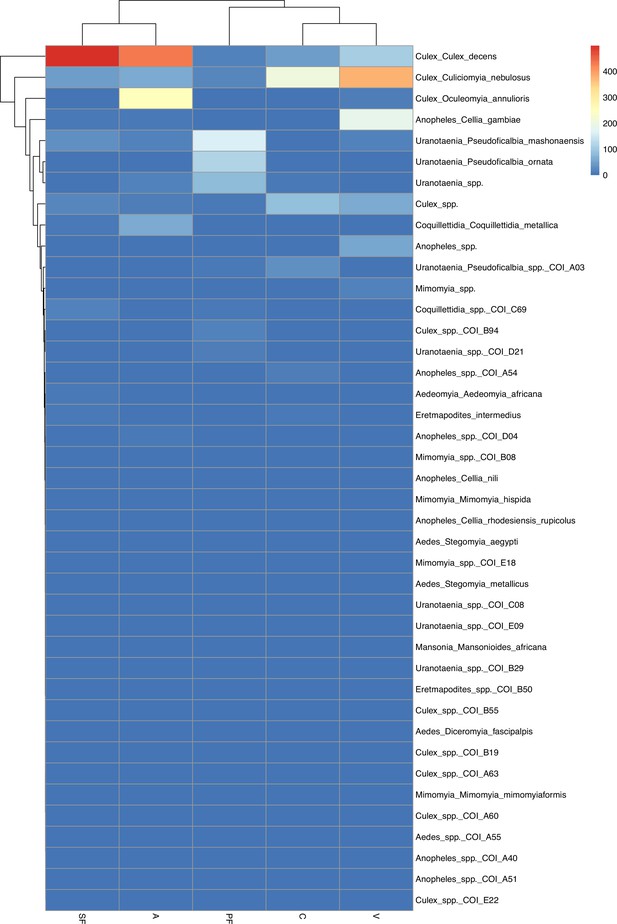
Hierarchical cluster analysis based on associations between mosquito species. Main mosquito groups were identified as CulAnn, CulDec, CulNeb, Cul_spec, Ura_spec, Ano_spec, and Coq_spec. CulAnn: Culex annulioris; CulDec: Culex decens; CulNeb: Culex nebulosus; Cul_spec: other Culex species; Ano_spec: Anopheles species; Ura_spec: Uranotaenia species; Coq_spec: Coquillettidia species, others: all other grouped species. PF: primary forest; SF: secondary forest; A: agriculture; C: camp; V: village.
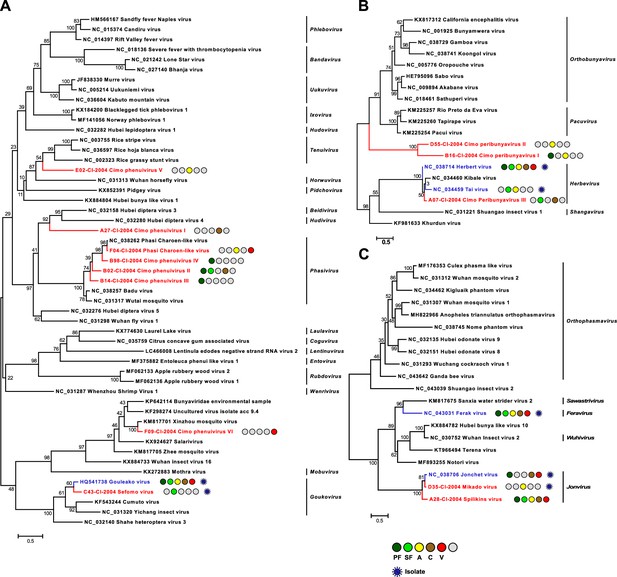
Phylogenetic analyses of detected bunyaviruses.
Phylogenetic trees were inferred with PhyML (LG substitution model) based on MAFFT-E protein alignments covering the conserved RdRp motifs of the families Phenuiviridae (A), Peribunyaviridae (B), and Phasmaviridae (C). The alignment length was approximately 290, 300, and 690 amino acids, respectively. Novel viruses from this study are indicated in red, and previously published viruses detected in our data set are indicated in blue. Sample origin from the different habitat types is indicated by colored circles while no detection is indicated by gray circles. Live virus isolates are marked with a blue virion. PF: primary forest; SF: secondary forest; A: agriculture; C: camp; V: village.

Phylogenetic analyses of detected phasmaviruses.
Phylogenetic trees were inferred with PhyML (WAG (A) and LG (B) substitution model) based on MAFFT-E protein alignments covering the glycoproteins (A) and nucleocapsid (B) proteins of the family Phasmaviridae. The alignment length was approximately 1700 and 280 amino acids, respectively. Novel viruses from this study are indicated in red, and previously published viruses detected in our data set are indicated in blue.
