Cell-to-cell heterogeneity in Sox2 and Bra expression guides progenitor motility and destiny
Figures
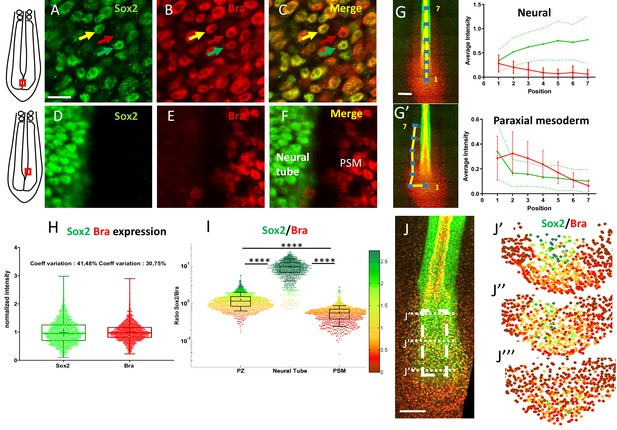
Posterior progenitors co-express Sox2 and Bra with a high degree of cell-to-cell heterogeneity.
(A–F) Immunodetection of Sox2 (green) and Bra (red) analyzed at the cellular scale in the caudal part of stage HH11 quail embryo, either in the PZ (A–C) or in the nascent NT and the PSM (D–F). Overlay images are presented in (C) and (F). Note cell-to-cell heterogeneity in Sox2 and Bra levels in the PZ, with neighboring cells expressing higher level of Bra (red arrow), higher level of Sox2 (green arrow), or comparable levels of both proteins (yellow arrow), a feature not apparent in the nascent NT and PSM tissues. (G, G’) Measurements of Sox2 and Bra levels along putative trajectories (yellow lines) of NT (G) and PSM (G’) cells. Fluorescence measurements (blue squares, left images), numbered from 1 to 7, red bars and green dashed lines are errors bars (variability between embryos). (H) Distribution of normalized cell-to-cell expression of Sox2 and Bra in the PZ (n=8 embryos). (I) Cell distribution of Sox2/Bra levels in the PZ (n=9 embryos), the NT (n=7 embryos), and the PSM (n=8 embryos); ratios have been color-coded according to a red (higher Bra) to green (higher Sox2) scale shown on the right side. (J–J’’’) Representation of the Sox2-to-Bra ratio (green to red same as (I)) in digital transversal sections (40 µm) made in the PZ (dashed lines in the double immunodetection image in (J)). Scale bars=10 µm in (A–F), 100 µm in (G) and (J). NT, neural tube; PSM, presomitic mesoderm; PZ, progenitor zone.
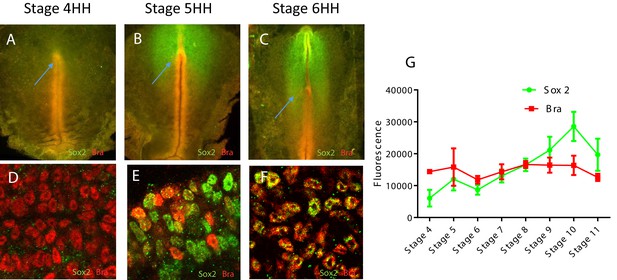
Sox2 and Bra protein co-expression and cell-to-cell heterogeneity start around stage HH5 in quail embryos.
(A–F) Double immunodetection of Sox2 and Bra viewed on whole-mount embryos (A–C) and at cell levels in the PZ (D–F) at stages HH4 (A, D), HH5 (B, E), and HH6 (C, F). (G) Sox2 and Bra average levels in posterior progenitors at different stages of development. PZ, progenitor zone.
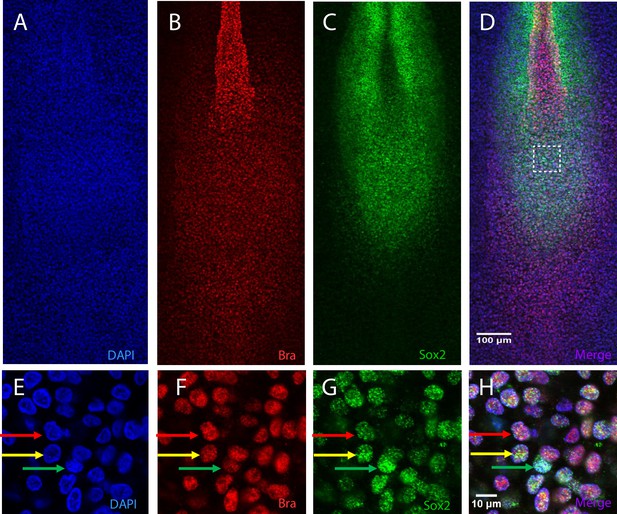
Chicken PZ cells co-express Sox2 and Bra proteins with a high degree of cell-to-cell heterogeneity.
