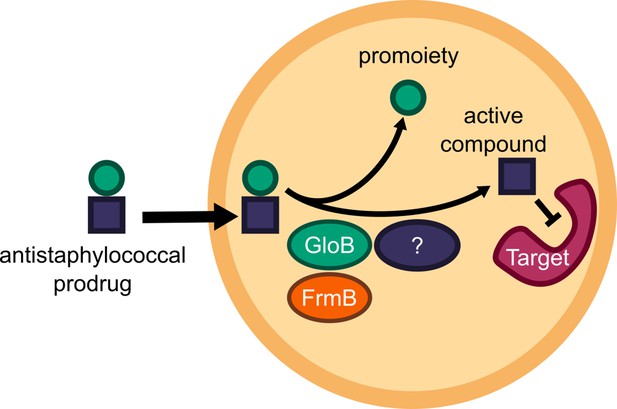
Structure-guided microbial targeting of antistaphylococcal prodrugs
Figures
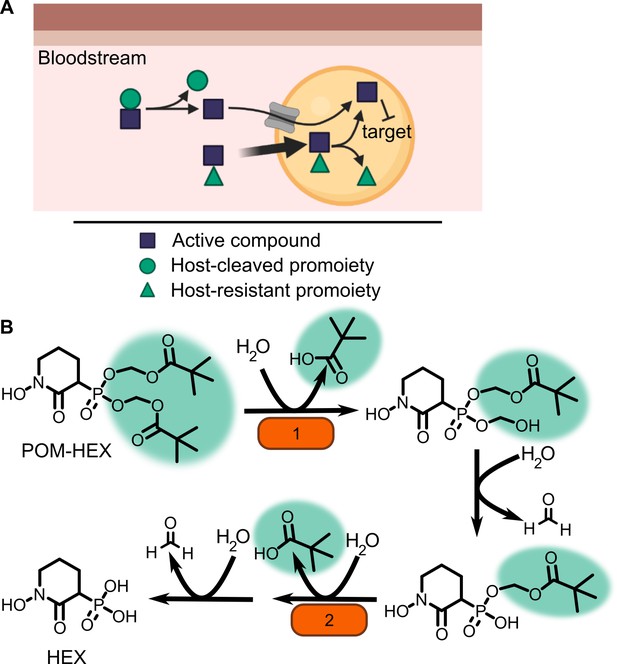
Prodrug activation model (A) and proposed enzymatic mechanism.
(B) Carboxy ester promoieties highlighted in green.
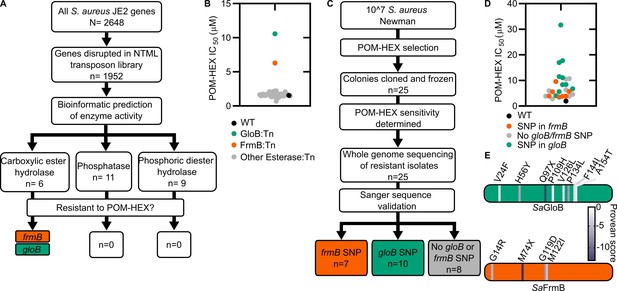
Forward and reverse genetics approaches identify FrmB and GloB as candidate POM-prodrug hydrolases in S. aureus.
(A) Reverse genetics identification of candidate prodrug activating enzymes. (B) POM-HEX susceptibility of identified candidate resistance genes from (A) as determined by IC50. Exact values and error reported in Figure 2—source data 1. (C) Forward genetic screen approach, all mutations listed in Figure 2—source data 2. (D) POM-HEX susceptibility of POM-HEX-resistant S. aureus. (E) Nonsynonymous point mutations identified by whole-genome sequencing in frmB and gloB. In all experiments, GloB is colored green and FrmB orange. Displayed are the means of three independent biological experiments.
-
Figure 2—source data 1
S. aureus transposon mutants tested and POM-HEX sensitivity.
Half-maximal inhibitory concentration (IC50) values for POM-HEX against predicted prodrug activating esterases. IC50 values are the result of three independent biological experiments with technical duplicates. p-values calculated as a one-way ANOVA with Dunnett’s correction for multiple comparisons.
- https://cdn.elifesciences.org/articles/66657/elife-66657-fig2-data1-v3.xlsx
-
Figure 2—source data 2
S. aureus Newman resistant isolate SNPs and POM-HEX sensitivity.
Genotype and phenotype of POM-HEX resistant S. aureus. Displayed are the whole-genome sequencing mutations that have been verified. Called mutations that were not observed via confirmatory Sanger sequencing are excluded. IC50 values are the result of three independent biologic replicates with technical duplicates.
- https://cdn.elifesciences.org/articles/66657/elife-66657-fig2-data2-v3.xlsx
-
Figure 2—source data 3
S. aureus transposon mutants with genes identified by whole-genome sequencing and POM-HEX sensitivity.
Half-maximal inhibitory concentration (IC50) values for POM-HEX against transposon mutations in genes identified by whole-genome sequencing. Assays performed in biological triplicate with technical duplicates. p-value calculated as a one-way ANOVA with Dunnett correction for multiple comparisons.
- https://cdn.elifesciences.org/articles/66657/elife-66657-fig2-data3-v3.xlsx
-
Figure 2—source data 4
Accession numbers for the isolates used in WhatsGNU analysis.
