Living with relatives offsets the harm caused by pathogens in natural populations
Figures
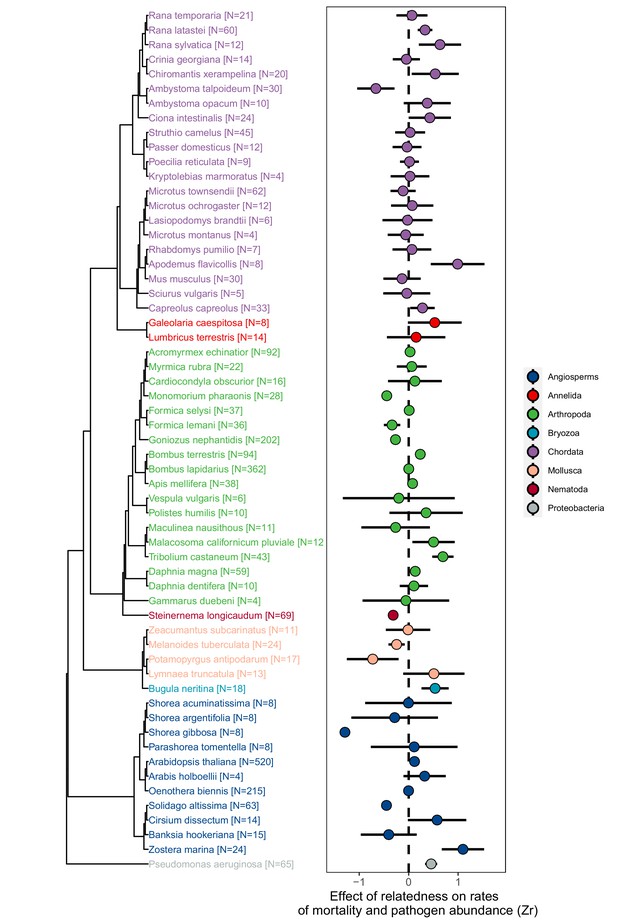
The effect of relatedness on rates of mortality and pathogen abundance across animals, plants, and bacteria.
Positive effect sizes (Zr) indicate that mortality and/or pathogen abundances increase with the levels of relatedness within groups, negative values show decreases, and values of zero (dotted line) are where there was no relationship. Points represent weighted means for each species and bars are 95% confidence intervals calculated from the sample sizes of the number of groups studied which are given in brackets. See Figure 1—figure supplements 1–3 for information on obtaining effect size information and testing for publication bias.
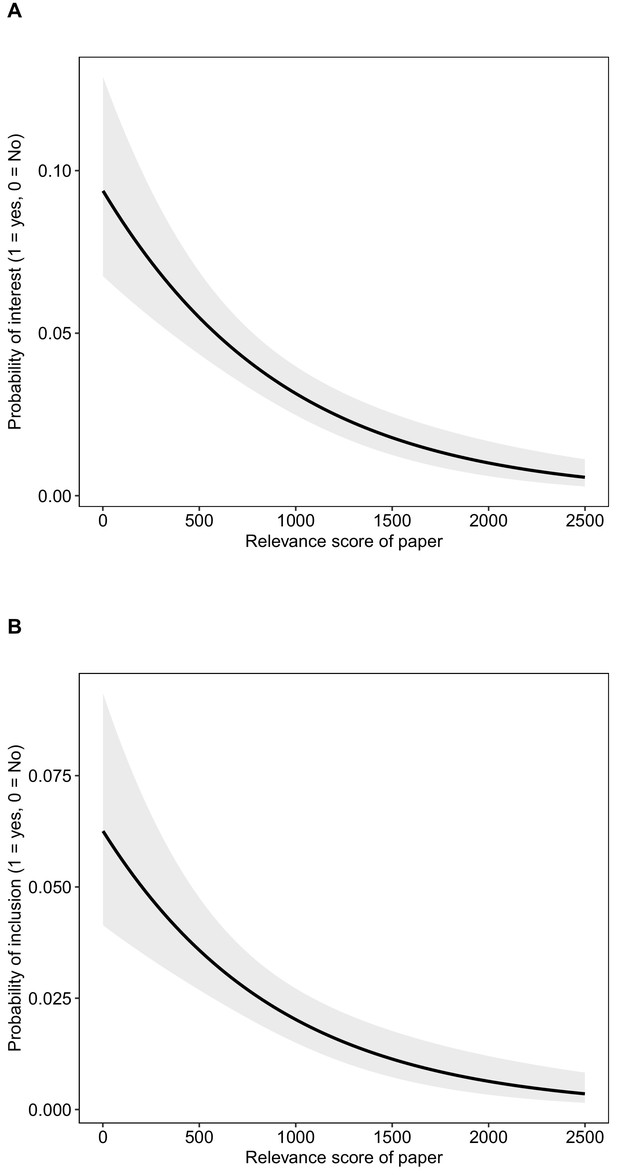
The probability that studies were of interest (A) and were included (B) in relation to the abstract relevance score (see ‘Literature searches’ in Materials and methods for details).