(A–H) Double immunodetection of Bra and Sox2, counterstained with DAPI, on stage HH10 chicken embryo. Horizontal sets present successively the DAPI staining (A, E), the Bra (B, F), and Sox2 (C, G) signals and the merged images (D, H) on ventral views of the posterior region (A–D) and on a close up of the PZ ((E–H), white square in (D)). Note variability in the Sox2 and Bra signal intensities in chicken PZ cells: a Bra high/Sox2 low nucleus (red arrow), a Bra low/Sox2 high nucleus (green arrow) and a nucleus with intermediate levels of Sox2 and Bra (yellow arrow). PZ, progenitor zone.
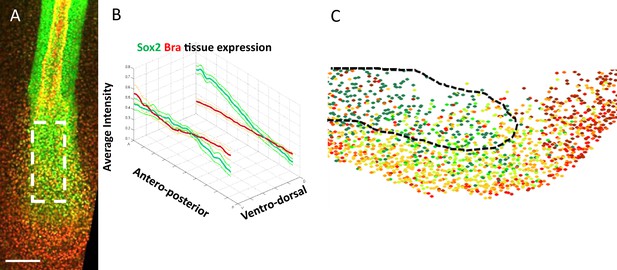
Sox2 and Bra expression patterns in PZ cells follow opposite gradients at the tissue level.
(A) Immunodetection of Sox2 (green) and Bra (red) viewed at the tissue scale in the caudal part of stage HH11 quail embryo. Dashed white rectangle delineates the PZ. (B) Quantification of Sox2 (green) and Bra (red) proteins at different antero-posterior and dorso-ventral levels (n=9 embryos, thick lines indicate average values, and thin lines demarcate the SEM). (C) Optical parasagittal section of Sox2-to-Bra ratios (corresponding to Figure 1J), anterior to the left. Prospective neural tissue is delineated with black dashed line. PZ, progenitor zone.
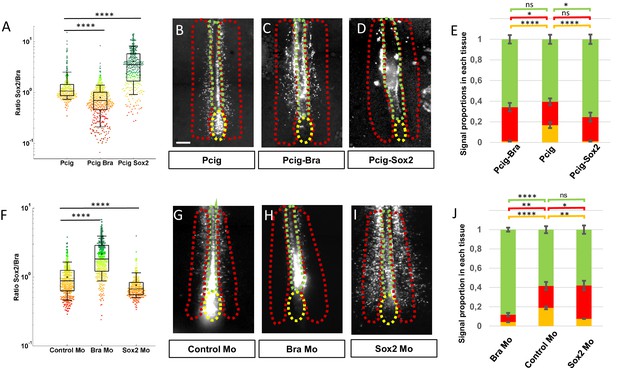
Sox2 and Bra levels are critical for progenitor maintenance and tissue distribution.
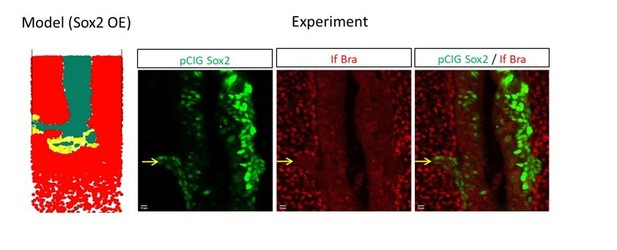
(A, F) Sox2-to-Bra ratios calculated following Bra and Sox2 double immunodetection in the PZ performed 7 hr after electroporation. Values were normalized to the average ratio of non-transfected cells of the same region. (A) Sox2/Bra values in cells transfected with Bra (Pcig-Bra) and Sox2 (Pcig-Sox2) expression vectors compared to cells transfected with the empty vector (Pcig). (F) Sox2/Bra values in cells transfected with morpholinos directed against Bra (Bra-Mo) or Sox2 (Sox2- Mo) compared to cells transfected with a Control-Mo. Ratios were calculated on the basis of 286–590 cells and 3–5 embryos per condition. (B–D), (G–I) Ventral views of embryos collected 20 hr after electroporation showing the GFP signals (white). The PZ, the PSM, and the NT are delineated by yellow, red, and green dash lines, respectively. Expression vectors or morpholinos used are indicated below each picture. Scale bar=100 µm. (E, J) Staked histograms displaying the proportion of cells in the PZ (yellow), the PSM (red), and the NT (green). For each experimental condition, proportion of cells in a given tissue was compared to the same tissue of control embryos by unpaired Student's test (n=27 embryos for Pcig-Bra, n=21 embryos control for Pcig, and n=23 embryos for Pcig-Sox2; n=28 embryos for Bra-Mo, n=27 embryos for Control-Mo, and n=28 embryos for Sox2-Mo). Error bars represent the SEM. NT, neural tube; PSM, presomitic mesoderm; PZ, progenitor zone.
