Intraocular dendritic cells characterize HLA-B27-associated acute anterior uveitis
Figures
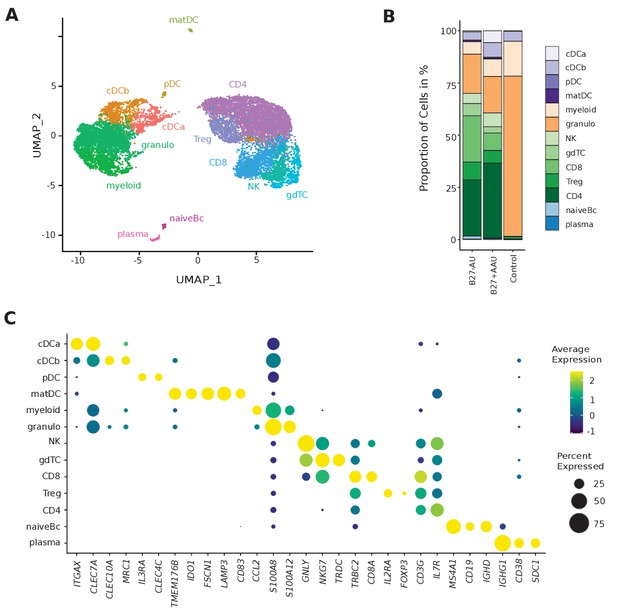
Single-cell transcriptomics reconstructs leukocyte subsets infiltrating the eye.
(A) Uniform manifold approximation and projection (UMAP) projection of seven pooled samples (control n = 1; B27-AU n = 2; B27+ AAU n = 4). The single-cell (sc) transcriptomes were manually annotated to cell types based on marker gene expression and distinguished in 13 cell clusters (color-coded; each dot represents one cell). (B) The mean proportion of cells (%) in each cluster per group is depicted in a stacked bar plot. (C) Dot plot of selected marker genes grouped by cluster. The average gene expression level is color-coded, and the circle size represents the percentage of cells expressing the gene. Threshold was set to a minimum of 10% of cells in the cluster expressing the gene. DC: dendritic cell, pDC: plasmacytoid DC, matDC: mature DC; granulo: granulocytes, NK cells: natural killer cells, gdTC: γδ T cells, Treg cells: regulatory T cells, Bc: B cells.
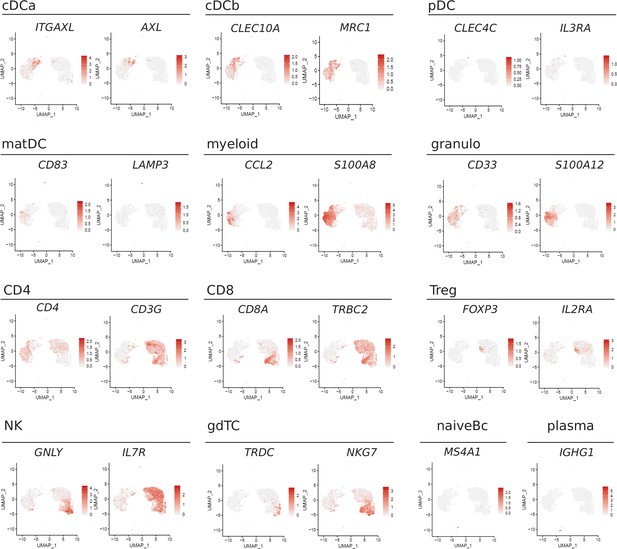
Feature plots of lineage marker genes.
Expression of lineage markers ITGAX, AXL, CLEC10A, MRC1, CLEC4C, IL3RA, CD83, LAMP3, CCL2, S100A8, CD33, S100A12, CD4, CD3G, CD8A, TRBC2, FOXP3, IL2RA, GNLY, IL7R, TRDC, NKG7, MS4A1, and IGHG1 is shown as feature plots. Each dot represents one cell.
