Mitochondrial genome sequencing of marine leukaemias reveals cancer contagion between clam species in the Seas of Southern Europe
Figures
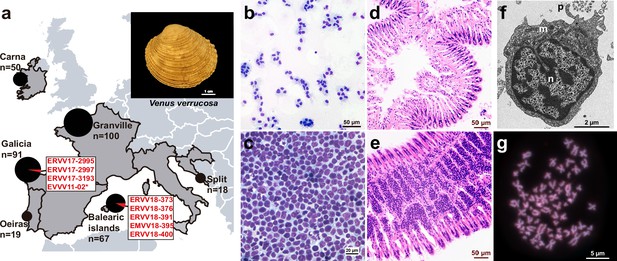
Geographical location of warty venus (V. verrucosa) specimens and diagnosis of hemic neoplasia.
(a) Locations of V. verrucosa clams collected for this study and specimens diagnosed with hemic neoplasia. Size of the pie charts correlates with the number of samples collected (number of samples ‘n’ is shown together with each pie chart). Pie charts show the proportion of samples with hemic neoplasia (black, no neoplastic specimens; red, neoplastic specimens). Codes of neoplastic samples are shown. Top-right corner shows a representative specimen of the species V. verrucosa. (b) Cytological examination of haemolymph smear (Hemacolor stain) from a healthy (N0) specimen, ERVV17-2963, shows normal haemocytes. (c) Haemolymph smear of a V. verrucosa specimen with high-grade (N3 stage) hemic neoplasia, ERVV17-3193, shows neoplastic cells that replaced normal haemocytes. (d) Detail of haematoxylin and eosin-stained of histological section from the gills of the healthy (N0) specimen ERVV17-2990. (e) Same for ERVV17-2995, a specimen infected with a high-grade (N3 stage) hemic neoplasia, showing neoplastic cells infiltrating the gills. (f) Transmission electron microscopy analysis of a V. verrucosa hemic neoplasia tumour cell shows a round shape, pseudopodia ‘p’, pleomorphic nucleus ‘n’ with scattered heterochromatin, and mitochondria ‘m’. (g) Metaphase chromosomes from a neoplastic cell found in the gills of the V. verrucosa specimen EVVV11-02, showing abnormal chromosome number (>19 pairs) and abnormal chromosome morphology. Chromosomes stained with 4′,6-DiAmidino-2-PhenylIndole (DAPI) and Propidium Iodide (PI).
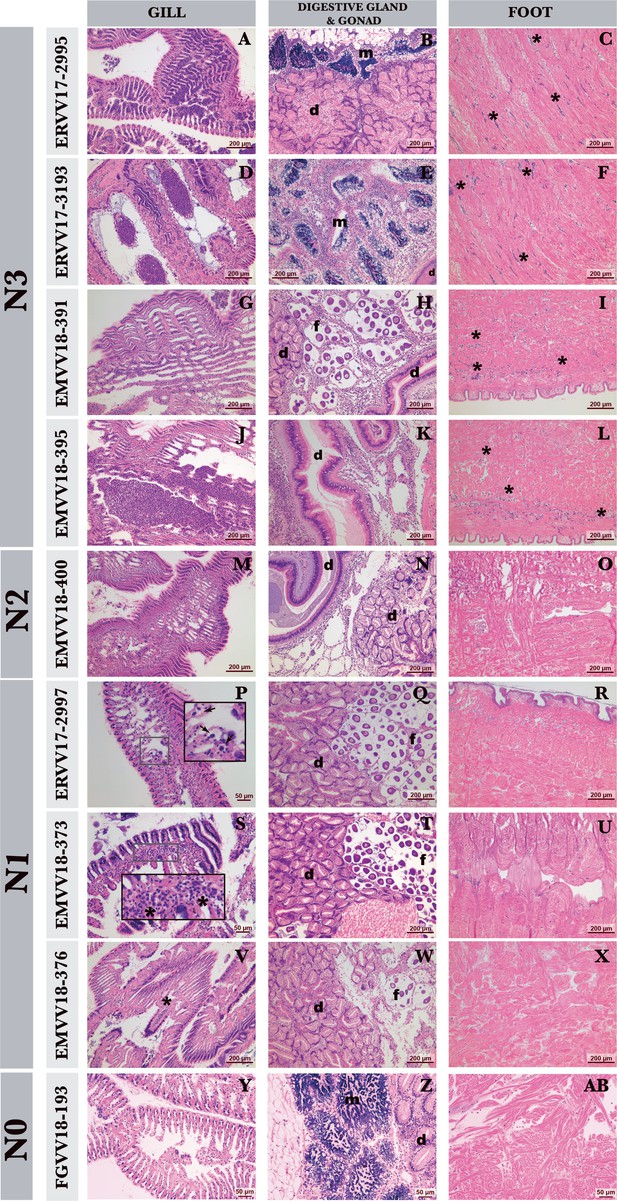
Histological diagnosis of hemic neoplasia in warty venus (V. verrucosa) specimens.