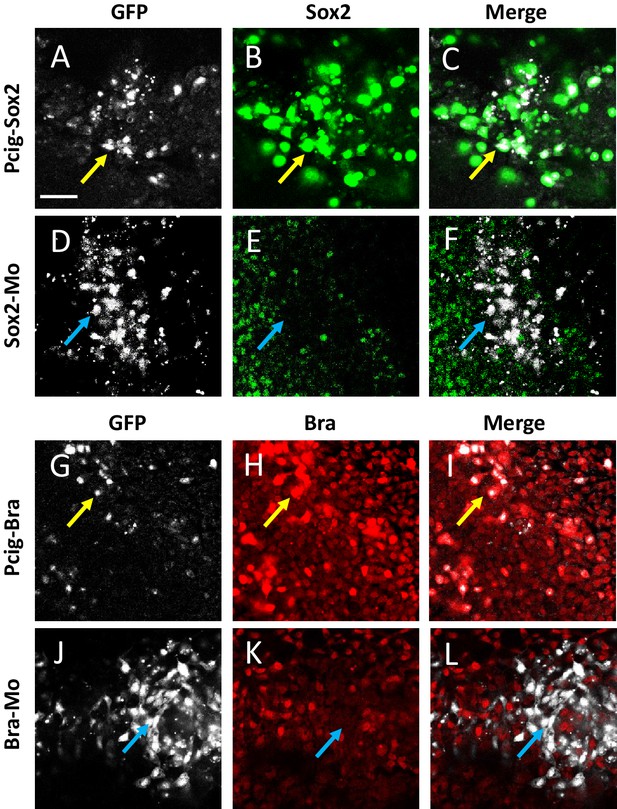
Efficient deregulation of Sox2 and Bra following electroporation of expression vectors and morpholinos.
(A–F) Immunodetection of Sox2 performed 7 hr after electroporation of the Pcig-Sox2 (A–C) and the Sox2-Mo (D–F). Horizontal sets show high magnification of the PZ and present successively the GFP signal (white (A, D)), the Sox2 signal (green (B, E)), and the merged images (C, F). Note co-localization of high Sox2 signal and GFP in Pcig-Sox2 transfected cells (yellow arrows in (A–C)) but not in Sox2-Mo transfected cells (blue arrows in (D–F)). (G–L) Immunodetection of Bra performed 7 hr after electroporation of the Pcig-Bra (G–I) and the Bra-Mo (J–L). Horizontal sets show high magnification of the PZ and present successively the GFP signal (white (G, J)), the Bra signal (red (H, K)), and the merged images (I, L). Note co-localization of high Bra signal and GFP in Pcig-Bra transfected cells (yellow arrows in (G–I)) but not in Bra-Mo transfected cells (blue arrows in (J–L)). Scale bar=50 µm. PZ, progenitor zone.
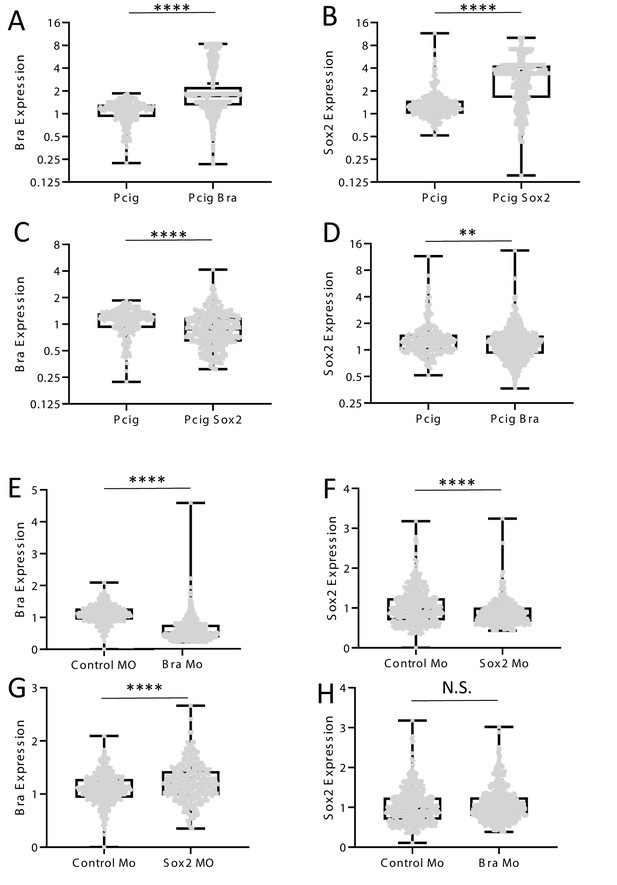
Quantification of Sox2 and Bra protein levels following electroporation of expression vectors and morpholinos.