- https://cdn.elifesciences.org/articles/66657/elife-66657-fig2-data4-v3.xlsx

Conservation of FrmB and GloB within S. aureus.
(A) WhatsGNU analysis of GloB and FrmB. Control proteins: ArgG, argininosuccinate synthase; Fba, fructose-bisphosphate aldolase; MenD, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1carboxylate synthase; and MenC, o-succinylbenzoate synthase. GNU stands for gene novelty unit and is a count of how many protein sequences in the database have an exact match to the queried sequence, with higher counts indicating sequence conservation. Strains across the x-axis are representative strains from the 18 s. aureus colony complexes which were used to query the S. aureus database. (B, C) MAFFT alignment of GloB (B) and FrmB (C) protein sequences across the S. aureus sequence database.
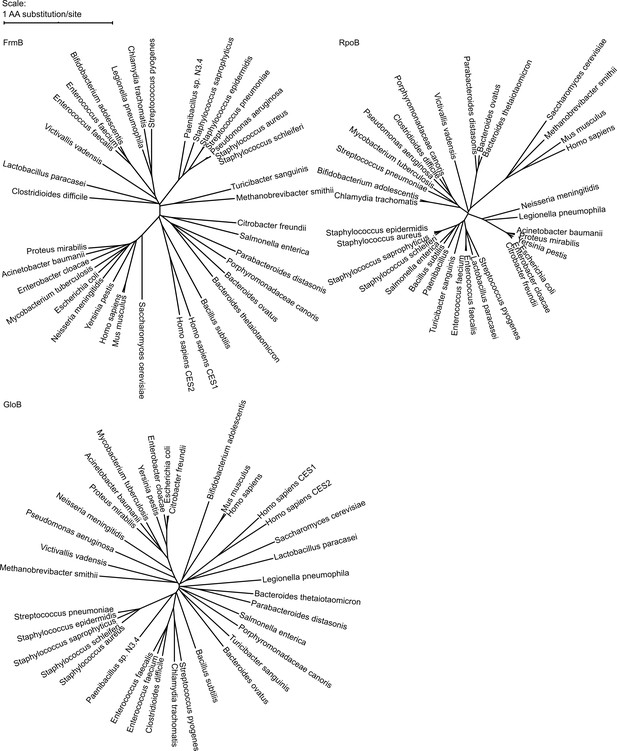
Phylogenetic tree of FrmB and GloB.
Sequences of GloB, FrmB, and RpoB (encoding the beta subunit of RNA polymerase, included for comparison) were retrieved from NCBI using BlastP against each organism. Sequence alignment performed using MUSCLE and alignment visualized using the interactive Tree of Life (iTOL).
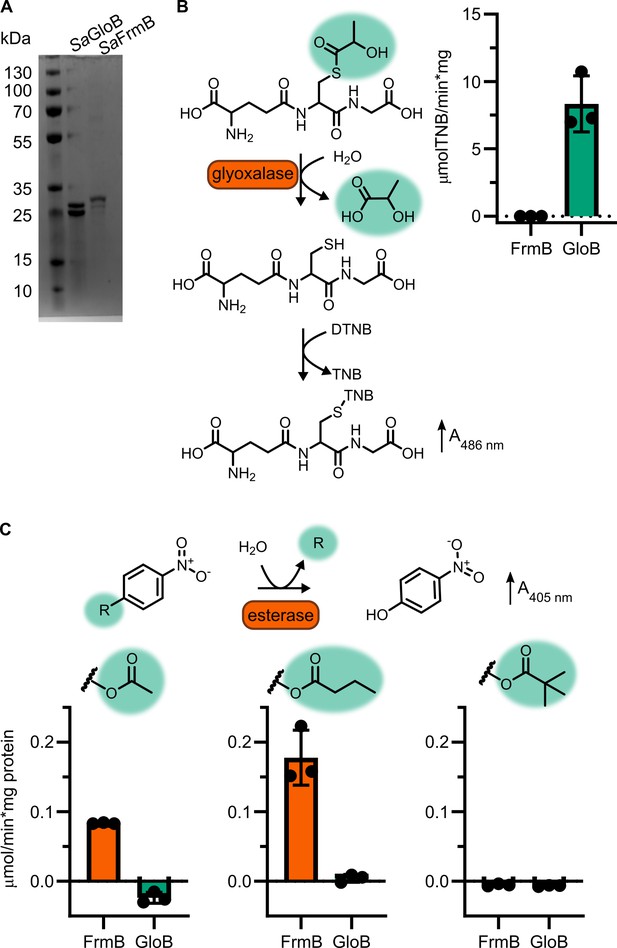
Enzymatic characterization of GloB and FrmB.
(A) SDS–PAGE gel of GloB and FrmB protein preparations. Expected molecular weights are 23.3 kDa and 29.5 kDa, respectively. (B) Glyoxalase II activity assay, enzymatic conversion of S-lactoylglutathione releases free glutathione and reacts with DTNB resulting in increased absorbance at 412 nm. (C) 4-Nitrophenyl activation results in increased absorbance at 405 nm. Left to right, activity when supplied 4-nitrophenyl acetate, 4-nitrophenyl butyrate, and 4-nitrophenyl trimethyl acetate. Displayed in points is the mean of two technical replicates for individual experiments, bars indicate mean ± SD of three independent biological experiments performed in technical duplicate.