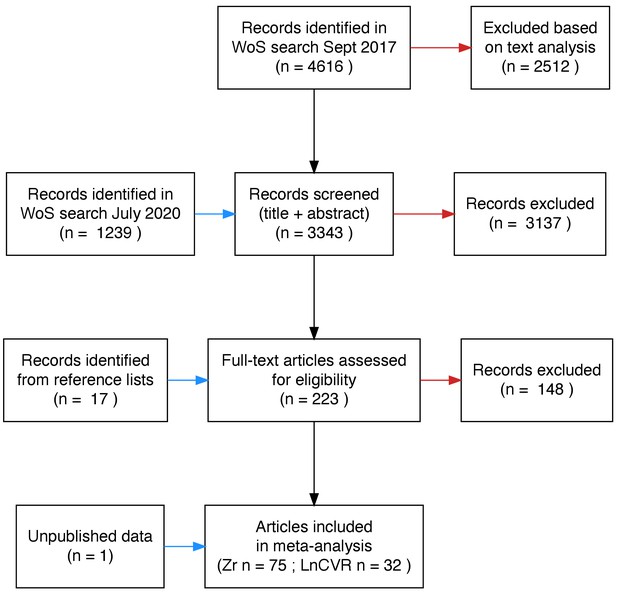
Preferred reporting items for meta-analyses (PRISMA) diagram of literature search.
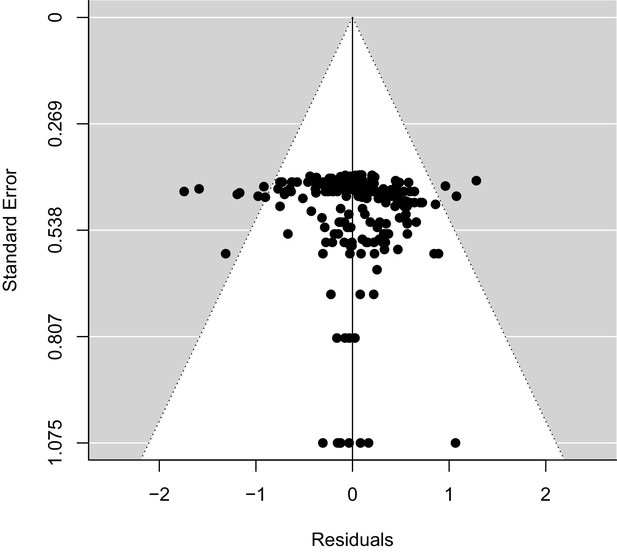
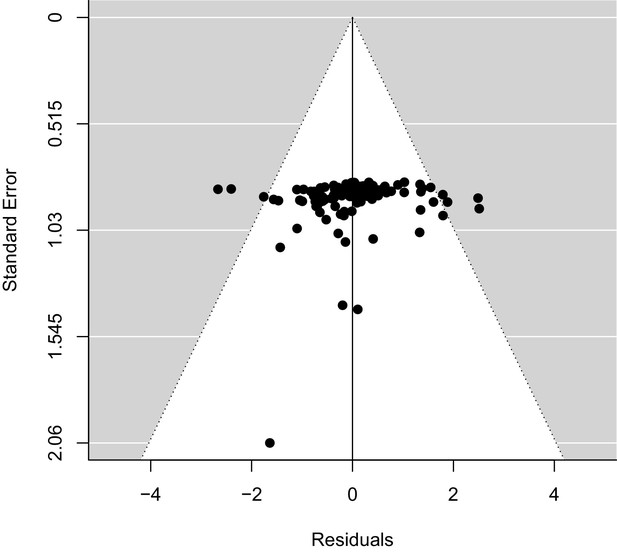
Examining evidence of publication bias in estimates of Zr.