(A–D) Quantification of Bra (A, C) and Sox2 (B, D) protein levels in cells electroporated with the Pcig-Bra (A, D) or the Pcig-Sox2 (B, C) expression vectors. Note the significant upregulation of Bra and Sox2 in cells electroporated with the Pcig-Bra and the Pcig-Sox2, respectively, compared to control cells (Pcig). Note also the significant downregulation of Bra and Sox2 in cells electroporated with the Pcig-Sox2 and the Pcig-Bra, respectively. (E–H) Quantification of Bra (E, G) and Sox2 (F, H) protein levels in cells electroporated with the Mo-Bra (E, H) or the Mo-Sox2 (F, G). Note the significant downregulation of Bra or Sox2 in cells electroporated with the Mo-Bra and the Mo-Sox2, respectively, compared to control cells (Control-Mo). Note also the significant upregulation of Bra in Mo-Sox2 electroporated cells but not that of Sox2 in Mo-Bra electroporated cells. Levels of protein expression are normalized to neighboring non-electroporated cells.
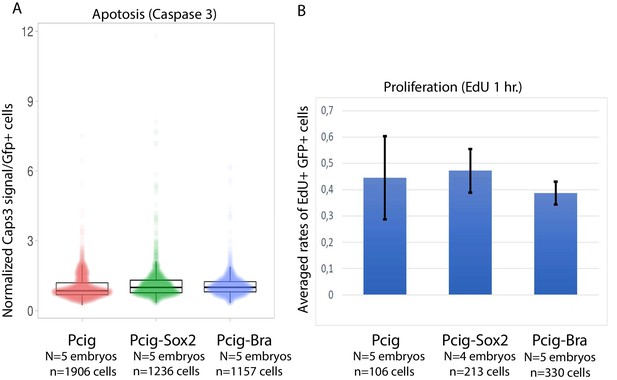
Effect of Sox2 and Bra overexpression on apoptosis and proliferation.
(A) Quantification of activated Caspase-3 immunostaining among in GFP positive (electroporated) progenitor cells 7 hr after Pcig, Pcig-Sox2, or Pcig-Bra transfection. (B) Quantification of the rates of EdU positive cells (1 hr pulse) among GFP positive (electroporated) progenitor cells 7 hr after Pcig, Pcig-Sox2, or Pcig-Bra transfection. Number of embryos and cells are indicated on the figure; error bars in (B) are standard deviation.
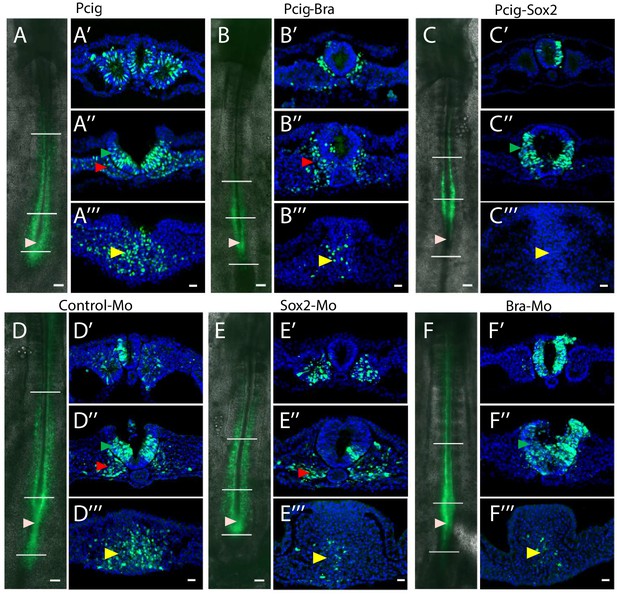
Tissue localization of transfected cells after Sox2 and Bra overexpression and downregulation.
Visualization of GFP-expressing cells (green) 20 hrs after electroporation viewed on whole embryos (ventral views, left panels from A to F) and on transverse sections performed at distinct antero-posteriors levels (indicated by whites bars by ' to ''' for each set of panels, ''' corresponding to the PZ level). Pcig (A–A’’’), Pcig-Bra (B–B’’’), Pcig-Sox2 (C–C’’’), Control-Mo (D–D’’’), Sox2-Mo (E–E’’’), and Bra-Mo (F–F’’’). Transverse section analysis confirms results of Figure 2: control cells located in the PZ (yellow arrowhead), in the PSM (red arrowhead), and in the NT (green arrowhead). Of note, all experimental conditions leads to depletion of electroporated cells in the ZP, Pcig-Bra, and Sox2-Mo leads to enrichment of cells in the paraxial tissue (red arrowheads), Pcig-Sox2, and Bra-Mo leads to enrichment of cells in the NT (green arrowheads). Beige arrowheads indicate the position of the posterior end of the notochord on whole-mount pictures. Scale bars represent 100 µm on whole mount and 20 µm on sections. NT, neural tube; PSM, presomitic mesoderm; PZ, progenitor zone.