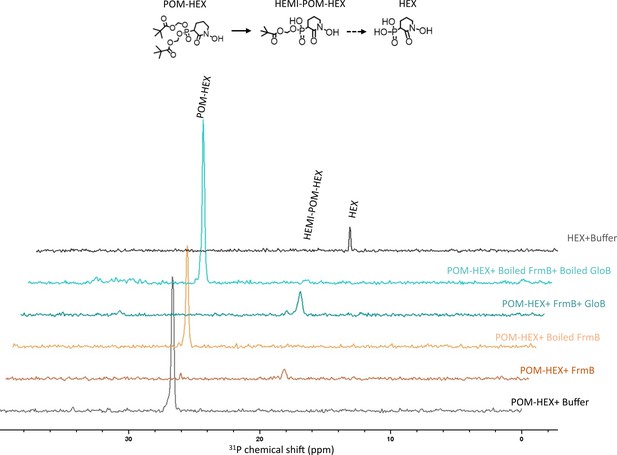
NMR characterization of POM-HEX activation by GloB and FrmB.
Two-dimensional (2D) 1H-31P HSQC NMR spectra of products following incubation of FrmB, GloB, catalytically inactive (boiled) GloB and FrmB, or buffer alone. Also included are the 1H-31P HSQC NMR spectra of POM-HEX and HEX. Displayed are representative traces of three independent experiments. HEMI-POM HEX peak inferred by predicted shift.
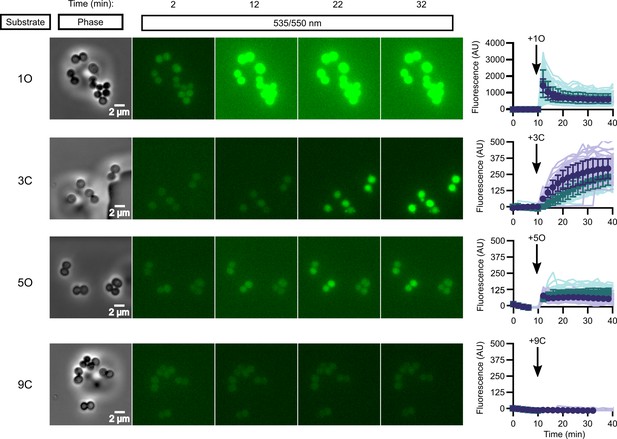
In vivo activation rates depend on ester promoiety selection.
Time series of activation of various fluorogenic substrates (Figure 3—figure supplement 1). Substrates were added into the microfluidics chamber at t = 10 minutes. On the right, quantification of individual cell or cell cluster fluorescence per area. Faint traces are individual cells and darker traces represent the mean of a given experiment. Each experiment was performed in biological duplicate, and each experiment is displayed in a different color (purple or green). Full movies viewable as Videos 1–4. Error bars denote SD.
-
Figure 3—source data 1
(1) Michaelis–Menten parameters for SaFrmB.
Displayed are the results of three independent biological replicates in technical duplicate. (2) Michaelis–Menten parameters for SaGloB. Displayed are the results of three independent biological replicates in technical duplicate.
- https://cdn.elifesciences.org/articles/66657/elife-66657-fig3-data1-v3.xlsx
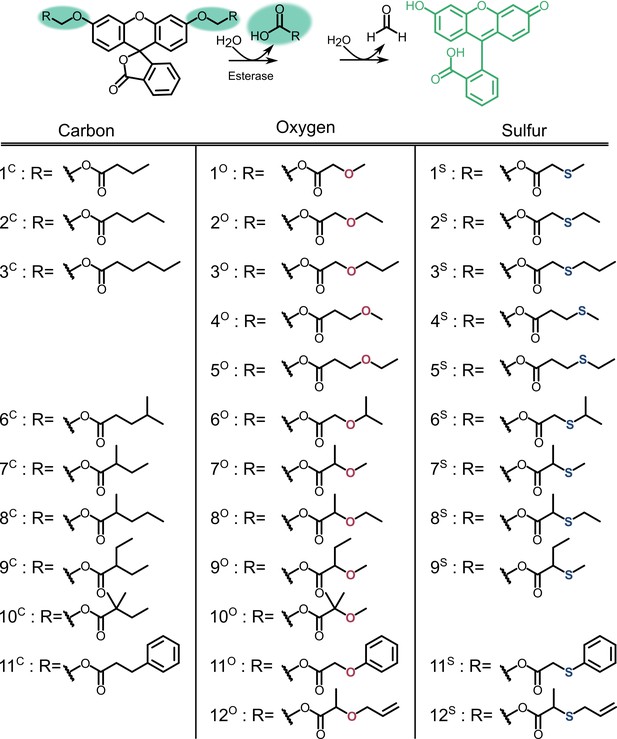
Profluorescent substrate library.
Activation of substrates via esterase action results in fluorescence.
