Genome-wide CRISPRi screening identifies OCIAD1 as a prohibitin client and regulatory determinant of mitochondrial Complex III assembly in human cells
Figures
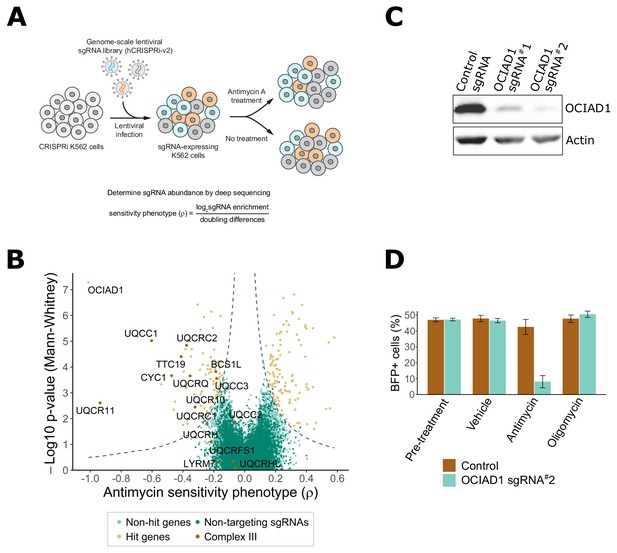
Genome-scale CRISPRi antimycin screen identifies genes regulating mitochondrial Complex III.
(A) Schematic overview of the genome-wide CRISPRi screen. K562 dCas9 cells stably expressing dCas9-KRAB were infected with a pooled genome-scale sgRNA library. After growth in galactose, cells were subjected to four pulses of antimycin A or vehicle treatment followed by a 48 hr recovery period. After the last antimycin A pulse, genomic DNA from each condition was isolated and sgRNA abundance was quantified by deep sequencing. (B) Volcano plot showing the statistical significance (y axis) versus phenotype scores (ρ, x axis) of control non-targeting and genome-wide targeting sgRNAs. Knockdown of Complex III structural proteins and assembly factors sensitized cells to antimycin A. Genes were considered a hit if they scored above a threshold of ρ z-score x−log10 p-value of 7 (dashed line). (C) CRISPRi knockdown of ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) expression. Western blot showing the expression level of OCIAD1 in K562 dCas9-KRAB cells stably expressing either a control non-targeting sgRNA or two different sgRNAs against OCIAD1. CRISPRi-based silencing reduced OCIAD1 protein expression by ~90%. (D) Validation of the OCIAD1 phenotype. K562 dCas9 cells were mixed with an equal number of K562 dCas9-KRAB BFP + cells stably expressing a non-targeting sgRNA (brown bars) or an sgRNA against OCIAD1 (light blue bars). Cell mixtures were then treated with the drug or a vehicle for 24 hr. The percentage of BFP + cells in the cell mixtures was measured by flow cytometry before and 24 hr after treatment. OCIAD1 silencing selectively sensitized cells to antimycin treatment.