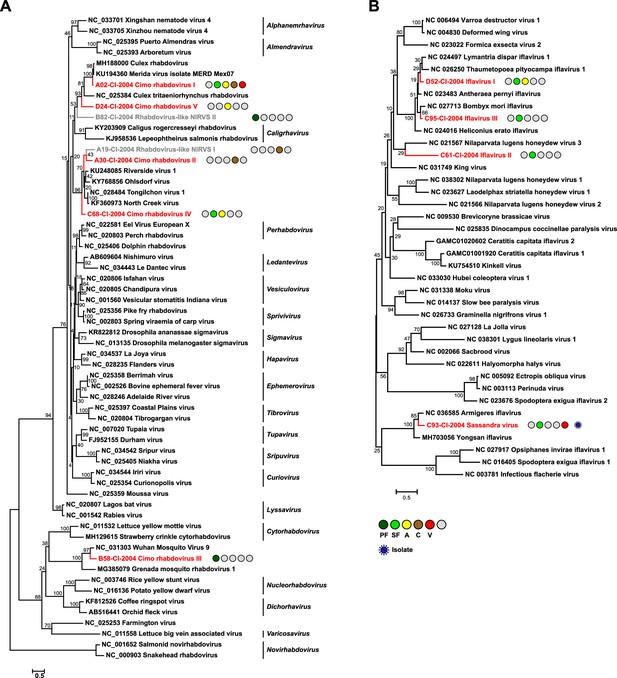
Phylogenetic analyses of detected rhabdoviruses and iflaviruses.
Phylogenetic trees were inferred with PhyML (LG substitution model) based on MAFFT-E protein alignments covering the conserved RdRp motifs of the families Rhabdoviridae (A) and Iflaviridae (B). The alignment length was approximately 270 and 250 amino acids, respectively. Novel viruses from this study are indicated in red, and detected virus-like sequences are indicated in gray. Sample origin from the different habitat types is indicated by colored circles while no detection is indicated by gray circles. Live virus isolates are marked with a blue virion. PF: primary forest; SF: secondary forest; A: agriculture; C: camp: V: village.

Phylogenetic analyses of detected flaviviruses and orbiviruses.
Phylogenetic trees were inferred with PhyML (GTR substitution model) based on MAFFT-E nucleotide alignments covering the conserved RdRp motifs of the genus Flavivirus (A) and with PhyML (LG substitution model) based on a MAFFT-E protein alignment of the polymerase of the genus Orbivirus (B). The alignment length was approximately 1170 nucleotides and 1190 amino acids, respectively. Novel viruses from this study are indicated in red, previously published viruses detected in our data set are indicated in blue, and detected virus-like sequences are indicated in gray. Sample origin from the different habitat types is indicated by colored circles while no detection is indicated by gray circles. Live virus isolates are marked with a blue virion. PF: primary forest; SF: secondary forest; A: agriculture; C: camp; V: village.
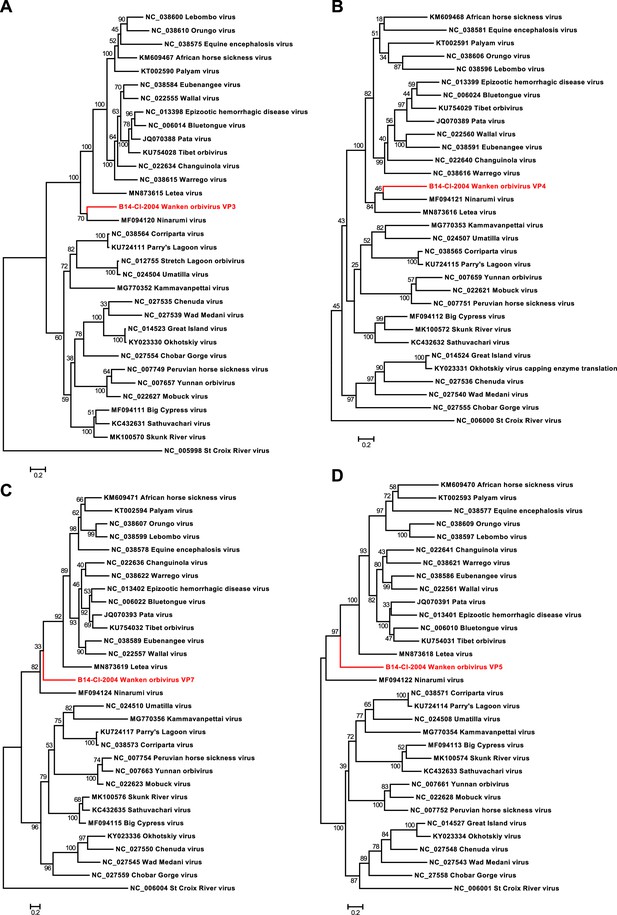
Phylogenetic analyses of the detected Wanken orbivirus (WKOV) and members of the genus Orbivirus.