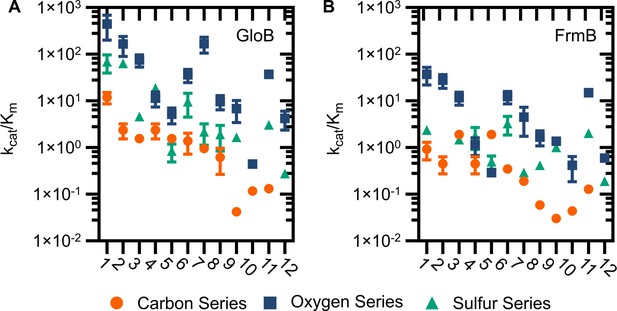
Catalytic efficiency of GloB (A) and FrmB (B).
Numbers correspond to the structures displayed in Figure S5, compounds in the carbon series denoted in orange, oxygen series in blue, and sulfur series in green.
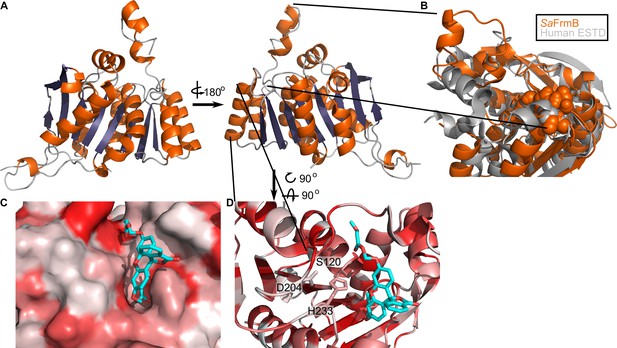
Three-dimensional structure of FrmB.
(A) Overall fold, a-helices colored in orange and β-strands colored in purple. (B) Comparison between SaFrmB (orange) and its closest human ortholog, ESTD (gray). Active site residues denoted in orange spheres. (C, D) Docking of substrate 1O (sticks) in the active site of FrmB. surface view, red indicates highly hydrophobic and white hydrophilic residues. Surface view (C) or stick view with catalytic triad (D).
-
Figure 4—source data 1
Summary of crystallographic data collection and refinement statistics.
- https://cdn.elifesciences.org/articles/66657/elife-66657-fig4-data1-v3.xlsx
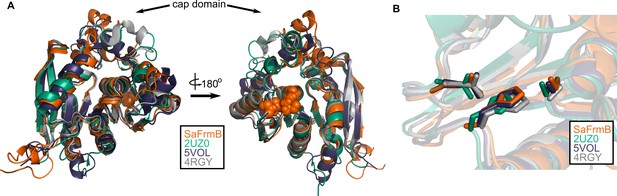
Structural conservation of FrmB.
(A) Overall structural alignment of FrmB (orange) with S. pneumonia EstA (PDB:2UZ0), B. intestinalis ferulic acid esterase (PDB:5VOL), and deep sea bacteria Est12 (PDB4RGY). (B) Conservation of the serine hydrolase catalytic triad in FrmB and related proteins.
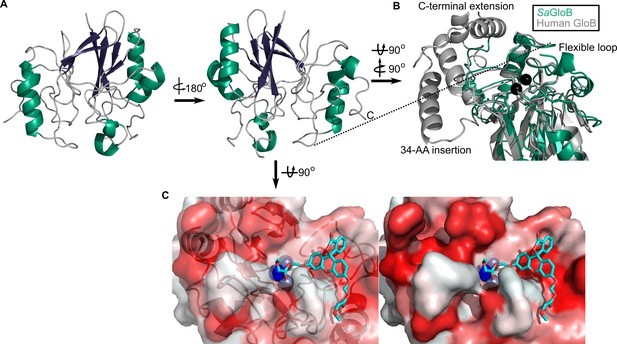
Three-dimensional structure of GloB.
(A) Overall fold, a-alpha helices colored in green and β-strands colored in purple. (B) Comparison of SaGloB (green) and human GloB (gray). (C) Docking of the substrate 1O (sticks) in the active site of GloB. Left, partial cartoon view; right, surface view. White represents hydrophilic residues, whereas red represents hydrophobic residues. Zn ions indicated as silver spheres; water indicated as blue sphere.
-
Figure 5—source data 1
Summary of crystallographic data collection and refinement statistics.
- https://cdn.elifesciences.org/articles/66657/elife-66657-fig5-data1-v3.xlsx
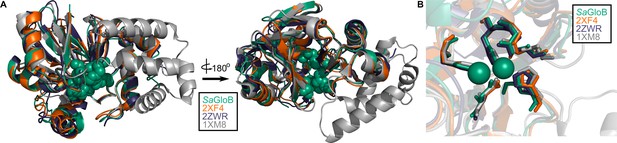
Structural conservation of GloB.
(A) Overall structural alignment of GloB (green) with S. enterica YcbI (PDB:2XF4), T. thermophilus TTHA1623 (PDB:2ZWR), and A. thaliana glyoxalase II (PDB:1XM8). Zinc coordinating residues are colored in green spheres. (B) Positioning of the zinc coordinating residues (green spheres).