Haematoxylin and eosin-stained photomicrographs of gill, digestive (d), gonad (male [m] and female [f]), and foot of warty venus (V. verrucosa) specimens diagnosed with different stages of hemic neoplasia: high (N3), medium (N2), light (N1), and healthy (N0). In the N3 stage, neoplastic cells infiltrate the connective tissue and vessels of different organs (A–L), and show low infiltration of foot (C, F, I, L). In N2 stage, cell groups are observed in different organs such as gills (M) or gonad and digestive gland (N) and are not detected in the foot (O). In N1 stage, groups of neoplastic or isolated cells are detected in gill sinuses (P, S, V) and not found in either gonad and digestive gland (Q, T, W) or foot (R, U, X). N0 stage is completely devoid of any trace of hemic neoplasia at either gill, digestive gland, and gonad and foot (Y, Z, AB). Arrows show isolated cells. Asterisks show groups of neoplastic cells. Scale bar 200 µm.
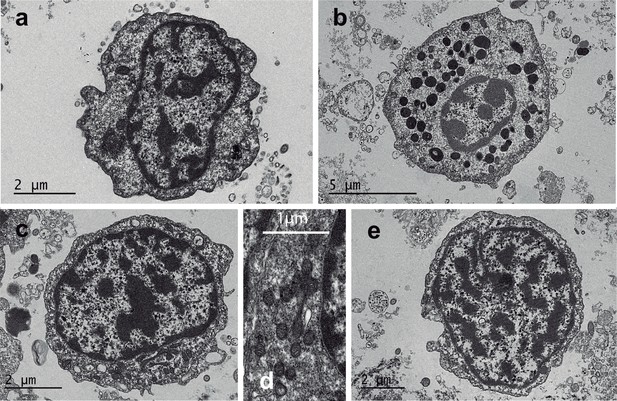
Transmission electron microscopy (TEM) of healthy and neoplastic V. verrucosa specimens.
TEM cellular ultrastructure of healthy warty venus haemocytes, hyalinocyte (a), and granulocyte (b) and neoplastic cells (c, d, e). (d) shows mitochondria of neoplastic cells in detail. Processed samples: ERVV17-2992 (a), ERVV17-2993 (b), ERVV17-2995 (c, , ), and ERVV17-3193 (d, e).
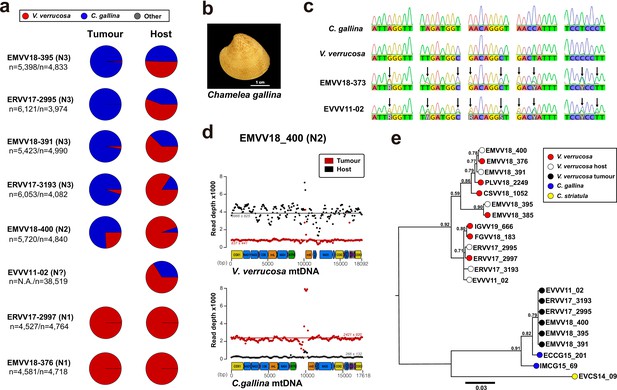
Mitochondrial DNA sequencing and phylogenetic analyses reveal cancer contagion between warty venus (V. verrucosa) and striped venus (C. gallina) clam species.