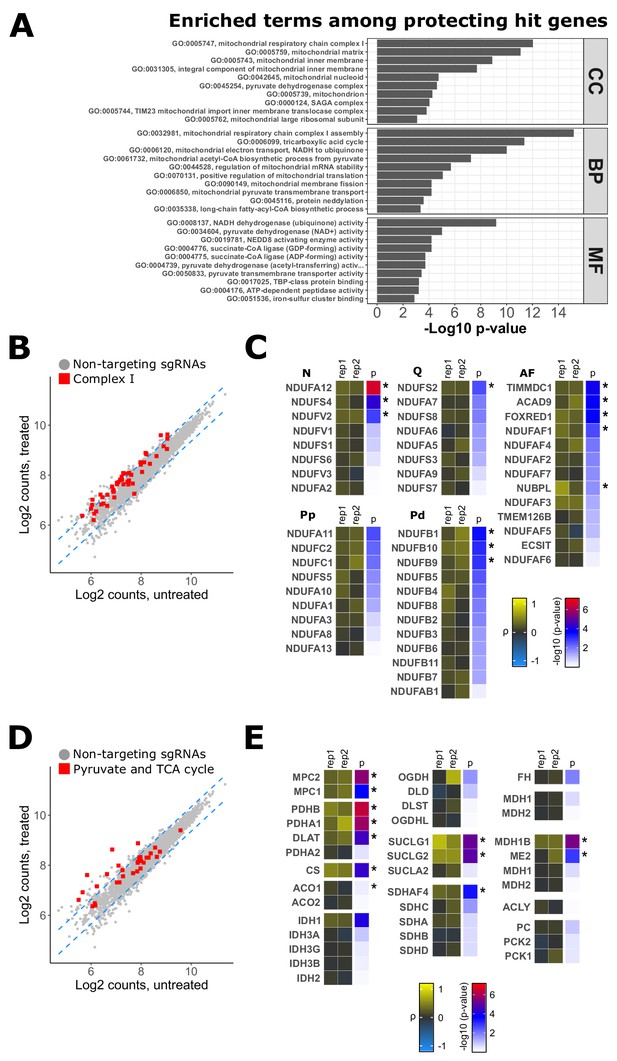
Silencing genes related to Complex I, pyruvate, and tricarboxylic acid (TCA) metabolism protect cells against chemical inhibition of Complex III.
(A) Top 10 categories from gene ontology (GO) enrichment analysis for protecting hit genes (ρ > 0). Mitochondrial terms related to Complex I, pyruvate metabolism, and the TCA cycle are specifically enriched in all three biological domains (CC: cellular component, BP: biological process, MF: molecular function). Terms are ordered by the maximum of −log10 p-value (elim algorithm with Fisher’s exact test). (B,D) Read count distribution of non-targeting control sgRNAs (gray circles) and sgRNAs related to Complex I structural subunits and assembly factors (B) or sgRNAs related to pyruvate metabolism and TCA cycle (D) in untreated and antimycin treated cells. Red squares represent the average read count of the top three targeting sgRNAs. Dashed blue lines represent the 95% prediction interval. (C,E) Tile plots displaying the phenotype scores (ρ) (first and middle columns) and associated p-values (right column) of both biological replicates for complex I related genes (C) and genes related to pyruvate and TCA metabolism (E). Complex I genes were grouped by module (N = N module; Q = Q module; Pp = proximal peripheral arm; Pd = distal peripheral arm) and assembly factors (AF). Significant genes are indicated by asterisks.
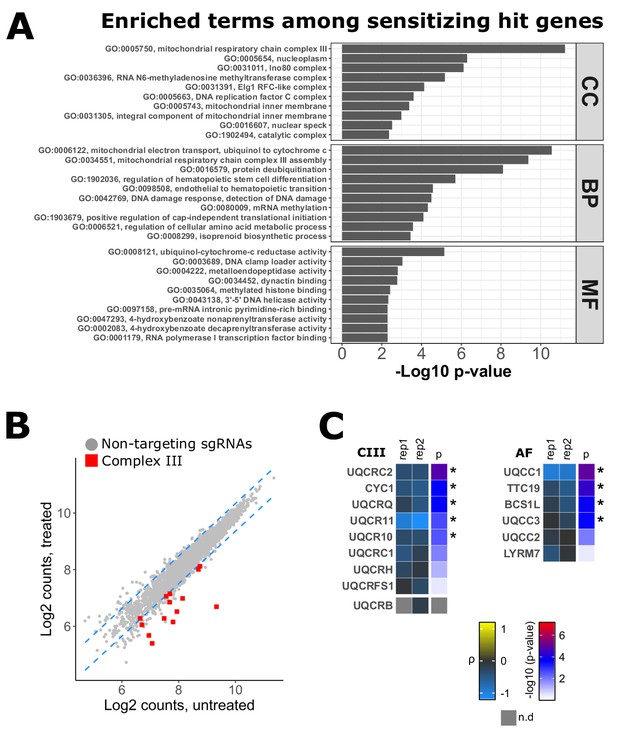
Silencing Complex III genes aggravate the cellular response to antimycin A.