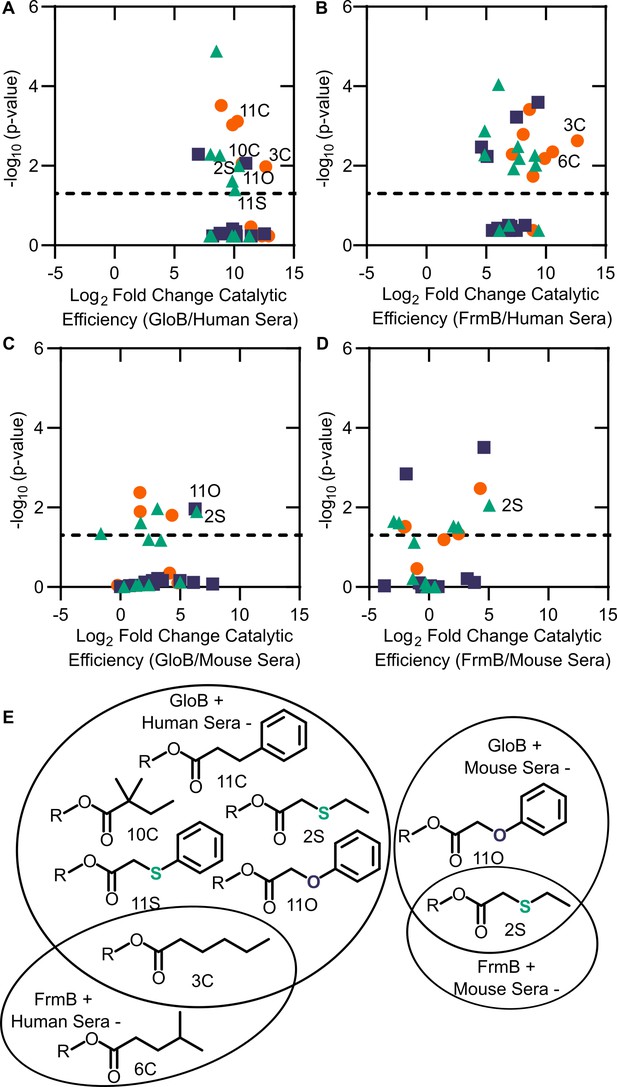
Comparison between microbial esterase and serum esterase catalytic efficiency.
(A–D) Volcano plots of catalytic efficiency. Displayed are the means of three independent experiments. p-values calculated as pairwise t-tests with Holm-Sidak correction for multiple comparisons. (A) Comparison between human sera and GloB, (B) human sera and FrmB, (C) mouse sera and GloB, and (D) mouse sera and FrmB. (E) Structures of ester substrates with 210 enrichment in catalytic efficiency for microbial esterases over human serum (left) or 25 enrichment over mouse serum. Dashed line indicates a p-value of 0.05.
-
Figure 6—source data 1
(1) Michaelis–Menten parameters for human sera.
Displayed are the results of three independent biological replicates in technical duplicate. (2) Michaelis–Menten parameters for mouse sera. Displayed are the results of three independent biological replicates in technical duplicate.
- https://cdn.elifesciences.org/articles/66657/elife-66657-fig6-data1-v3.xlsx
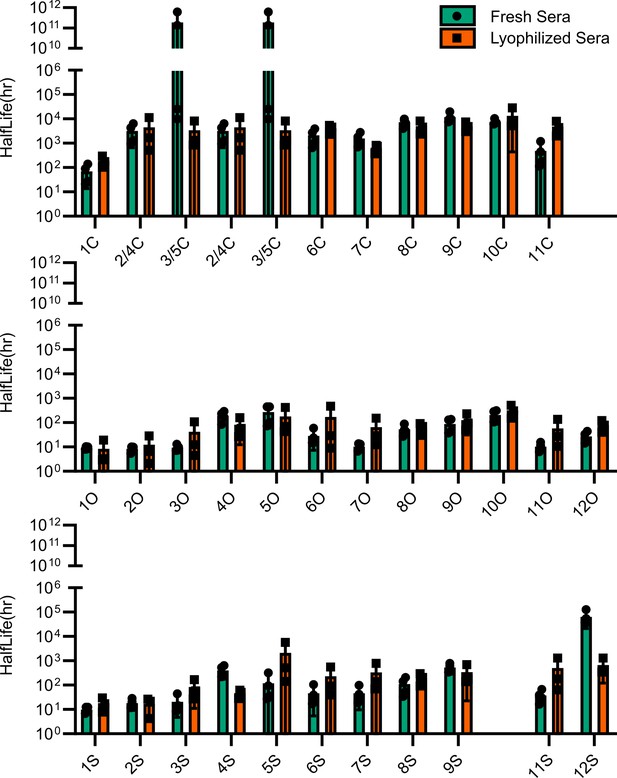
Comparison of esterase activity between fresh and lyophilized human sera.
Points represent individual experiments; bars represent the mean ± SD of the four replicates.