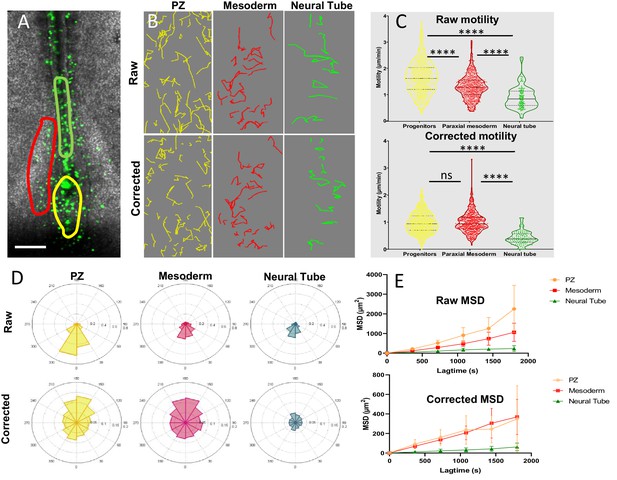
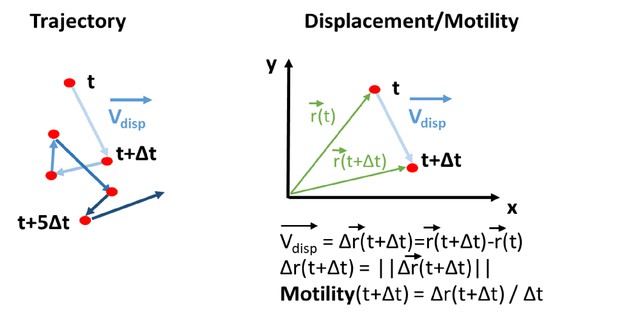
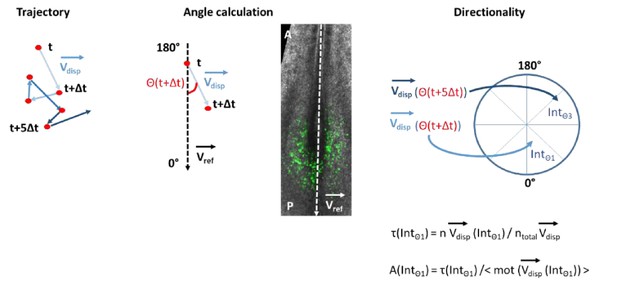
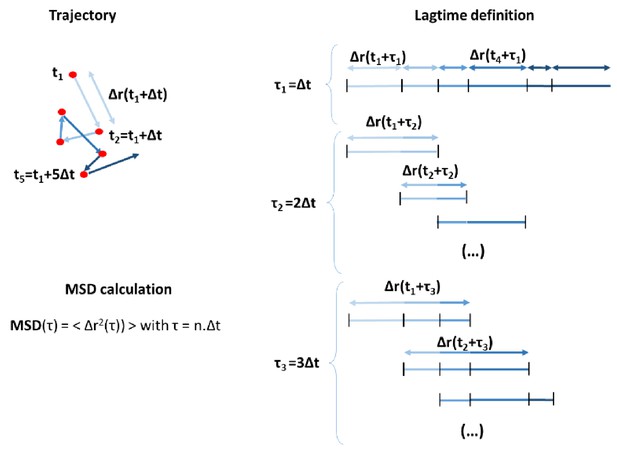
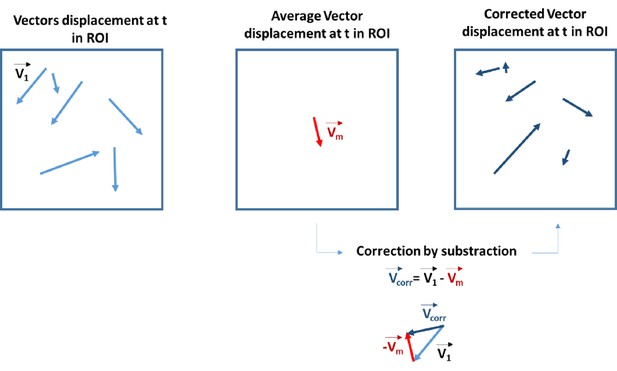
Progenitors display high motility without strong directionality.
(A) Representative image of an H2B-GFP electroporated quail embryo (ventral view) analyzed by live imaging . Transfected cells are detected by the GFP signal (green) . The PZ, the nascent PSM, and the NT are delineated by yellow, red, and green lines respectively. (B) Examples of cell trajectories before (raw) and after tissue motion subtraction (corrected). (C) Distribution of the raw (top) and corrected (bottom) cell motilities computed in the different regions. (D) Directionality of motion assessed by the distribution of angles weighed by the velocity for the different regions, before and after tissue subtraction. (E) Assessment of diffusion by analysis of the mean squared displacement in function of time for the different regions (n=7 embryos, 538 cell trajectories analyzed in the PZ, 496 in the PSM, and 128 in the NT). Scale bar=100 µm. NT, neural tube; PSM, presomitic mesoderm; PZ, progenitor zone.