Residuals from an intercept-only model of Zr fitted in metafor.
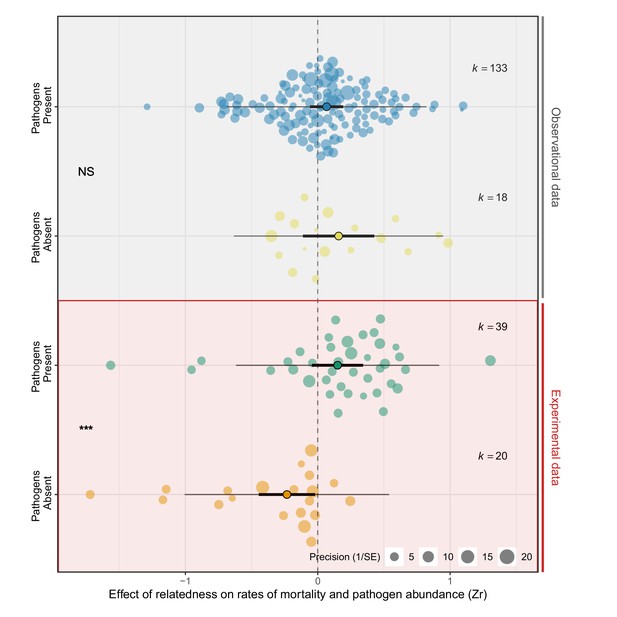
Experimental manipulations are key to detecting the effects of pathogens on groups of relatives.
Positive effect sizes (Zr) indicate that mortality and/or pathogen abundances increase with the levels of relatedness within groups, negative values show decreases, and values of zero (dotted line) are where there was no relationship. Studies that experimentally manipulated pathogen presence showed that groups of relatives had higher rates of mortality when pathogens were present, but lower mortality when pathogens were absent. Points with black edges represent means, thick bars are 95% CIs, thin bars are prediction intervals, and k is the number of effect sizes. Each dot is an individual effect size and with size scaled to 1/SE (orchard plots: Nakagawa et al., 2021). Statistical differences are from Bayesian Phylogenetic Multi-level Meta-regressions (BPMMs) and placed mid-way between comparison groups denoted with symbols: NS = non-significant, *pMCMC < 0.05, **pMCMC < 0.01, ***pMCMC < 0.001.
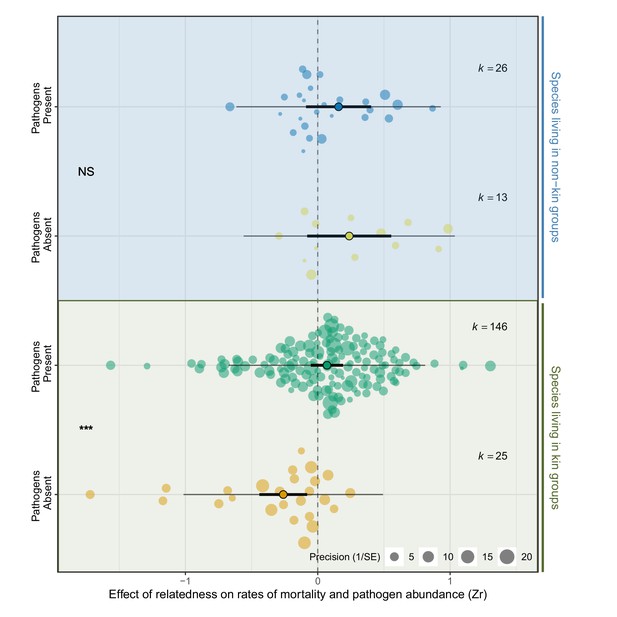
Species that live in kin groups responded differently to experimental manipulation of pathogens compared to species that live with non-kin.