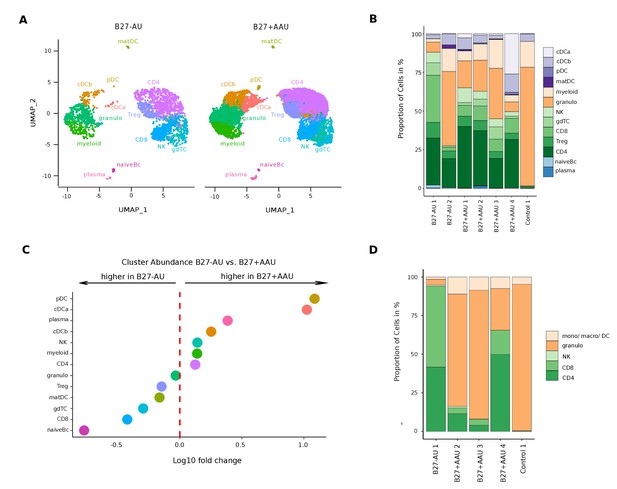
A unique intraocular leukocyte pattern characterizes individual uveitis causes.
(A) UMAP projection of pooled B27-AU (n = 2) versus pooled B27+ AAU (n = 4) samples. The single-cell (sc) transcriptomes were manually annotated to cell types based on marker gene expression and distinguished in 13 cell clusters (color-coded; each dot represents one cell). (B) The proportion of cells (%) in each cluster is depicted in a stacked bar plot for individual samples. (C) Dot plot of cluster abundance of B27-AU versus B27+ AAU. The x axis represents the decadic logarithm of fold change of proportional cluster size. (D) Box plots of proportion of cells (%) of cDCa and pDC from B27-AU and B27+ AAU. The boxes show the median, and the lower and upper quartile. Whiskers include 1.5 times the interquartile range of the box. The overlaid dots represent individual observations. (E) Leukocytes of aqueous humor (AqH) samples were analyzed according to their frequency (%) of granulocytes, monocytes/macrophages/DCs, CD4+ and CD8+ T cells, and NK cells. The proportion of each cell population identified via flow cytometry is depicted in a stacked bar plot. DC: dendritic cell, pDC: plasmacytoid DC, matDC: mature DC; granulo: granulocytes, NK cells: natural killer cells, gdTC: γδ T cells, Treg cells: regulatory T cells, Bc: B cells, mono: monocyte, macro: macrophage.
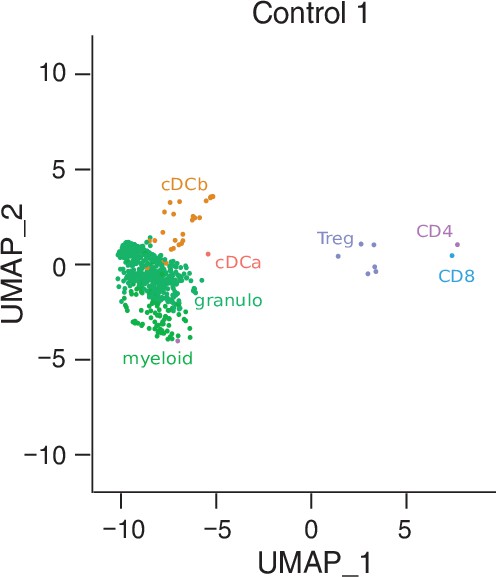
Individualized scRNA-seq results of anterior chamber-derived leukocytes.
UMAP projection as in Figure 2A of only the endophthalmitis control patient. The single-cell (sc) transcriptomes were manually annotated to cell types based on marker gene expression and distinguished in 13 cell clusters (color-coded; each dot represents one cell).