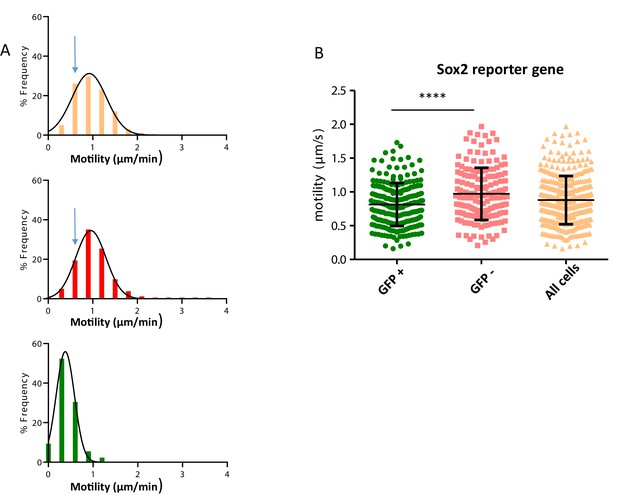
Distribution of motility frequencies.
(A) Representation of the distribution of frequency for different classes of speeds ranging from slow -moving cells to fast-moving cells corresponding to Figure 3C corrected motility. Top panel (yellow) corresponds to the PZ, middle panel (red) to the PSM, and bottom panel (green) to the NT. Note that the distributions of speed are different between the three groups, the NT containing more slow cells than the two other tissues and the progenitor zone having slightly more slower cells than the PSM (higher histogram pointed by the blue arrow). (B) Cell motilities for progenitors positive or negative for the Sox2 reporter (N1N2-eGFP-Pest). Tracking has been made in the PZ, on cell nuclei constitutively expressing NLS-Scarlet signal (co-electroporated). Note that GFP positive progenitors display significantly slower motility than negative cells (n=5 embryos). NT, neural tube; PSM, presomitic mesoderm; PZ, progenitor zone.
Cellular migration in PZ, NT and PSM (corresponding to Figure 3).
H2B-GFP electroporated quail embryos. The movie shows a ventral view of the posterior region of a representative embryo (10x). GFP signal is in green, bright field in blue. Moving regions of interest are delineated as follow: yellow for the progenitor, red for the nascent paraxial mesoderm and green for the NT. Cell trajectories are marked and colored accordingly.
Progenitor migration expressing NLS-Scarlet and Sox2 reporter (N1N2-eGfp-Dest) (corresponding to Figure 3—figure supplement 1B).
The movie shows a ventral view of the posterior region of a representative embryo (10x). The Sox2 signal is in green, the constitutive nuclear signal is in red.
Posterior progenitor migration (corresponding to Figure 3).
H2B-GFP electroporated quail embryos. The movie shows a ventral view of the posterior region of a representative embryo (10x). The GFP signal is in white. The right movie show the raw signal and the left movie shows some example of cell tracking (yellow). Note that nuclei are often changing neighbors.
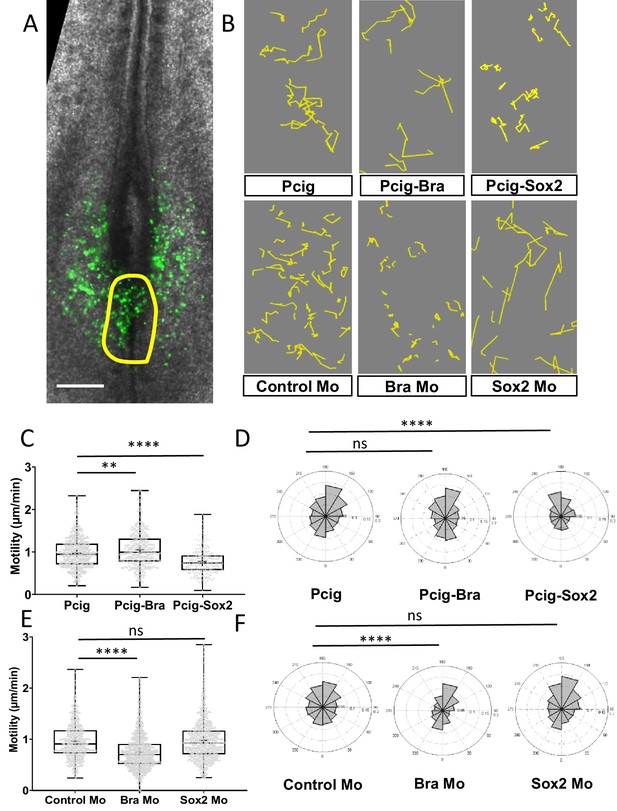
Sox2 and Bra deregulations affect progenitor motility.