(a) In eight warty venus specimens sequenced with Illumina paired-ends, the pie charts show the proportion of reads mapping Cox1 reference sequences from 137 different Verenidae species, including V. verrucosa (red), C. gallina (blue), and the remaining species (grey). Two different tissues were sequenced: the tumour tissue (left pie chart), typically haemolymph, and the host/matched-normal tissue (right pie chart), typically foot. Note that for specimen EVVV11-02 only the host/matched-normal tissue (gills) was available. ‘n’ denotes the total number of reads mapping the Cox1 reference for the tumour tissue (left), and the host tissue (right). (b) Representative specimen of the species C. gallina. (c) Capillary sequencing electropherograms of mitochondrial Cox1 gene fragments from two neoplastic V. verrucosa specimens (EMVV18-373 and EVVV11-02) and two healthy reference specimens from V. verrucosa and C. gallina. The results show overlapping peaks (arrows) in the sequenced tissues from the neoplastic animals, which suggest coexistence of mitochondrial DNA (mtDNA) haplotypes from two clam species. (d) In V. verrucosa neoplastic (N2-stage) specimen EMVV18-400, mtDNA read depth shows different proportion of warty venus and striped venus mtDNA haplotypes in the tumour tissue (haemolymph) and the matched-normal tissue (foot). (e) Molecular phylogeny using Bayesian inference inferred on the alignment of all mitochondria coding genes and rRNA gene sequences (15 loci) that includes six neoplastic V. verrucosa specimens with evidence of cancer contagion from C. gallina. Bootstrap values are shown above the branches.
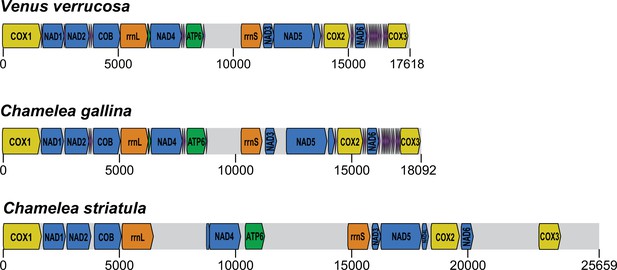
Draft reference mitochondrial DNA (mtDNA) genome assemblies reconstructed for V. verrucosa, C. gallina, and C. striatula.
Preliminary ‘reference’ mtDNA genome assembly obtained from paired-end sequencing data, together with gene annotation.
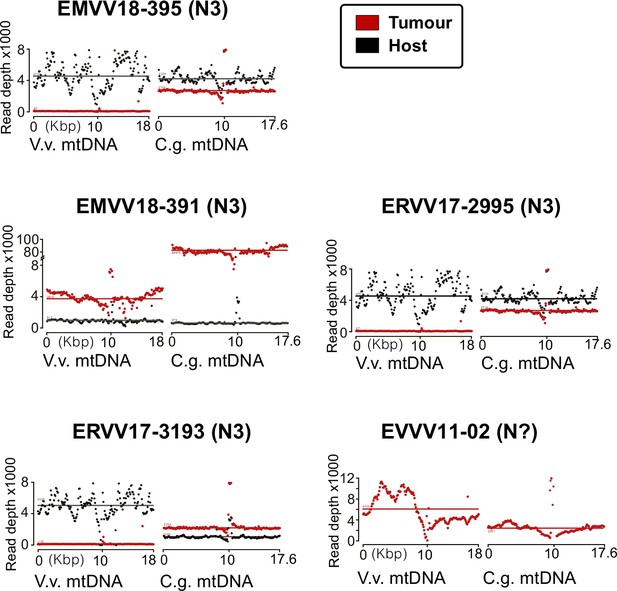
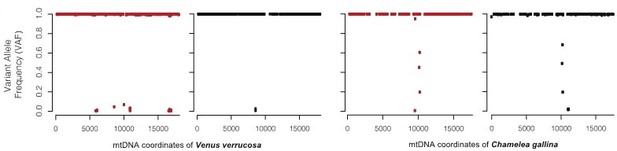
Read depth analysis of the mitochondrial DNA (mtDNA) confirms coexistence of two different clam species haplotypes.
mtDNA read depth shows different proportion of warty venus and striped venus mtDNA haplotypes in the tumour tissue (haemolymph) and the matched-normal tissue (foot) in the remaining five tumours analysed.