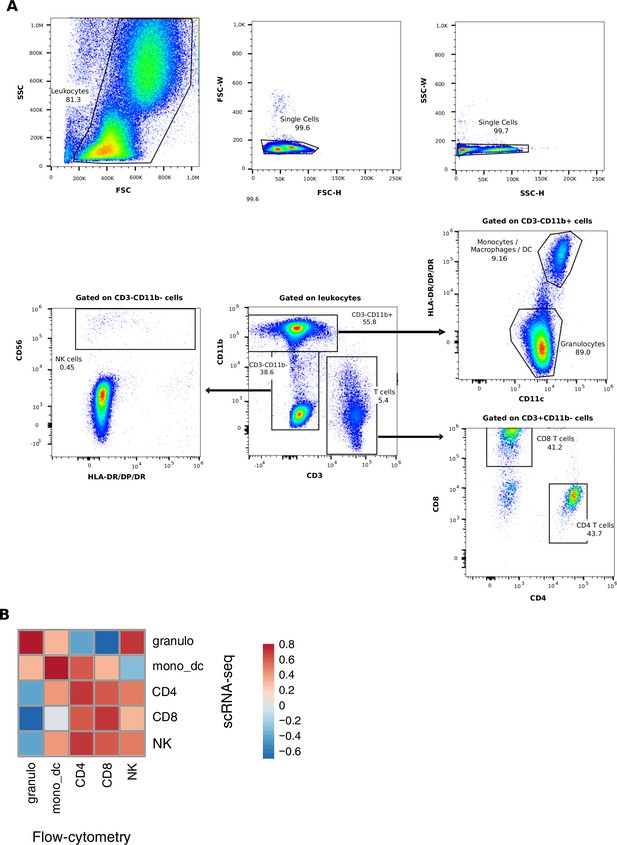
Flow cytometric analysis of aqueous humor samples.
(A) Leukocytes in side scatter (SSC)/forward scatter (FSC) scatter were identified, doublets were excluded, and leukocytes were gated on CD3-CD11b+ myeloid cells, CD3+ CD11b- T cells, and CD3-CD11b- cells. Myeloid cells were further classified into CD11c+ HLA-DR- granulocytes and CD11c+ HLA-DR+ monocytes/macrophages/dendritic cells (DCs). T cells were further subdivided into CD4+- and CD8+-expressing cells. CD3-CD11b- cells were analyzed according to their frequency of CD56+ natural killer (NK) cells. Representative analysis is shown. (B) Correlation plot (Spearman’s correlation coefficient) between flow proportions (columns) and single-cell RNA-sequencing (scRNA-seq) (rows) proportions is shown as a heatmap, with high correlation coefficients shown in red.
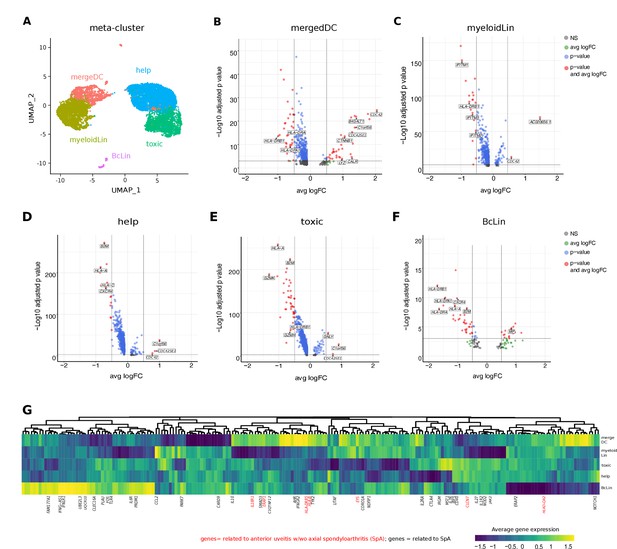
Intraocular leukocytes express a subtype-specific transcriptional phenotype.
(A) UMAP projection of pooled B27-AU (n = 2) versus pooled B27+ AAU (n = 4) samples. The single-cell (sc) transcriptomes were manually annotated to cell types based on marker gene expression and distinguished in five meta-clusters (color-coded; each dot represents one cell). Differentially expressed (DE) genes of B27-AU vs B27+ AAU of each meta-cluster (B) mergeDC (matDC, pDC, DCa, cDCb), (C) myeloidLin (myeloid, granulo), (D) help (Treg, CD4), (E) toxic (CD8, NK, gdTC), (F) and BcLin (naive Bc, plasma) are depicted as volcano plots. The threshold for average natural logarithmic fold change (avg logFC) was set to 0.5 and for adjusted p-value, to 0.001. Selected genes are labeled. (G) Heatmap showing differences in average gene expression (B27+ AAU minus B27-AU) of genome-wide association study (GWAS) risk genes (Supplementary file 1g). Data were scaled column wise. Columns were clustered using euclidean distance measure and complete linkage. Yellow color indicates a higher expression in B27+ AAU samples, and blue color indicates a higher expression in B27-AU samples. Risk genes for anterior uveitis and for spondyloarthropathies (SpA) are labeled in red and black, respectively. DC: dendritic cell, pDC: plasmacytoid DC, matDC: mature DC; granulo: granulocytes, NK cells: natural killer cells, gdTC: γδ T cells, Treg cells: regulatory T cells, Bc: B cells, BcLin, B-cell lineage.