Phylogenetic trees were inferred with PhyML (LG substitution model) based on MAFFT-E protein alignments of VP3 (A), VP4 (B), VP7 (C), and VP5 (D). The alignment length was approximately 890, 640, 350, and 510 amino acids, respectively. WKOV is indicated in red.

Phylogenetic analyses of detected reoviruses, alphaviruses, and mesoniviruses.
Phylogenetic trees were inferred with PhyML (LG substitution model) based on a MAFFT-E protein alignment of the polymerase of members of the family Reoviridae (A) or with PhyML (GTR substitution model) based on MAFFT-E nucleotide alignments covering the E2-6K-E1 region of members of the genus Alphavirus (B) of the polymerase of members of the family Mesoniviridae (C). The alignment length was approximately 290 amino acids and 2720 and 7430 nucleotides, respectively. Novel viruses from this study are indicated in red, previously published viruses detected in our data set are indicated in blue, and detected virus-like sequences are indicated in gray. Sample origin from the different habitat types is indicated by colored circles while no detection is indicated by gray circles. Live virus isolates are marked with a blue virion. PF: primary forest; SF: secondary forest; A: agriculture; C: camp; V: village.
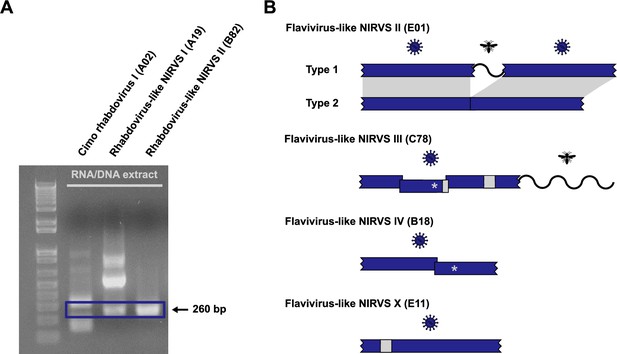
Potential non-retroviral integrated RNA virus sequences (NIRVS).
(A) PCR amplicons of the generic rhabdovirus PCR assay. RNA/DNA extracts without reverse transcription were used for the PCR. Nested PCR amplicons of Cimo rhabdovirus I and two rhabdovirus-like NIRVS were visualized by ethidium bromide-stained agarose gel electrophoresis. Amplicons with the expected size of 260 bp are framed by a blue box. (B) Schematic representation of selected flavivirus-like NIRVS. Stop codons are indicated by an asterisk, deletions are shown as light gray boxes, frame shifts are indicated by overlapping blue boxes, and insertions with similarity to insect genes are shown as waved lines.
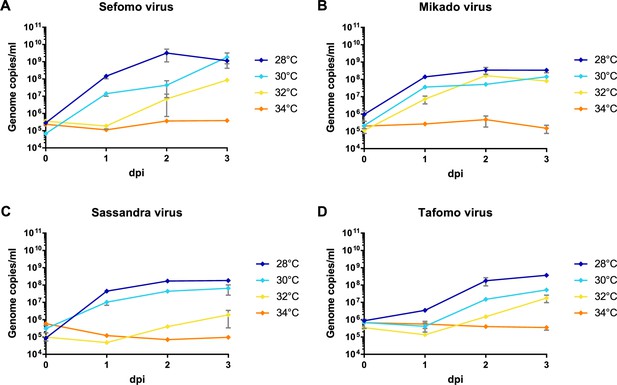
Temperature-dependent replication of novel virus isolates.
C6/36 cells were infected with an MOI of 0.1 with Sefomo virus (A), Mikado virus (B), Sassandra virus (C), and Tafomo virus (D). Replication was measured for 3 dpi at 28, 30, 32, and 34°C.
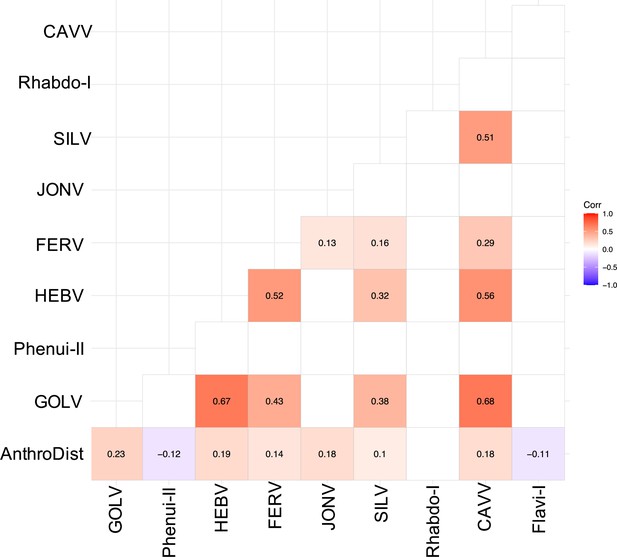
Spearman’s rank correlation rho for the most abundant viruses.