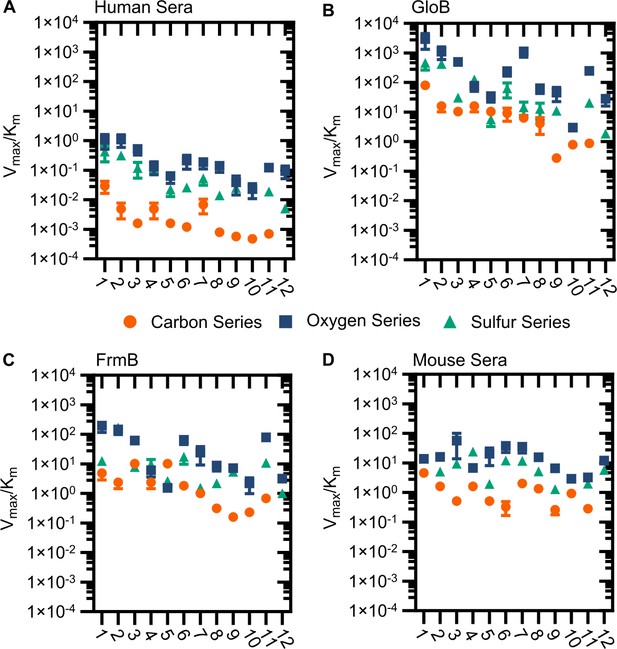
Modified catalytic efficiency (pmol fluorescein produced * min−1*µg−1 protein) of (A) human sera, (B) GloB, (C) FrmB, and (D) mouse sera.
X-axis corresponds to compound identities in Figure S5. Carbon containing compounds indicated in orange, oxygen in blue, and sulfur in green. Displayed are the means ± SD of three independent biological experiments.
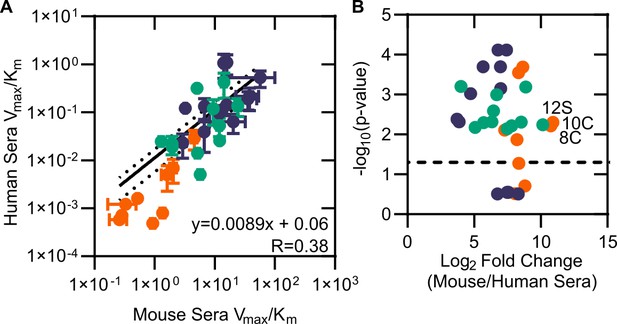
Comparison of mouse and human sera.
(A) Modified catalytic efficiency (pmol fluorescein produced * min-1*µg-1 protein) of human and mouse sera. Displayed is a linear regression of the fit between mouse and human sera. (B) Volcano plot of catalytic efficiency. Displayed are the means of three independent experiments. p-values calculated as pairwise t-tests with Holm–Sidak correction for multiple comparisons. Dashed line indicates a p-value of 0.05.
Videos
In vivo activation rates depend on ester promoiety selection.
Time series of fluorogenic ester substrate 1O activation. Substrate added at t = 10 min. Experiments were performed in biological duplicate.
In vivo activation rates depend on ester promoiety selection.
Time series of fluorogenic ester substrate 3C activation. Substrate added at t = 10 min. Experiments were performed in biological duplicate.
In vivo activation rates depend on ester promoiety selection.
Time series of fluorogenic ester substrate 5O activation. Substrate added at t = 10 min. Experiments were performed in biological duplicate.
In vivo activation rates depend on ester promoiety selection.
Time series of fluorogenic ester substrate 9C activation. Substrate added at t = 10 min. Experiments were performed in biological duplicate.
Tables
Mutational frequencies of S. aureus.
Newman for POM-HEX and several clinically utilized antibiotics. Experiments performed in technical duplicate and biological triplicate. Displayed are the means ± SD.
| Antibiotic (MIC) | 4× MIC | 10× MIC |
|---|---|---|
| Vancomycin (2 µg/mL) | <1 E-10 | <1 E-10 |
| Clindamycin (0.25 µg/mL) | <1 E-10 | <1 E-10 |
| Rifampicin (7.5 ng/mL) | 8.96 ± 0.20 E-08 | 7.64 ± 2.10 E-08 |
| Nafcillin (0.5 µg/mL) | <1 E-10 | <1 E-10 |
| Linezolid (1.67 µg/mL) | <1 E-10 | <1 E-10 |
| POM-HEX (3.13 µg/mL) | 5.96 ± 1.60 E-07 | 2.92 ± 0.57 E-07 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Staphylococcus aureus) | GloB | GenBank | WP_001223008.1 | GloII |
| Gene (Staphylococcus aureus) | FrmB | GenBank | estA | |
| Strain, strain background (Staphylococcus aureus) | JE2 | BEI Resources | NR-46543 | |
| Strain, strain background (Staphylococcus aureus) | NE64 | Nebraska Transposon Mutant Library | Hypothetical protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE145 | Nebraska Transposon Mutant Library | protoporphyrinogen oxidase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE202 | Nebraska Transposon Mutant Library | ABC transporter, ATP-binding protein, MsbA family | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE223 | Nebraska Transposon Mutant Library | hydroxyacylglutathione hydrolase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE293 | Nebraska Transposon Mutant Library | staphylococcal accessory regulator Rot | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE355 | Nebraska Transposon Mutant Library | tributyrin esterase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE364 | Nebraska Transposon Mutant Library | NAD-dependent epimerase/dehydratase family protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE377 | Nebraska Transposon Mutant Library | pyrroline-5-carboxylate reductase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE386 | Nebraska Transposon Mutant Library | Hypothetical protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE478 | Nebraska Transposon Mutant Library | peptide ABC transporter, permease protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE503 | Nebraska Transposon Mutant Library | conserved hypothetical protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE520 | Nebraska Transposon Mutant Library | sensor histidine kinase SaeS | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE532 | Nebraska Transposon Mutant Library | PTS system, mannitol specific IIBC component | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE541 | Nebraska Transposon Mutant Library | alkaline phosphatase synthesis transcriptional regulatory protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE621 | Nebraska Transposon