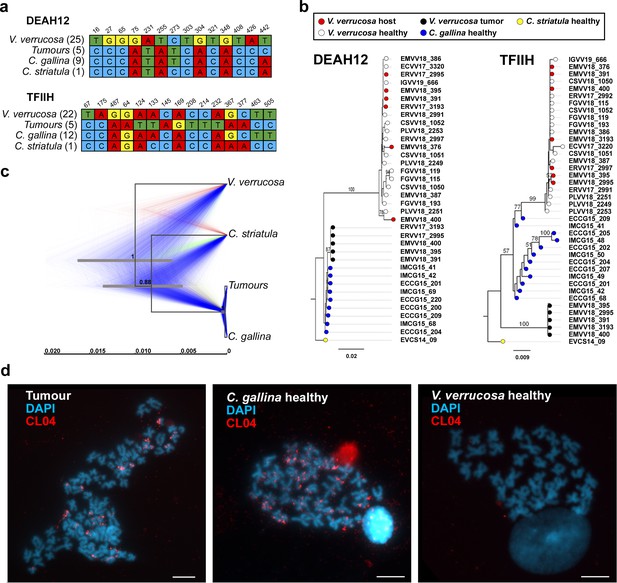
Nuclear DNA sequencing and phylogenetic analyses confirm a single cancer lineage spreading in populations of the warty venus (V. verrucosa) that originated in the striped venus (C. gallina).
(a) Single-nucleotide variants discriminating between V. verrucosa tumours and the three canonical species (V. verrucosa, C. gallina, and C. striatula) along a 441- and a 559-bp long fragments of nuclear genes DEAH12 and TFIIH, respectively. (b) Maximum Likelihood molecular phylogenies based on the two fragments of the nuclear DNA markers DEAH12 and TFIIH. Bootstrap support values (500 replicates) from Maximum Likelihood analyses above 50 are shown on the corresponding branches. (c) Multispecies coalescent (MSC) tree of V. verrucosa, their tumours and Chamelea sp. based on the entire mitochondrial DNA (mtDNA) and the two nuclear markers, DEAH12 and TFIIH. A maximum clade credibility (MCC) tree is shown, with posterior probabilities below the branches, and 95% highest probability density (HPD) intervals of node heights as grey bars. The trees distribution shown includes 1000 trees and represents the range of alternative topologies, in which blue is the most common set of topologies, red the second most common one, and green the remaining. (d) Fluorescence in situ hybridization (FISH) to specifically detect the satellite DNA CL4 in one V. verrucosa tumour and healthy specimens from the species C. gallina and V. verrucosa shows probes accumulate in heterochromatic regions, mainly in subcentromeric and subtelomeric positions, from the chromosomes of the tumour and the healthy C. gallina tested but not in healthy V. verrucosa.
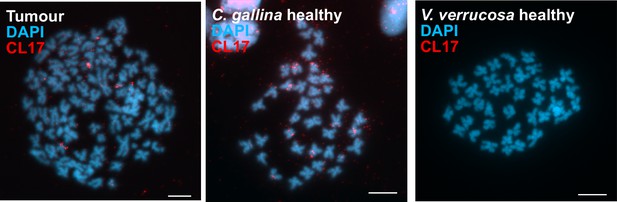
CL17 satellite DNA supports tumour transmission from C. gallina.
Fluorescence in situ hybridization (FISH) to specifically detect the satellite DNA CL17 in one V. verrucosa tumour and healthy specimens from the species C. gallina and V. verrucosa shows probes accumulate in heterochromatic regions, mainly in subcentromeric and subtelomeric positions, from the chromosomes of the tumour and the healthy C. gallina tested but not in healthy V. verrucosa.
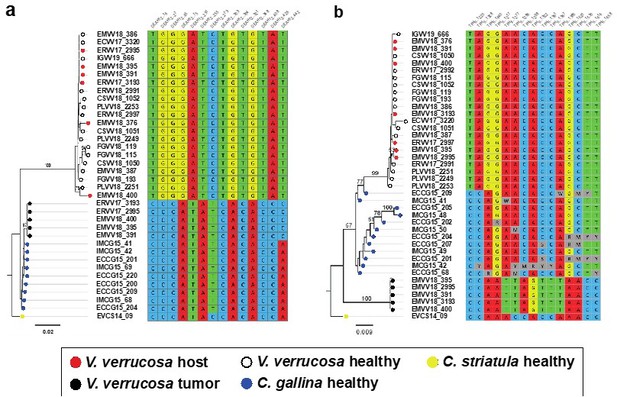
Molecular phylogenies based on the two selected nuclear markers.
(a) DEAH12 gene and (b) TFIIH gene, and diagnostic loci discriminating among species and tumour. Bootstrap support values (500 replicates) from ML analyses above 50 are shown above the corresponding branches. Note all diagnostic nucleotides are identical between tumours (black dots).