Only significant correlations are shown. AnthroDist refers to the gradient of anthropogenic disturbance from low to high. PF: primary forest; SF: secondary forest; A: agriculture; C: camp; V: village.
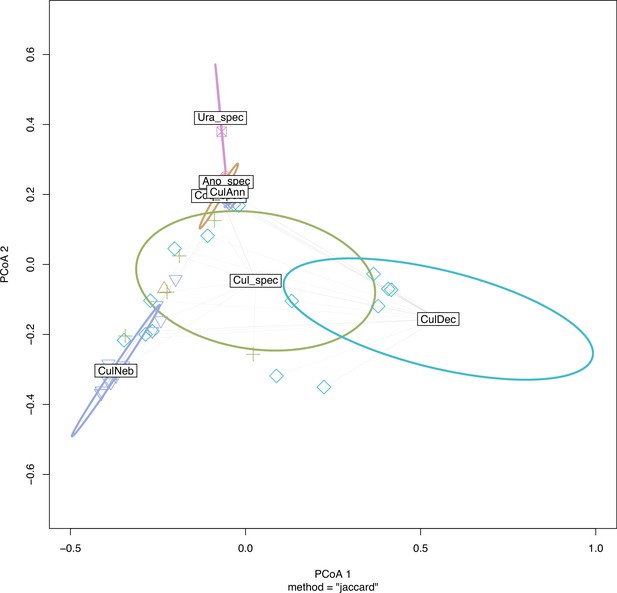
Ordination plot of the principal coordinate analysis (PCoA) showing that the viral community primarily partitions by Culex nebulosus (CulNeb), Uranotaenia species (Ura_spec), and Culex decens (CulDec).

Ordination plot of the principle coordinate analysis (PCoA) showing that the viral community primarily partitions by Culex nebulosus (CulNeb), Uranotaenia species (Ura_spec) and Culex decens (CulDec) as shown in Figure 6—figure supplement 1, which can be related to their main habitats in primary forest, agriculture and villages.
CulAnn: Culex annulioris, Cul_spec: other Culex species; Ano_spec: Anopheles species; Ura_spec: Uranotaenia species.
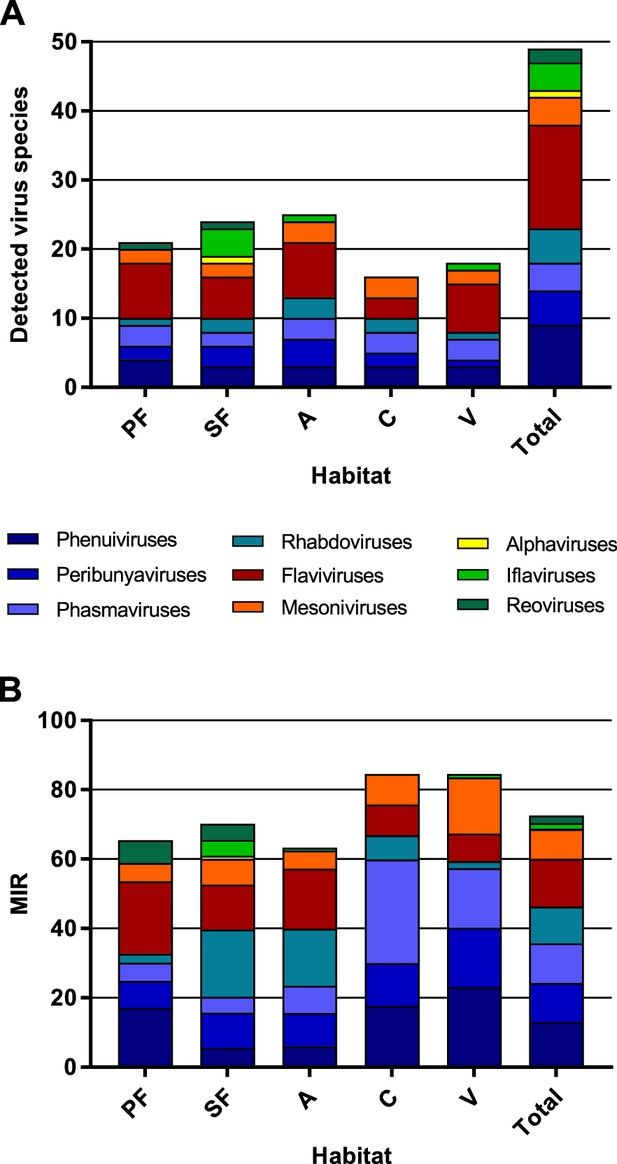
Richness and cumulative minimum infection rate (MIR) across all tested virus taxa.
The number of distinct viruses (A) and the cumulated MIR per 1000 mosquitoes (B) were calculated for all habitat types and for the complete data set. Different virus taxa are shown in different colors. PF: primary forest; SF: secondary forest; A: agriculture; C: camp; V: village.
