Y chromosome functions in mammalian spermatogenesis
Figures
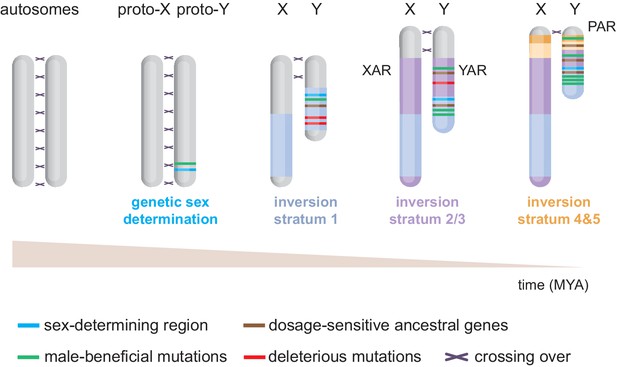
Evolution of the eutherian sex chromosomes.
The X and Y chromosomes evolved from a pair of autosomes. First, the testis-determining gene Sry evolved on the proto-Y. Then, successive stratification events occurred, wherein X-Y meiotic recombination arrests along specific regions of the Y, likely due to inversions. The first stratum (i.e. discrete non-recombining region) includes Sry and originated in the last common therian ancestor ~166 million years ago (MYA). After the split from marsupials, the X/Y-added regions (XAR/YAR) fused from an autosome to the sex chromosomes in the eutherian ancestor. A second stratification event occurred independently in both marsupial and eutherian ancestors ~ 97–117 MYA. Concomitantly, in eutherians, a third stratum was formed encompassing the YAR. A fourth and fifth stratum evolved in the ancestor of old-world monkeys ~ 25–44 MYA. Recombination now only occurs within the pseudoautosomal region (PAR).
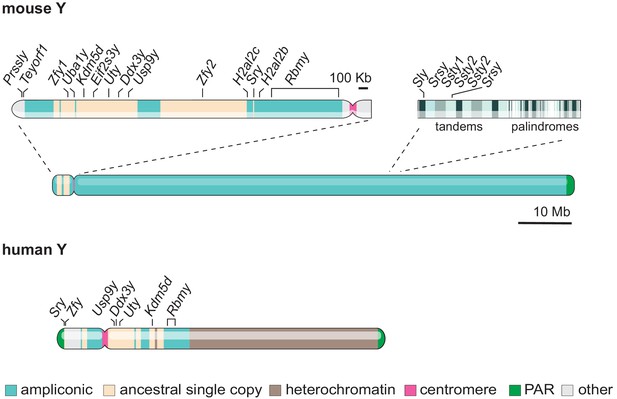
Gene content and structure of the mouse and human Y chromosome.
The short arm of the mouse Y chromosome (Yp) has retained ancestral non-ampliconic genes. The mouse Y chromosome long arm (Yq) contains alternating tandem and palindromic repeats of ampliconic core blocks throughout. Ampliconic core blocks encompass rodent-specific gene clusters. The human Y shares seven ancestral protein-coding genes with the murine Y (labelled). Whereas the human Y chromosome is roughly 60 % heterochromatic, the mouse Y chromosome is 99.9% euchromatic (Skaletsky et al., 2003; Soh et al., 2014). The 10 Mb scale bar applies to both mouse and human Y chromosomes.
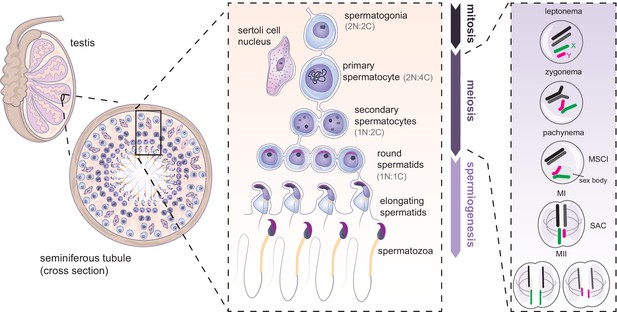
Mammalian spermatogenesis in seminiferous tubules of the testis.
Spermatogenesis occurs in three main phases: mitosis, meiosis and spermiogenesis. First, undifferentiated spermatogonial cells undergo multiple mitotic divisions. Subsequently, germ cells commit to meiosis, undergoing meiotic DNA replication (becoming 2 N:4 C) synapsis and recombination between homologues (shown in black and dark gray), and silencing of the X (green) and Y (magenta) chromosomes. Spermatocytes then undergo two rounds of cell division (becoming 1N:1C). The resulting round spermatids later undergo spermiogenesis, further specialising into mature spermatozoa. N = number of chromosomes, C = number of chromatids. SAC: spindle assembly checkpoint.
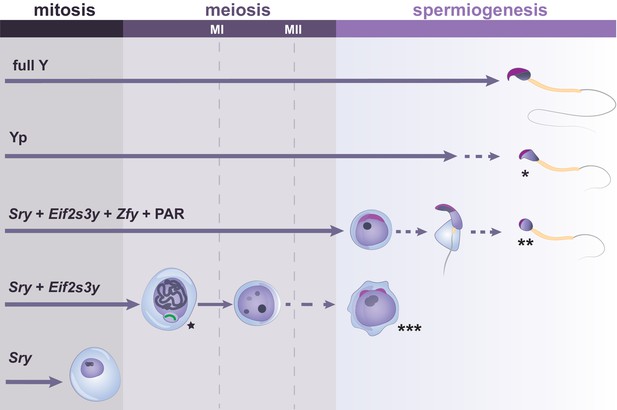
Y-linked gene complements and developmental progression through spermatogenesis.