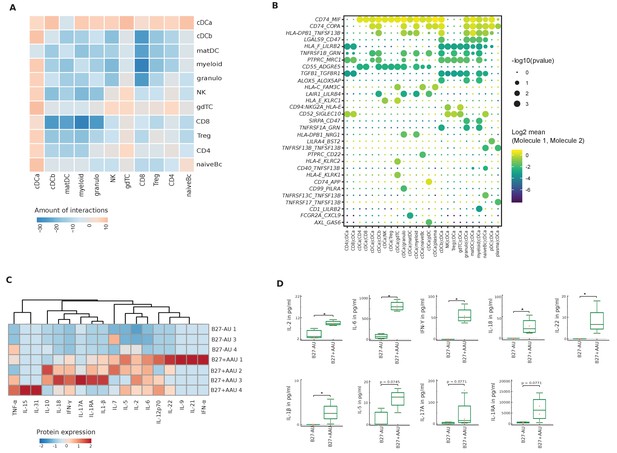
Altered local inter-cellular signaling in uveitis.
(A) The total count of receptor-ligand interactions between cell clusters of B27-AU and B27+ AAU single-cell (sc) transcriptomes was obtained with CellPhoneDBv2.0 (see Figure 4—figure supplement 1 for separate analyses). The heatmap shows the differences between B27+ AAU and B27-AU (amount of predicted interactions of all cell types, excluding plasma and plasmacytoid DC (pDC) due to few cells (<10) of B27+ AAU minus those of B27-AU). (B) Overview of all ligand–receptor interactions of the cDCa cluster with at least one significant interaction. Circle size indicates the p-values. The means of the average expression level of interacting molecule 1 in cluster 1 and interacting molecule 2 in cluster 2 are color-coded. (C) Heatmap showing aqueous humor (AqH) cytokine level of B27-AU (n = 3) and B27+ AAU (n = 4) patients (see Supplementary file 1h showing single values). Data were scaled column wise. Columns were clustered using euclidean distance measure and complete linkage. (D) Box plots of interleukin (IL)-2, IL-6, interferon (IFN)-γ, IL-18, IL-22, IL-1β, IL-5, IL-17A, and IL-1 receptor antagonist (IL-1RA) (pg/ml) in the AqH of patients with B27-AU and B27+ AAU. Dots represent individual data. Mann-Whitney U-test (*p<0.05).
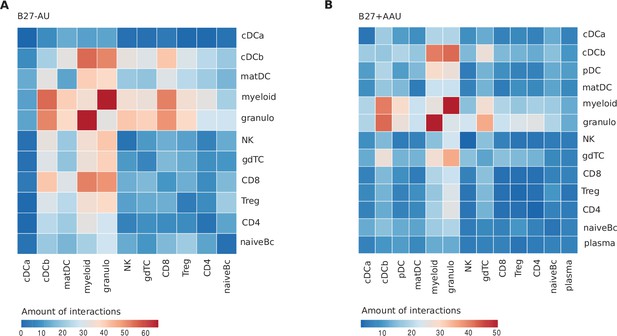
Detailed results of the interactome prediction analysis.
Predicted receptor-ligand interactions between cell clusters in (A) B27-AU and (B) B27+ AAU were obtained with CellPhoneDBv2.0. The legend represents the amount of interactions.
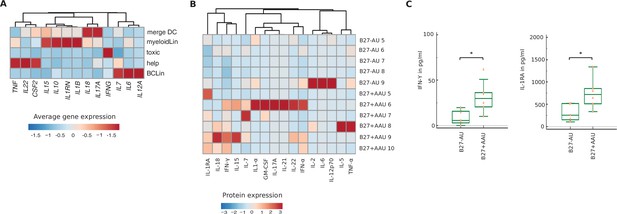
Cytokine expression in meta-cluster and cytokine serum level.