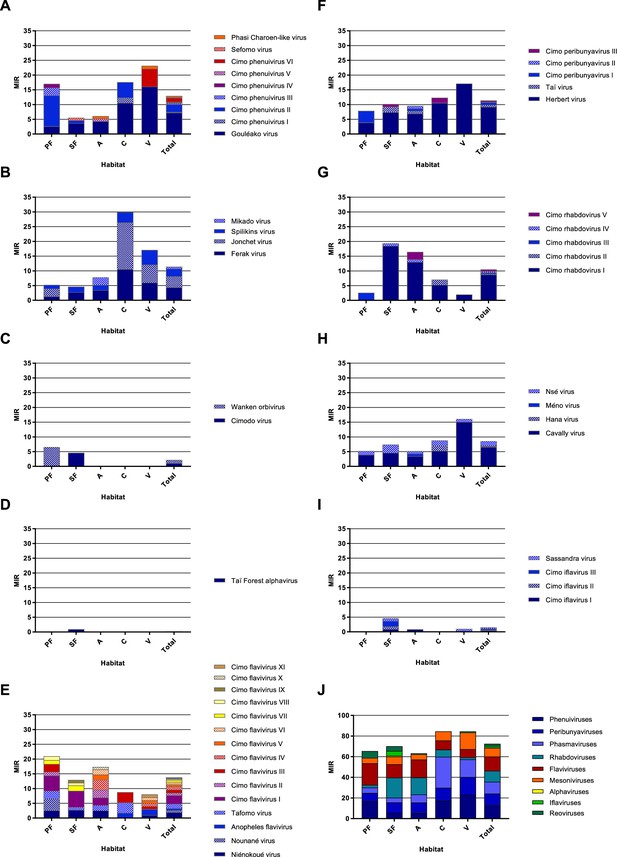
Cumulative minimum infection rate (MIR) per virus taxon.
The cumulated MIR per 1000 mosquitoes was calculated for the analyzed taxa Phenuiviridae (A), Phasmaviridae (B), Reoviridae (C), Alphavirus (D), Flavivirus (E), Peribunyaviridae (F), Rhabdoviridae (G), Mesoniviridae (H), and Iflaviridae (I), as well as for all detected viruses (J) in the different habitat types and for the complete data set. The different viruses or taxa are shown in different colors.
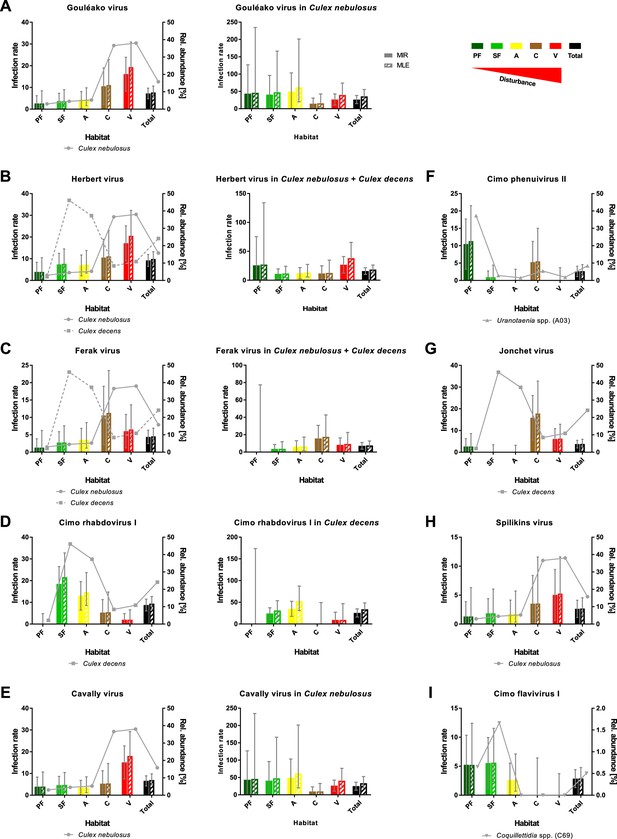
Prevalence patterns of selected viruses along the disturbance gradient.
For all viruses that were detected in >10 pools, Gouléako virus (GOLV) (A), Herbert virus (HEBV) (B), Ferak virus (FERV) (C), Cimo rhabdovirus I (D), Cavally virus (CAVV) (E), Cimo phenuivirus II (F), Jonchet virus (G), Spilikins virus (H), and Cimo flavivirus I (I), the minimum infection rate (MIR) and maximum likelihood estimation (MLE) per 1000 mosquitoes of the whole data set were calculated for all habitat types (left graphs for A–E). The abundance of the main mosquito host species was plotted. The five viruses GOLV (A), HEBV (B), FERV (C), Cimo rhabdovirus I (D), and CAVV (E) occurred frequently enough in their main mosquito host species (>10 positive pools) to analyze their prevalence in these species. For these viruses, the MIR and MLE per 1000 mosquitoes of the respective species were calculated for all habitat types (right graphs). Significant differences in the infection probability with the most abundant viruses in the different habitats are shown in Supplementary file 2.