Mutant Library | Hypothetical protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE812 | Nebraska Transposon Mutant Library | tetrahydrodipicolinate acetyltransferase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE874 | Nebraska Transposon Mutant Library | Hypothetical protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE929 | Nebraska Transposon Mutant Library | acetyl-CoA carboxylase, biotin carboxylase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE937 | Nebraska Transposon Mutant Library | tandem lipoprotein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE949 | Nebraska Transposon Mutant Library | Putative Transposase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1039 | Nebraska Transposon Mutant Library | excinuclease ABC, A subunit | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1051 | Nebraska Transposon Mutant Library | Hypothetical protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1071 | Nebraska Transposon Mutant Library | Hypothetical protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1118 | Nebraska Transposon Mutant Library | dihydrolipoamide dehydrogenase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1127 | Nebraska Transposon Mutant Library | gamma-hemolysin component B | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1173 | Nebraska Transposon Mutant Library | tRNA pseudouridine synthase A | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1225 | Nebraska Transposon Mutant Library | ABC transporter, ATP-binding protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1238 | Nebraska Transposon Mutant Library | transcriptional regulator, TetR family | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1283 | Nebraska Transposon Mutant Library | Hypothetical protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1296 | Nebraska Transposon Mutant Library | hydroxyacylglutathione hydrolase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1486 | Nebraska Transposon Mutant Library | phosphonate ABC transporter, permease protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1505 | Nebraska Transposon Mutant Library | tributyrin esterase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1519 | Nebraska Transposon Mutant Library | Hypothetical Alkaline Phosphatase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1547 | Nebraska Transposon Mutant Library | Hypothetical protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1610 | Nebraska Transposon Mutant Library | Hypothetical protein | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1682 | Nebraska Transposon Mutant Library | PfkB family kinase | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Staphylococcus aureus) | NE1723 | Nebraska Transposon Mutant Library | type I restriction-modification system, M subunit | Available from BEI Resources as: NR-48501 |
| Strain, strain background (Escherichia coli) | BL21 (DE3) | Sigma | CMC0014 | Chemically Competent |
| Biological sample (Homo sapiens) | Fresh Human Serum | This paper | Whole blood was collected from a willing volunteer into untreated BD vacutainer tubes (BD, BD366430), allowed to clot, and aggregates were removed via centrifugation | |
| Biological sample (Homo sapiens) | Lyophilized Human Serum | Rockland Inc | D314-5 | |
| Biological sample (Mus musculus) | Lyophilized Mouse Serum | Rockland Inc | D308-5 | |
| Recombinant DNA reagent | pET28a-SaFrmB | This paper | E. coli expression plasmid for SaFrmB with cleavable HIS tag. | |
| Recombinant DNA reagent | pET28a-SaGloB | This paper | E. coli expression plasmid for SaGloB with cleavable HIS tag. | |
| Recombinant DNA reagent | BG1861-SaGloB | This paper | E. coli expression plasmid for SaGloB with HIS tag. | |
| Recombinant DNA reagent | BG1861-SaFrmB | This paper | E. coli expression plasmid for SaFrmB with HIS tag. | |
| Sequence-based reagent | NWMN_0144_F | This paper | PCR Primer | TTTTCCTGATCCTGATTCAC |
| Sequence-based reagent | NWMN_0144_R | This paper | PCR Primer | ATGATGCTTCCATGTTTGTT |
| Sequence-based reagent | NWMN_0306_F | This paper | PCR Primer | AATACACCGGGTAACACAAC |
| Sequence-based reagent | NWMN_0306_R | This Paper | PCR Primer | CGTTTTGTTGAGCTAATTCC |
| Sequence-based reagent | NWMN_0309_F | This paper | PCR Primer | ACCATGCTTAAAGGGATTTT |
| Sequence-based reagent | NWMN_0309_R | This Paper | PCR Primer | TGTCACCTAAGTCAACACCA |
| Sequence-based reagent | NWMN_0407 (lpl4nm) _F | This paper | PCR Primer | CCGTTGGAGATAGGAAGTTA |