Positive effect sizes (Zr) indicate that mortality and/or pathogen abundances increase with the levels of relatedness within groups, negative values show decreases, and values of zero (dotted line) are where there was no relationship. When pathogens were experimentally removed species that live with kin had higher survival, which was reversed when pathogens were present. In contrast, there was no effect of relatedness on mortality when pathogens were present or absent in species that live with non-kin. The components of the orchard plots are the same as in Figure 2. Statistical differences are from Bayesian Phylogenetic Multi-level Meta-regressions (BPMMs) and placed mid-way between comparison groups denoted with symbols: NS = non-significant, *pMCMC < 0.05, **pMCMC < 0.01, ***pMCMC < 0.001.
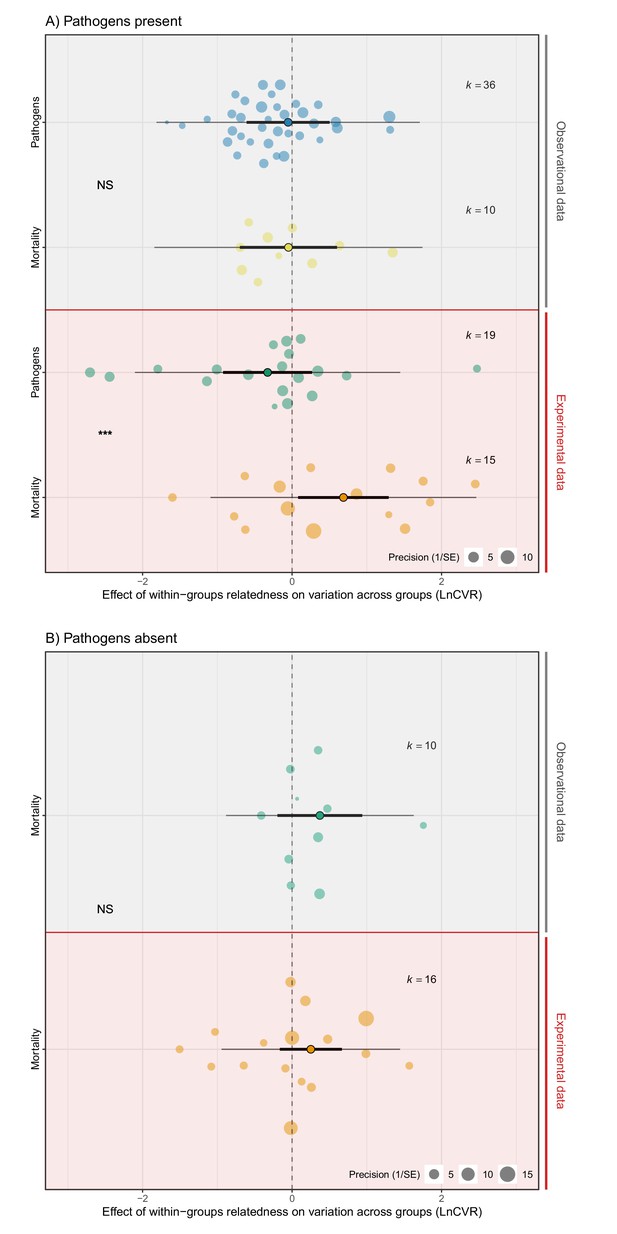
The effect of within-group relatedness on variance in mortality and pathogen abundance.