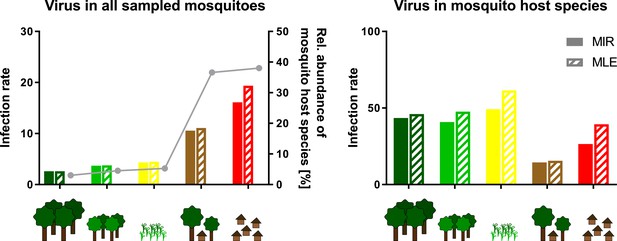
Schematic presentation of the abundance effect.
Infection rates are shown for Gouléako virus (GOLV) in all sampled mosquitoes (representing minimum infection rate (MIR) and maximum likelihood estimation (MLE) values as shown in Figure 8) and only in the main mosquito host species, Culex nebulosus. The abundance of the main mosquito host species, Culex nebulosus is indicated by a gray line.
Tables
Distribution and host association of detected viruses.
Number of positive pools per habitat and mosquito host species of all detected viruses and virus-like sequences. The main mosquito host species are indicated in bold letters. PF: primary forest; SF: secondary forest; A: agriculture; C: camp; V: village; nd: not determined.
| Virus family | Virus | No. of positive pools | Mosquito species | Live virus isolate | Representative pool (GenBank accession number)* | Sequence length (nt)† | References | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ∑ | PF | SF | A | C | V | |||||||
| Phenuiviridae | Gouléako virus | 33 | 2 | 4 | 5 | 6 | 16 | Culex nebulosus, Culex decens, Culex spp., nd | x | A05 (NC_043051) | Full genome | Marklewitz et al., 2011 |
| Cimo phenuivirus I | 1 | 0 | 0 | 0 | 1 | 0 | nd | A27 (MZ202291) | 892 | This study | ||
| Cimo phenuivirus II | 12 | 8 | 1 | 0 | 3 | 0 | Uranotaenia mashonaensis, Uranotaenia ornata, Uranotaenia spp., nd | B02 (MZ202292) | 1096 | This study | ||
| Cimo phenuivirus III | 2 | 2 | 0 | 0 | 0 | 0 | nd | B14 (MZ202293) | 1099 | This study | ||
| Cimo phenuivirus IV | 1 | 1 | 0 | 0 | 0 | 0 | nd | B98 (MZ202294) | 1096 | This study | ||
| Cimo phenuivirus V | 1 | 0 | 0 | 1 | 0 | 0 | Anopheles spp. | E02 (MZ202298) | 1091 | This study | ||
| Cimo phenuivirus VI | 6 | 0 | 0 | 0 | 0 | 6 | Anopheles gambiae, Anopheles nili, Anopheles spp., nd | F09 (MZ202300) | 1210 | This study | ||
| Sefomo virus | 1 | 0 | 1 | 0 | 0 | 0 | Culex decens | x | C43 (MZ202295) | Full genome | This study | |
| Phasi Charoen-like virus | 2 | 0 | 0 | 1 | 0 | 1 | Aedes aegypti, nd | F04 (MZ202299) | 1092 | This study | ||
| Peribunyaviridae | Herbert virus | 42 | 3 | 8 | 8 | 6 | 17 | Culex nebulosus, Culex decens, Culex spp., Mimomyia mimomyiaformis, Coquillettidia spp., nd | x | F23 (NC_038714) | Full genome | Marklewitz et al., 2013 |
| Taï virus | 3 | 0 | 2 | 1 | 0 | 0 | Culex (Culex) decens, nd | x | F47 (NC_034459) | Full genome | Marklewitz et al., 2013 | |
| Cimo peribunyavirus I | 4 | 3 | 0 | 1 | 0 | 0 | Uranotaenia mashonaensis, nd | B04 (MZ202287) | 2278 | This study | ||
| Cimo peribunyavirus II | 1 | 0 | 0 | 1 | 0 | 0 | nd | D55 (MZ202289) | 3596; 2228 (M segment) | This study | ||
| Cimo peribunyavirus III | 2 | 0 | 1 | 0 | 1 | 0 | Culex nebulosus, Culex decens | A07 (MZ202286) | 1488 | This study | ||
| Phasmaviridae | Ferak virus | 20 | 1 | 3 | 4 | 6 | 6 | Culex nebulosus, Culex decens, nd | x | C51 (NC_043031) | Full genome | Marklewitz et al., 2015 |
| Jonchet virus | 17 | 2 | 0 | 0 | 9 | 6 | Culex decens, Culex nebulosus, Culex spp., nd | x | B81 (NC_038706) | Full genome | Marklewitz et al., 2015 | |
| Spilikins virus | 12 | 1 | 2 | 2 | 2 | 5 | Culex nebulosus, Culex spp., nd | A28 (MZ202269) | Complete CDS | This study | ||
| Mikado virus | 3 | 0 | 0 | 3 | 0 | 0 | Culex annulioris | x | D35 (MZ202272) | Complete CDS | This study | |