| Sequence-based reagent | NWMN_0407 (lpl4nm) _R | This paper | PCR Primer | TTTGTGCTTCTTTTGAACCT |
| Sequence-based reagent | NWMN_0654_F | This paper | PCR Primer | GAAAATGGAAGACTGATTGC |
| Sequence-based reagent | NWMN_0654_R | This paper | PCR Primer | TAATGCATCTGACAAAGTCG |
| Sequence-based reagent | NWMN_0762_F | This paper | PCR Primer | GGTGAAGTTTTGGACGATAA |
| Sequence-based reagent | NWMN_0762_R | This paper | PCR Primer | TTTTCATCTGTCCGACTTTT |
| Sequence-based reagent | NWMN_1101_F | This paper | PCR Primer | TCCACCTATTGGAATTATCG |
| Sequence-based reagent | NWMN_1101_R | This paper | PCR Primer | AGACGTTCAATTTCAGTGCT |
| Sequence-based reagent | NWMN_1192 (pgsA) _F | This paper | PCR Primer | TGGGACGAAGTAATTACAGTT |
| Sequence-based reagent | NWMN_1192 (pgsA) _R | This paper | PCR Primer | ATATCCCCCTTGTATCGTTT |
| Sequence-based reagent | NWMN_1308 (dapD) _F | This paper | PCR Primer | TCTATTCGTGGAGGTACGAT |
| Sequence-based reagent | NWMN_1308 (dapD) _R | This paper | PCR Primer | ATCGTATGTGAGCCATTACC |
| Sequence-based reagent | NWMN_1410_F | This paper | PCR Primer | CGATAAACCTAAACCACTCG |
| Sequence-based reagent | NWMN_1410_R | This paper | PCR Primer | ATAAACAATGCTTGCCAAAT |
| Sequence-based reagent | NWMN_1505_F | This paper | PCR Primer | TGAAGGTGAATTAAGCGATG |
| Sequence-based reagent | NWMN_1505_R | This paper | PCR Primer | TGCTATTCCCAATTTGTTCA |
| Sequence-based reagent | NWMN_1655_F | This paper | PCR Primer | GAATTGTTGCAATTTAATGGT |
| Sequence-based reagent | NWMN_1655_R | This paper | PCR Primer | AACGTAATCATGCTCCATTC |
| Sequence-based reagent | NWMN_1679_F | This paper | PCR Primer | CCATGGGAAAAATTAGACAA |
| Sequence-based reagent | NWMN_1679_R | This paper | PCR Primer | AAATATCGCCTCACCTTTTT |
| Sequence-based reagent | NWMN_1723 (hemY) _F | This paper | PCR Primer | GCCGAATACACATCCATTAT |
| Sequence-based reagent | NWMN_1723 (hemY) _R | This paper | PCR Primer | AACCTTTGTCTCTGCTTCAA |
| Sequence-based reagent | NWMN_1851 (nadC) _F | This paper | PCR Primer | AGCCATTTTAGCACCATAAA |
| Sequence-based reagent | NWMN_1851 (nadC)_R | This paper | PCR Primer | TAGAATCCTGTCCTCCTGAA |
| Sequence-based reagent | NWMN_2057 (mtlF)_F | This paper | PCR Primer | TGTACAACGGTGTTGTTTTG |
| Sequence-based reagent | NWMN_2057 (mtlF)_R | This paper | PCR Primer | CGGTGAATAGTACGAGAGGA |
| Sequence-based reagent | NWMN_2528_F | This paper | PCR Primer | ACTGATGCTTTACCAGAAAC |
| Sequence-based reagent | NWMN_2528_R | This paper | PCR Primer | TCAGCGGTAGTAATAAAGGT |
| Chemical compound, drug | POM-HEX | White et al., 2018 | ||
| Chemical compound, drug | Hemi-HEX | Lin et al., 2020 | ||
| Chemical compound, drug | HEX | Lin et al. 2018 | ||
| Chemical compound, drug | Fluorescent Prosubstrates | White et al., 2018 | ||
| Chemical compound, drug | S-D-lactoylglutathione | Sigma-Aldrich | L7140 | |
| Chemical compound, drug | 4-Nitrophenyl acetate | Sigma-Aldrich | N8130 | |
| Chemical compound, drug | 4-Nitrophenyl butyrate | Sigma-Aldrich | N9876 | |
| Chemical compound, drug | 4-Nitrophenyl trimethyl acetate | Sigma-Aldrich | 135046 | |
| Software, algorithm | WhatsGNU | Moustafa and Planet, 2020 | https://github.com/ahmedmagds/WhatsGNU | |
| Software, algorithm | MUSCLE | Letunic and Bork, 2019 | https://www.ebi.ac.uk/Tools/msa/muscle/ | |
| Software, algorithm | iTOL | Madeira et al., 2019 | https://itol.embl.de/ | |
| Software, algorithm | HKL-3000 | Minor et al., 2006 | https://hkl-xray.com/hkl-3000 | |
| Software, algorithm | PHASER | McCoy et al., 2007 | ||
| Software, algorithm | COOT | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | |
| Software, algorithm | PHENIX | Adams et al., 2010 | http://www.phenix-online.org/ | |
| Software, algorithm | ChemDraw3D | https://www.cambridgesoft.com/Ensemble_for_Chemistry/details/Default.aspx?fid=13&pid=668 | ||
| Software, algorithm | AutoDock Tools 1.5.7 | Morris et al., 2009 | http://autodock.scripps.edu/resources/adt | |
| Software, algorithm | AutoDock Vina | Trott and Olson, 2010 | http://vina.scripps.edu/ | |
| Software, algorithm | GraphPad Prism | https://www.graphpad.com/scientific-software/prism/ | ||
| Other | CellASIC ONIX2 microfluidic plate | EMD-Millipore | B04A-03-5PK |
Additional files
-
Supplementary file 1
Primers used during this study.
- https://cdn.elifesciences.org/articles/66657/elife-66657-supp1-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66657/elife-66657-transrepform-v3.docx