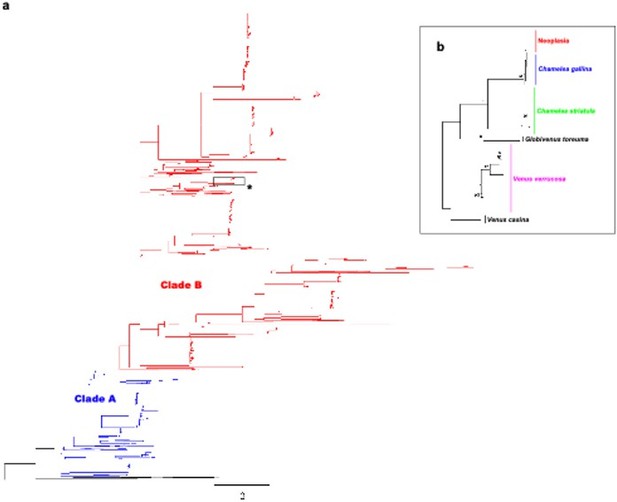
Molecular phylogenetic analysis by Maximum Likelihood method across Venerids.
a) Maximum likelihood (ML) molecular phylogenetic tree of Veneridae based on a 323 bp alignment including 2045 non-redundant COI gene sequences recovered from Genbank/Boldsystems and all sequences derived from this work. Highlighted clades A and B according to Chen et al. 2011. Bootstrap support values (500 replicates) from ML analyses above 50 are shown above the corresponding branches. b) Zoom-in to the cluster of the venerid harbouring the neoplastic haplotype, both Chamelea species and Venus verrucosa in the previous tree (asterisk).
Tables
Clam specimens and tissues sequenced with Illumina paired-ends.
Sixteen specimens (eight neoplastic and eight non-neoplastic) from three different clam species (V. verrucosa, C. gallina, and C. striatula) were sequenced with Illumina paired-ends. Columns 5 and 6 show the number of reads generated for the host tissue (when neoplastic, matched-normal tissue was foot) and the tumoural haemolymph, respectively. (*) denotes the only available tissue from this neoplastic animal, collected in 2011, were gills. (#) denotes hemic neoplasia stage was not determined because cytohistological examination was not possible in this individual, which was diagnosed by cytogenetics.
| Clam species | Specimen origin | Specimen code | Diagnosis | Foot reads | Haemolymph reads |
|---|---|---|---|---|---|
| V. verrucosa | Galicia, Spain | ERVV17-2995 | N3 | 833 M | 919 M |
| V. verrucosa | Galicia, Spain | ERVV17-2997 | N1 | 766 M | 598 M |
| V. verrucosa | Galicia, Spain | ERVV17-3193 | N3 | 739 M | 850 M |
| V. verrucosa | Balearic Islands, Spain | EMVV18-376 | N1 | 784 M | 849 M |
| V. verrucosa | Balearic Islands, Spain | EMVV18-391 | N3 | 617 M | 623 M |
| V. verrucosa | Balearic Islands, Spain | EMVV18-395 | N3 | 697 M | 679 M |
| V. verrucosa | Balearic Islands, Spain | EMVV18-400 | N1 | 782 M | 1133 M |
| V. verrucosa | Galicia, Spain | EVVV11-02 | N# | 743 M* | –* |
| V. verrucosa | Split, Croatia | CSVV18-1052 | Healthy | 161 M | – |
| V. verrucosa | Balearic Islands, Spain | EMVV18-385 | Healthy | 143 M | – |
| V. verrucosa | Granville, France | FGVV18-183 | Healthy | 752 M | – |
| V. verrucosa | Carna, Ireland | IGVV19-666 | Healthy | 155 M | – |
| V. verrucosa | Oeiras, Portugal | PLVV18-2249 | Healthy | 163 M | – |
| C. gallina | S.Benedetto, Italy | IMCG15-69 | Healthy | 147 M | – |
| C. gallina | Cadiz, Spain | ECCG15-201 | Healthy | 752 M | – |
| C. striatula | Galicia, Spain | EVCS14-09 | Healthy | 706 M | – |
Additional files
-
Supplementary file 1
Sampling data of 570 specimens analysed in this study.
- https://cdn.elifesciences.org/articles/66946/elife-66946-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66946/elife-66946-transrepform1-v1.docx