| Rhabdoviridae | Cimo rhabdovirus I | 40 | 0 | 20 | 15 | 3 | 2 | Culex decens, Culex spp., nd | A02 (MZ202301) | 1146 | This study | |
| Cimo rhabdovirus II | 1 | 0 | 0 | 0 | 1 | 0 | nd | A30 (MZ202302) | 988 | This study | ||
| Cimo rhabdovirus III | 2 | 2 | 0 | 0 | 0 | 0 | nd | B58 (MZ202303) | 769 | This study | ||
| Cimo rhabdovirus IV | 2 | 0 | 1 | 1 | 0 | 0 | nd | C68 (MZ202304) | 980 | This study | ||
| Cimo rhabdovirus V | 3 | 0 | 0 | 3 | 0 | 0 | Coquillettidia metallica, nd | D24 (MZ202305) | 898 | This study | ||
| Potential Rhabdovirus-like NIRVS | Rhabdovirus-like NIRVS I | 1 | 0 | 0 | 0 | 1 | 0 | nd | A19 (MZ399708) | 638 | This study | |
| Rhabdovirus-like NIRVS II | 1 | 1 | 0 | 0 | 0 | 0 | nd | B82 (MZ399709) | 566 | This study | ||
| Reoviridae | Cimodo virus | 5 | 0 | 5 | 0 | 0 | 0 | Culex decens, nd | x | C74 (NC_023420) | Full genome | Hermanns et al., 2014 |
| Wanken orbivirus | 5 | 5 | 0 | 0 | 0 | 0 | Uranotaenia mashonaensis, nd | B14 (MZ202276) | Full genome | This study | ||
| Mesoniviridae | Cavally virus | 30 | 3 | 5 | 4 | 3 | 15 | Culex nebulosus, Culex decens, Culex spp., nd | x | C79 (NC_015668) | Full genome | Zirkel et al., 2011 |
| Hana virus | 1 | 0 | 0 | 0 | 1 | 0 | Culex spp. | x | A04 (NC_020899) | Full genome | Zirkel et al., 2013 | |
| Méno virus | 1 | 0 | 0 | 1 | 0 | 0 | Uranotaenia spp. | x | E09 (NC_020900) | Full genome | Zirkel et al., 2013 | |
| Nsé virus | 7 | 1 | 3 | 1 | 1 | 1 | Culex nebulosus, Culex decens | x | F24 (NC_020901) | Full genome | Zirkel et al., 2013 | |
| Togaviridae | Taï Forest alphavirus | 1 | 0 | 1 | 0 | 0 | 0 | Culex decens | C21 (NC_032681) | Full genome | Hermanns et al., 2017 | |
| Iflaviridae | Cimo iflavirus I | 2 | 0 | 1 | 1 | 0 | 0 | Culex decens, Culex nebulosus | D52 (MZ202268) | 1105 | This study | |
| Cimo iflavirus II | 1 | 0 | 1 | 0 | 0 | 0 | Culex decens | C61 (MZ202265) | 186 | This study | ||
| Cimo iflavirus III | 2 | 0 | 2 | 0 | 0 | 0 | Culex decens | C95 (MZ202267) | 730 | This study | ||
| Sassandra virus | 2 | 0 | 1 | 0 | 0 | 1 | Culex spp., nd | x | C93 (MZ202266) | Full genome | This study | |
| Flaviviridae | Niénokoué virus | 9 | 2 | 3 | 3 | 0 | 1 | Coquillettidia metallica, Culex spp., nd | x | B51 (NC_024299) | Full genome | Junglen et al., 2017 |
| Nounané virus | 3 | 3 | 0 | 0 | 0 | 0 | Uranotaenia mashonaensis | x | B31 (NC_033715) | Complete CDS | Junglen et al., 2009a | |
| Anopheles flavivirus | 3 | 0 | 0 | 0 | 1 | 2 | Anopheles gambiae, Anopheles spp. | F10 (MZ202263) | 1172 | This study | ||
| Tafomo virus | 7 | 2 | 1 | 2 | 2 | 0 | Culex spp., nd | x | B22 (MZ202252) | Full genome | This study | |
| Cimo flavivirus I | 13 | 4 | 6 | 3 | 0 | 0 | Coquillettidia spp. (unknown COI-type C69), nd | B36 (MZ202253) | 702 | This study | ||
| Cimo flavivirus II | 4 | 1 | 0 | 3 | 0 | 0 | Uranotaenia spp., nd | D01 (MZ202258) | 790 | This study | ||
| Cimo flavivirus III | 5 | 2 | 0 | 0 | 2 | 1 | Uranotaenia mashonaensis, Uranotaenia spp., nd | B01 (MZ202251) | 790 | This study | ||
| Cimo flavivirus IV | 5 | 0 | 0 | 4 | 0 | 1 | Mimomyia spp., nd | E08 (MZ202261) | 514 | This study | ||
| Cimo flavivirus V | 3 | 0 | 0 | 2 | 0 | 1 | Mimomyia hispida, nd | D20 (MZ202259) | 1192 | This study | ||
| Cimo flavivirus VI | 3 | 0 | 0 | 2 | 0 | 1 | Anopheles rhodesiensis rupicolus, Anopheles spp. | E02 (MZ202260) | 516 | This study | ||