(A) Heatmap showing gene expression of cytokines that were used for luminex analysis within each ´meta-cluster´. Data were scaled column wise. Columns were clustered using euclidean distance measure and complete linkage. (B) Heatmap showing the serum cytokine level of B27-AU (n = 5) and B27+ AAU (n = 6) patients (see Supplementary file 1i showing single values). Data were scaled row wise. Columns were clustered hierarchically using euclidean distance measure and complete linkage. (C) Box plots of interferon (IFN)-γ and interleukin-1 receptor antagonist (IL-1RA) (pg/ml) in the sera of patients with B27-AU and B27+ AAU. Dots represent individual data. Mann-Whitney U-test (*p<0.05).
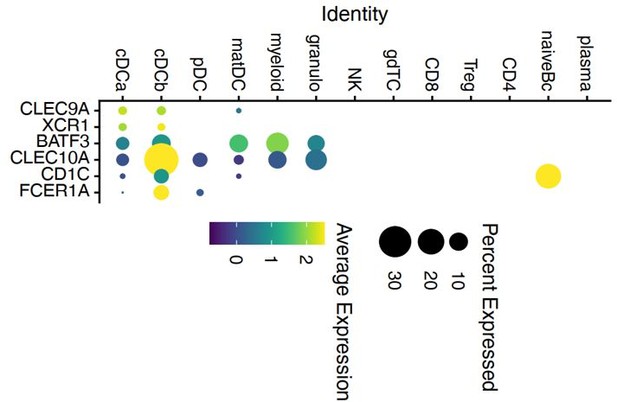
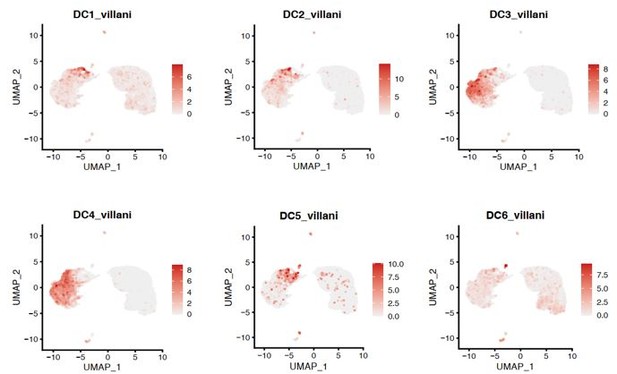
Dot plot of selected marker genes for DC defined in a previous study of Heming et al.
2020 grouped by cluster. The average gene expression level is color-coded and the circle-size represents the percentage of cells expressing the gene. Threshold was set to a minimum of 10% of cells in the cluster expressing the gene. DC: dendritic cell, pDC: plasmacytoid cell, matDC: mature DC; granulo: granulocytes, NK: natural killer cells, gdTC: γδ T cells, Treg: regulatory T cells, Bc: B cells.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Biological sample (Homo sapiens) | Aqueous humor (AqH) | Department of Ophthalmology at St Franziskus Hospital, Münster, Germany | Deidentified | |
| Biological sample (H. sapiens) | Serum | Department of Ophthalmology at St Franziskus Hospital, Münster, Germany | Deidentified | |
| Antibody | Anti-CD3 (mouse monoclonal;OKT3; PerCP-Cy5.5) | Biolegend | Cat# 317336; RRID:AB_2561628 | Dilution (1:20) |
| Antibody | Anti-CD4 (mouse monoclonal; OKT4;BV510) | Biolegend | Cat# 317444; RRID:AB_2561866 | Dilution (1:20) |
| Antibody | Anti-CD8a (mouse monoclonal; SK1; APC) | Biolegend | Cat# 344722; RRID:AB_2075388 | Dilution (1:20) |
| Antibody | Anti-CD11b (rat monoclonal; M1/70; FITC) | Biolegend | Cat# 101205; RRID:AB_312788 | Dilution (1:200) |
| Antibody | Anti-CD11c (mouse monoclonal; 3.9; Pacific Blue) | Biolegend | Cat# 301625; RRID:AB_10662901 | Dilution (1:20) |
| Antibody | Anti-CD56 (mouse monoclonal; N901; PC7) | Beckman Coulter | Cat# A21692; RRID:AB_2892144 | Dilution (1:100) |
| Antibody | Anti-HLA-DR (mouse monoclonal; Immu-357;ECD) | Beckman Coulter | Cat# IM3636;RRID:AB_10643231 | Dilution (1:100) |
| Antibody | FcR-blocking reagent, human | Miltenyi | Cat# 130-059-901; RRID:AB_2892112 | 20 µl/Test |
| Commercial assay or kit | ProcartaPlex Human Cytokine Panel 1B (25 plex) 96 tests Kit | Thermo Fisher Scientific | Cat# PX250-12166-901; RRID:AB_2576119 | Luminex analysis |
| Commercial assay or kit | Chromium Single Cell 3' Library & Gel Bead Kit v2 and v3 | 10x Genomics | Cat# PN-120237Cat# PN-1000075 | RNA-seq analysis |