Specific Y genes control the developmental progress of germ cells in spermatogenesis. The presence of the Y short arm (Yp) allows formation of sperm which have abnormal heads, exhibiting reduced curvature and poor chromatin compaction (indicated by *). The presence of Sry, Eif2s3y, Zfy genes and a PAR allows progression to the round spermatid stage, but spermatid elongation is abnormal and delayed, with rare production of sperm with poorly structured heads (indicated by **). Germ cells expressing only Sry and Eif2s3y have incomplete MSCI (indicated by the star next to the X chromosome shown in green). Moreover, most germ cells of this model arrest before the second meiotic division (MII), with occasional progression to form mostly diploid and abnormally shaped round spermatids (indicated by ***). When only Sry is expressed, spermatogonial cells fail to proliferate and to enter meiosis. Arrowed lines show germ cell progression through spermatogenesis, with dotted lines indicating abnormal cell differentiation at a low frequency.
Tables
Dosage compensation of the sex chromosomes.
Without dosage compensation, the transcriptional output of each cell is 0.5 for individual chromosomes, and one for chromosome pairs. X-chromosome upregulation (XCU) doubles each X-chromosome’s output (shown in red). X-chromosome inactivation (XCI) silences output of one X in females (shown in blue).
| No compensation | XCU | XCI | |
|---|---|---|---|
| female | AA = 1XX = 1 | AA = 1 = 2 | AA = 1 = 1 |
| male | AA = 1XY = 0.5 | AA = 1Y = 1 | AA = 1Y = 1 |
Summary of mouse Y chromosome genes and their known functions.
A gene is classified as ‘ancestral’ if it is predicted to have been present in the last common eutherian ancestor. The list of animals where each gene is conserved only includes organisms where a high-quality Y chromosome sequence is available, and the presented list is not exhaustive. *Rbmy was initially thought to be important in spermatid morphogenesis, but this was later questioned (Szot et al., 2003).
| Y gene | Sequence class | X-homologue | Y copy number | Conserved in | Reported functions in the testis | Key gene ontology process | Expression pattern |
|---|---|---|---|---|---|---|---|
| Sry | Ancestral | Sox3 | 1 | Opossum, bull, rat, mouse, marmoset, rhesus, chimp, human | Testis determination | DNA-binding transcription factor activity | Somatic cells in genital ridge, germ cell specific in adult |
| Rbmy array | Ancestral | Rbmx | ~30 | Opossum, bull, rat, mouse, marmoset, rhesus, chimp, human | Unknown * | RNA splicing | Testis biased |
| Zfy1,Zfy2 | Ancestral | Zfx | 2 | Bull, rat, mouse, marmoset, rhesus, chimp, human | Apoptotic elimination of univalent spermatocytes at metaphase I, meiosis II completion, MSCI initiation and maintenance, spermatid head and tail morphogenesis | Transcription activator | Spermatogenic cells specific |
| Kdm5d | Ancestral | Kdm5c | 1 | Opossum, rat, mouse, marmoset, rhesus, chimp, human | Unknown | Histone demethylase that specifically demethylates 'Lys-4' of histone H3 | Ubiquitous |
| Uty | Ancestral | Utx | 1 | Bull, rat, mouse, marmoset, rhesus, chimp, human | Unknown | Histone demethylase activity (H3-K27 specific) | Ubiquitous |
| Ddx3y | ancestral | Ddx3x | 1 | Bull, rat, mouse, marmoset, rhesus, chimp, human | Unknown | ATP-dependent RNA helicase | Ubiquitous |
| Usp9y | Ancestral | Usp9x | 1 | Bull, rat, mouse, marmoset, rhesus, chimp, human | Unknown | Ubiquitination regulator, peptidase C19 | Testis biased |
| Uba1y | Ancestral | Uba1x | 1 | Opossum, bull, rat, mouse | Unknown | U1 ubiquitin activator | Testis biased, mostly spermatogonia and round spermatid |
| Eif2s3y | Ancestral | Eif2s3x | 1 | Bull, rat, mouse | Spermatogonial proliferation and differentiation | Translation initiation | Ubiquitous |
| Teyorf | Acquired | – | 1 | Mouse | Unknown | Claudin transmembrane | Testis specific |
| Prslly | Acquired | – | 1 | Mouse | Unknown | Serine-type endopeptidase activity | Testis specific |
| H2al2b, H2al2c | Acquired | H2al1 | 2 | Mouse | Unknown | DNA packaging, pericentric heterochromatin regulation | Testis specific, expressed from round spermatid stage |
| Sly | Acquired | Slx,Slxl1 | 126 | Mouse | Interacts with SSTY to recruit SMRT/N-Cor in turn mediating spermatid-specific gene expression | Chromatin remodelling | Testis biased |
| Srsy | Acquired | Srsx | 197 | Mouse | Unknown | Unknown | Testis biased |
| Ssty1&2 | Acquired | Sstx | 85&221 | Mouse | H3K4me3-reader at the promoter of spermatid-specific genes, recruits SLY and SLX/SLX1 | Methylated histone binding | Testis biased |
| Rbm31y | Acquired | Rbm31x | 2 | Mouse | Unknown | RNA binding | Testis biased |