(A) Representative image of a Pcig electroporated quail embryo (ventral view) used to perform progenitor tracking and motility analysis. Transfected cells are detected by the GFP-signal (green) and the PZ is delineated by the yellow line. (B) Examples of cell tracks after correction in embryos electroporated with expression vectors or morpholinos indicated on each panel. (C, E) Distribution of PZ cell motilities after tissue motion subtraction in gain of function (C) and in downregulation (E) experiments. (D, F) Directionality of cell motion after tissue motion subtraction assessed by the distribution of angles in gain of function (D) and in downregulation (F) experiments (n=7 embryos and 541 trajectories for Pcig, n=5 embryos and 307 trajectories for Pcig-Bra, and n=5 embryos and 234 trajectories for Pcig-Sox2; n=5 embryos and 590 trajectories for Control-Mo, n=7 embryos and 753 trajectories for Bra-Mo, and n=5 Embryos and 874 trajectories for Sox2-Mo). Scale bar=100 µm. PZ, progenitor zone.
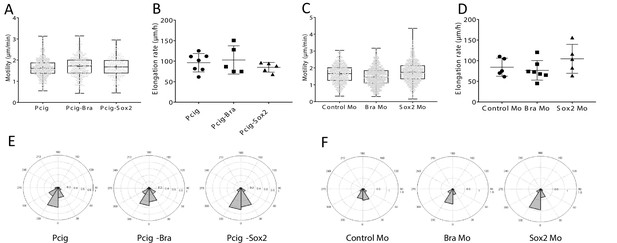
Effects of Sox2 and Bra on raw cell and tissue movements.
Raw cell movements (A, C), elongation(B, D), and raw angle distribution (E, F) measurements for Sox2 and Bra overexpression (A, B, E) and downregulation (C, D, F).
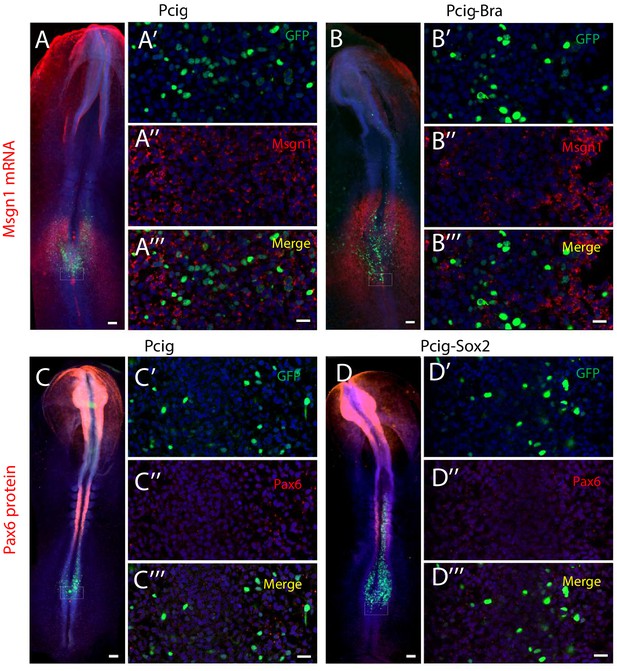
Cell differentiation in response to Sox2 and Bra overexpression.
Analysis of msgn1 mRNA expression (A–B’’’) and Pax6 Protein (C-D’’’) (red signals) 7 hr after transfection, transfected nuclei are visualized in green. Whole-mounts (A–D) and high-resolution images in the ZP (white dash rectangle on whole mount) (A’–A’’’, B–B’’’, C’–C’’’, D’–D’’’); Pcig (n=6 and 5, A–A’’’, C–C’’’), Pcig-Bra (n=8, B–B’’’), and Pcig-Sox2 (n=9, D–D’’’). Note that Bra overexpression does not induce an upregulation of Msgn1 after 7 hr, and that Sox2 overexpression does not induce an upregulation of Pax6 after 7 hr. Scale bars represent 100 µm on whole mount and 20 µm on high magnifications. PZ, progenitor zone.
Progenitor migration in Sox2 and Bra overexpression experiments (Corresponding to Figure 4).
The movie shows ventral views of posterior regions of three representative embryos, focused on the PZ that has been set as a reference point (10x). GFP signal is in green, bright field is in blue. Cell trajectories are marked and colored in yellow. Bra overexpression is on the left, Sox2 overexpression on the right and control embryo is in the middle.
Progenitor migration in Sox2 and Bra Mo experiments (Corresponding to Figure 4).
The movie shows ventral views of posterior regions of three representative embryos, focused on the PZ that has been set as a reference point (10x). GFP signal is in green, bright field is in blue. Cell trajectories are marked and colored in yellow. Bra Mo is on the left, Sox2 Mo is on the right and control embryo is in the middle.