Positive effect sizes (LnCVR) show that variation in rates of mortality and/or pathogen abundances across groups (accounting for mean differences – see Figure 4—figure supplement 1 for mean variance relationship) increases with within-group relatedness, negative values show decreases in variance, and zero values show no change in variance (dotted line). (A) In the presence of pathogens, relatedness increased variance in mortality, but decreased variance in pathogen abundance. (B) When pathogens were absent, relatedness did not influence variance in mortality. The components of the orchard plots are the same as in Figure 2. Statistical differences are from Bayesian Phylogenetic Multi-level Meta-regressions (BPMMs) and placed mid-way between comparison groups denoted with symbols: NS = non-significant, *pMCMC < 0.05, ** pMCMC < 0.01, ***pMCMC < 0.001. Figure 4—figure supplement 2 for examination of publication bias.
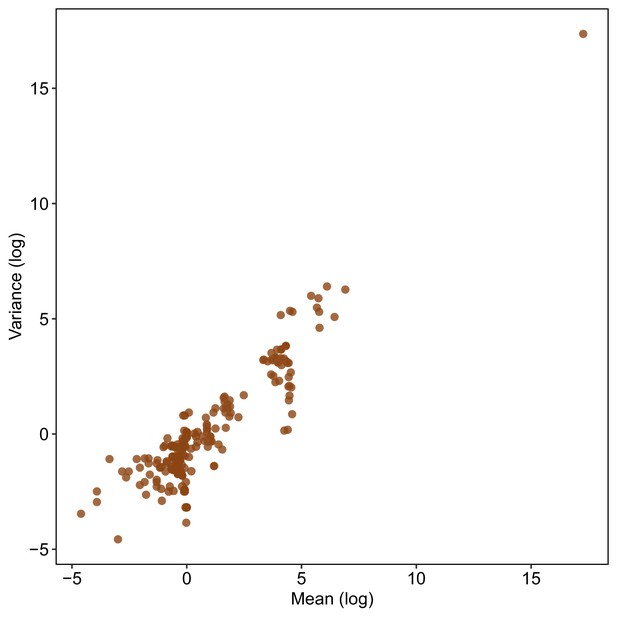
Relationship between the mean (log) and SD (log) across studies used to estimate LnCVR.
Additional files
-
Supplementary file 1
Supplementary Tables S1-S19 and R session information in html format.
Tables S1-19 can be found in excel format at the open science framework DOI 10.17605/OSF.IO/Q3ANE. Table S1: The references screened for effect sizes of within-group relatedness on mortality and pathogen abundance and whether they were included in analyses. Table S2: Data used for analysis of the effect of within-group relatedness on mortality and pathogen abundance (r). Table S3: Data on whether species typically live with kin or non-kin. Table S4: Data used for analysis of the effect of within-group relatedness on variance in mortality and pathogen abundance (LnCVR). Table S5: The overall effect of within-group relatedness on mortality and pathogen abundance on Zr. Table S6: The effect of pathogen presence and whether mortality or pathogen abundance was measured on Zr. Table S7: The effect of pathogen manipulation on Zr. Table S8: The effect of relatedness manipulations and pathogen presence on Zr. Table S9: The effect of kin structure and pathogen presence measures on Zr. Table S10: The overall effect of within-group relatedness on variance (LnCVR) mortality and pathogen abundances. Table S11: The effect of pathogen manipulation on LnCVR. Table S12: The effect of pathogen manipulation and whether mortality or pathogen abundance was measured on LnCVR. Table S13: The effect of relatedness manipulations and pathogen presence on LnCVR. Table S14: The effect of kin structure and pathogen presence measures on Zr excluding potential effects of inbreeding. Table S15: Effect of parasite manipulations and fitness measures on LnCVR excluding potential effects of inbreeding. Table S16: Laboratory effects on Zr. Table S17: Laboratory effects on LnCVR. Table S18: The sensitivity of analyses on Zr to the statistical techniques used in primary studies. Table S19: Data on within-group relatedness on mortality in ostrich chicks.
- https://cdn.elifesciences.org/articles/66649/elife-66649-supp1-v1.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66649/elife-66649-transrepform-v1.pdf