| Cimo flavivirus VII | 3 | 1 | 2 | 0 | 0 | 0 | nd | B36 (MZ202254) | 1188 | This study | ||
| Cimo flavivirus VIII | 2 | 1 | 1 | 0 | 0 | 0 | Eretmapodites intermedius, nd | B85 (MZ202255) | 1188 | This study | ||
| Cimo flavivirus IX | 1 | 0 | 1 | 0 | 0 | 0 | nd | C51 (MZ202257) | 1188 | This study | ||
| Cimo flavivirus X | 1 | 0 | 0 | 1 | 0 | 0 | nd | E08 (MZ202262) | 736 | This study | ||
| Cimo flavivirus XI | 1 | 0 | 0 | 0 | 0 | 1 | Culex nebulosus | F41 (MZ202264) | 1193 | This study | ||
| Potential Flavivirus-like NIRVS | Flavivirus-like NIRVS I | 10 | 2 | 3 | 4 | 0 | 1 | Coquillettidia metallica,nd | B60 (MZ399699) | 943 | This study | |
| Flavivirus-like NIRVS II | 2 | 0 | 0 | 1 | 0 | 1 | Aedes aegypti, Aedes spp. | E01 (MZ399703) | 846 | This study | ||
| Flavivirus-like NIRVS III | 3 | 0 | 2 | 0 | 1 | 0 | Eretmapodites spp., nd | C78 (MZ399701) | 668 | This study | ||
| Flavivirus-like NIRVS IV | 2 | 2 | 0 | 0 | 0 | 0 | nd | B18 (MZ399698) | 515 | This study | ||
| Flavivirus-like NIRVS V | 2 | 1 | 0 | 0 | 1 | 0 | nd | B68 (MZ399700) | 512 | This study | ||
| Flavivirus-like NIRVS VI | 2 | 0 | 0 | 0 | 0 | 2 | Mimomyia spp., nd | F16 (MZ399707) | 383 | This study | ||
| Flavivirus-like NIRVS VII | 3 | 0 | 2 | 1 | 0 | 0 | nd | D57 (MZ399702) | 332 | This study | ||
| Flavivirus-like NIRVS VIII | 13 | 8 | 1 | 2 | 2 | 0 | Uranotaenia spp., nd | A25 (MZ399697) | 835 | This study | ||
| Flavivirus-like NIRVS IX | 8 | 2 | 1 | 2 | 2 | 1 | Uranotaenia spp., Uranotaenia mashonaensis, nd | E26 (MZ399706) | 650 | This study | ||
| Flavivirus-like NIRVS X | 15 | 4 | 3 | 6 | 1 | 1 | Coquillettidia metallica, nd | E11 (MZ399705) | 640 | This study | ||
-
*
RdRp encoding sequence (previously published sequences are indicated in italic).
-
†
RdRp encoding segment or sequence, unless otherwise stated.
Additional files
-
Supplementary file 1
Model estimates (base: habitat primary forest and mosquito group Anopheles sp.) for the counts of mosquito individuals per mosquito group and habitat.
Significant predictors show the difference to the baseline combination. CulAnn: Culex annuloris; CulDec: Culex decens; CulNeb: Culex nebulosus; Cul_spec: other Culex species; Ura_spec: Uranotaenia species; Coq_spec: Coquillettidia species; Ano_spec: Anopheles species; others: all other grouped species. PF: primary forest; SF: secondary forest; A: agriculture; C: camp; V: village.
- https://cdn.elifesciences.org/articles/66550/elife-66550-supp1-v2.xlsx
-
Supplementary file 2
Significant differences in the infection probability with the most abundant viruses in the different habitats (only Tukey contrasts are shown; bold: p<0.05; italic: p<0.1).
No significant differences were detected for FERV (n = 20), JONV (n = 17), Spilikins virus (n = 12), and Cimo flavivirus I (n = 13; no findings in camp and village). Combinations with zero virus detections shown in light gray. All virus detections were positively associated with the number of mosquitoes per pool (model estimates not shown).
- https://cdn.elifesciences.org/articles/66550/elife-66550-supp2-v2.docx
-
Supplementary file 3
Primer pairs used for generic RT-PCR assays.
- https://cdn.elifesciences.org/articles/66550/elife-66550-supp3-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/66550/elife-66550-mdarchecklist1-v2.docx