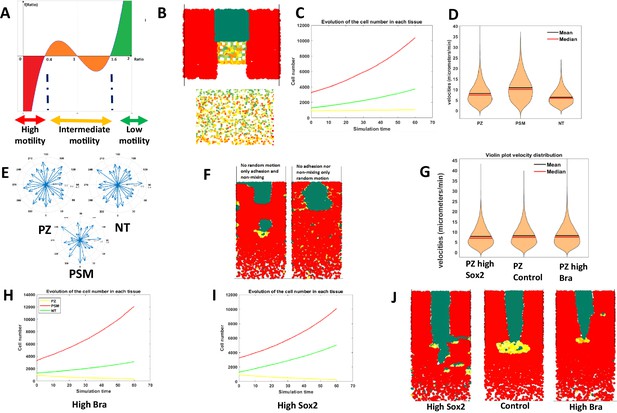
Mathematical modeling of progenitor behaviors downstream of Sox2 and Bra heterogeneous expression.
(A) Graphical representation of the mathematical function defining the Sox2-to-Bra ratio dynamics. The Sox2/Bra value oscillates randomly from 0.4 to 1.6 and noise in the system ensures that some cells pass below 0.4 to be specified into PSM cells (red) while some cells pass above 1.6 to become NT cells (green). Low ratios (below 0.4) confer high motility , high ratios (above 1.6) inhibit motility and ratios between 0.4 and 1.6 confer intermediate levels of motility. (B) Posterior region showing the spatial heterogeneity of Sox2/Bra levels with a close-up on the PZ on the bottom panel. (C) Evolution of the number of each cell type over time. (D) Distribution of cell motilities. (E) Directionality of migration in the three tissues. (F) Simulation without random motility (left) or without non-mixing and adhesion (right) for progenitors. (G) Effects of deregulations of the Sox2/Bra values on cell motility (G), cell numbers (H, I), and on tissue evolution at 10 hr (J). NT, neural tube; PSM, presomitic mesoderm; PZ, progenitor zone.
Mathematical simulations: combined random and graded expression of Sox2 and Bra (mixed).
Mathematical simulation of Sox2 and Bra overexpression on the mixed model.
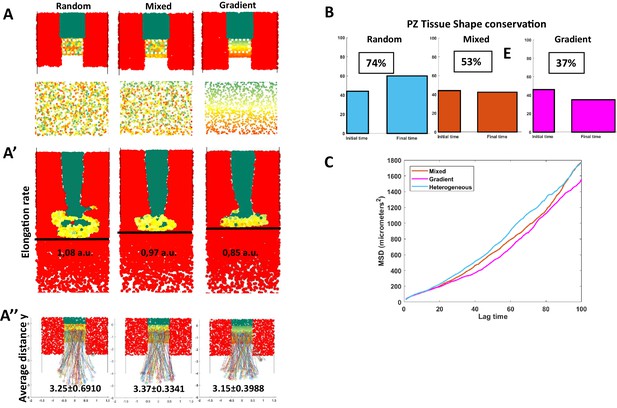
Comparison of spatial organizations by modeling.
(A) Random model (left), mixed (defined in Figure 5), and the graded model following opposite gradients (right). (A) Initial conditions and close-up of the PZ showing the spatial organization of Sox2/Bra levels (bottom panels). (A’) Elongation rates measured with the distance traveled by the posterior part of the PZ (black line) in 10 hr. (A’’) Y displacement of resident progenitors located at the center (along the anteroposterior axis) of the PZ for each model. (B) Initial (left) and final (right) shapes of the PZ in the different models. Conservation of proportions (length/width) is noted in percentage (100% would correspond to an unchanged shape). (C) MSD calculated for progenitors in the three models. MSD, mean squared displacement; PZ, progenitor zone.
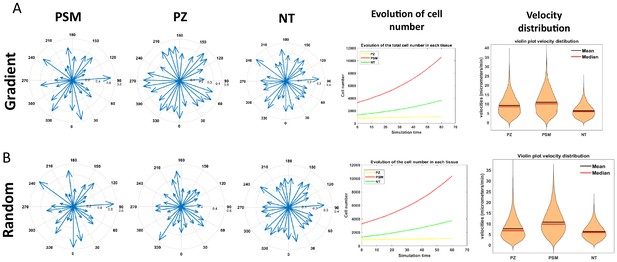
Directionality of migration, evolution of cell number, and velocity distribution in the random and gradient models.
The top panel corresponds to measurements of the gradient model; the bottom panel corresponds to the random model. (A, B) (left): Directionality of migration in the three tissues in the gradient model (A, left) and the random model (B, left). (A, B) (middle): Evolution of the number of each cell type over time in the gradient model (A, middle) and the random model (B, middle). (A, B) (right): Distribution of cell motilities in the gradient model (A, right) and the random model (B, right).
Mathematical simulations of random, graded and mixed models.