| Commercial assay or kit | AMPure XP beads | Beckman Coulter | Cat# A63881 | RNA-seq analysis |
| Commercial assay or kit | NextSeq 500/550 High Output Kit v2.5 (150 cycles) | Illumina | Cat# 20024907 | RNA-seq analysis |
| Commercial assay or kit | NovaSeq 6,000 S4 Reagent Kit v1.5 (300 cycles) | Illumina | Cat# 20028312 | RNA-seq analysis |
| Software, algorithm | cellranger v3.0.2 | 10x Genomics; https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger | RRID:SCR_017344 | RNA-seq analysis |
| Software, algorithm | R Project for Statistical Computing; R v4.0.2 | https://www.r-project.org/ | RRID:SCR_001905 | RNA-seq analysis; statistical analysis |
| Software, algorithm | Seurat v3.1.5 | Stuart et al., 2019; http://seurat.r-forge.r-project.org/ | RRID:SCR_007322 | RNA-seq analysis |
| Software, algorithm | HARMONY | Korsunsky et al., 2019; https://github.com/immunogenomics/harmony | RNA-seq analysis | |
| Software, algorithm | CellPhoneDB | Efremova et al., 2020; https://www.cellphonedb.org/ | RRID:SCR_017054 | RNA-seq analysis |
| Software, algorithm | EnhancedVolcano. | Blighe et al., 2018; https://github.com/kevinblighe/EnhancedVolcano | RRID:SCR_018931 | RNA-seq analysis |
| Software, algorithm | cerebroApp | Hillje et al., 2020; https://github.com/romanhaa/cerebroApp | RNA-seq analysis | |
| Software, algorithm | FACS Kaluza software v2.1.1 | Beckman Coulter; https://www.beckman.com/coulter-flow-cytometers/software/kaluza | RRID:SCR_016182 | Flow cytometry |
| Software, algorithm | FlowJo v10.6.1 | BD Biosciences; https://www.flowjo.com/solutions/flowjo | RRID:SCR_008520 | Flow cytometry |
| Software, algorithm | ProcartaPlex Analyst 1.0 software | Thermo Fisher Scientific; https://www.thermofisher.com/de/de/home/global/forms/life-science/procartaplex-analyst-software.html | Luminex analysis | |
| Software, algorithm | MedCalc Statistical Software version 19.3.1 | MedCalc Software Ltd, Ostend, Belgium; https://www.medcalc.org; 2020 | RRID:SCR_015044 | Statistical analysis |
Additional files
-
Supplementary file 1
Supplementary tables.
(a) Clinical data and laboratory analysis. (b) Summary of technical information regarding library preparation and sequencing. (c) List of top DE genes per cluster in Figure 1—figure supplement 1Figure 1. (d) Absolute and relative cluster size in Figure 2. (e) List of DE genes per meta-cluster in Figure 3. (f) List of the DE genes per cluster in Figure 2. (g) GWAS risk genes per meta-cluster in Figure 3G. (h) AqH cytokine level in Figure 4. (i) Cytokine serum level in Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/67396/elife-67396-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67396/elife-67396-transrepform1-v1.docx