(A) Sensitizing antimycin A genes. Top 10 categories from gene ontology (GO) enrichment analysis (ρ <0). Mitochondrial terms related to Complex III are enriched in all three biological domains (CC: cellular component, BP: biological process, MF: molecular function). Terms are ordered by the maximum of −log10 p-value (elim algorithm with Fisher’s exact test). (B) Read count distribution of non-targeting control sgRNAs (gray circles) and Complex III structural subunit and assembly factor sgRNAs (red squares, average of top three sgRNAs) in untreated and antimycin treated cells. Dashed blue lines represent the 95% prediction interval. (C) Tile plots displaying the phenotype scores (ρ) of each biological replicate (first and middle columns) and associated p-values (right column) for CIII2 structural genes (CIII) and assembly factors (AF). Significant genes are indicated by asterisks (n.d. = not determined).
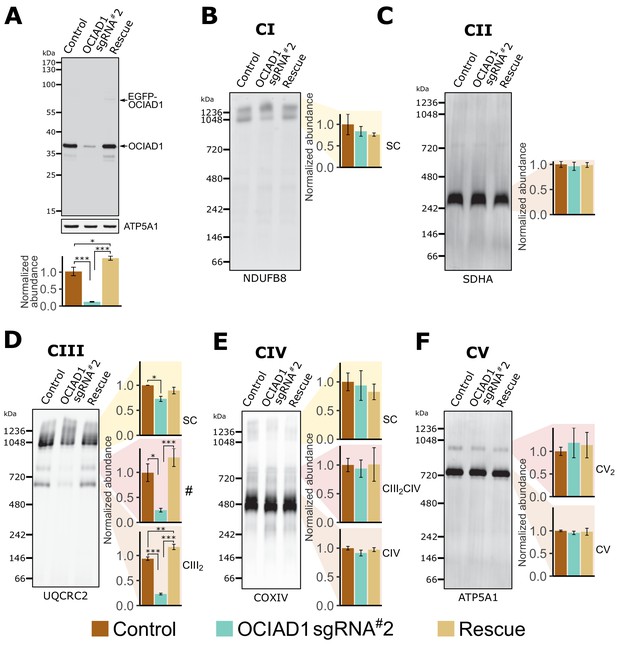
Ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) is required for CIII2 assembly.
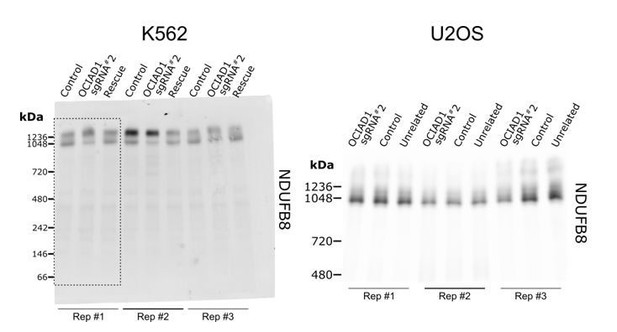
(A) Western blot showing CRISPRi silencing of OCIAD1 protein expression (12.47 ± 1.06% of control) in K562 cells. Rescue of OCIAD1 (141.20 ± 6.07% of control) by lentivirus transduction with a P2A multicistronic vector with high cleavage efficiency (98.89 ± 0.12%). The upper band (EGFP-OCIAD1) represents intact fusion gene product. ATP5A1 served as loading control. (B–F) OCIAD1 is selectively required for Complex III assembly. Blue-native polyacrylamide gel electrophoresis (BN-PAGE) analysis of digitonin-solubilized mitochondria followed by Western blotting using NDUFB8 (Complex I), SDHA (Complex II), UQCRC2 (Complex III), COXIV (Complex IV), and ATP5A1 (ATP synthase). The ATP5A1 signal from monomeric CV (F) was used as a loading control to quantify UQCRC2 intensities (D) as both proteins were probed on the same membrane. In panel B, both bands were used for quantification. Values represent normalized intensity ± SEM (n = 3 biological replicates). Asterisks (*p<0.05, **p<0.01, or ***p<0.001) correspond to the adjusted (false discovery rate [FDR]) p-values from the post-ANOVA (analysis of variance) pairwise t-test.
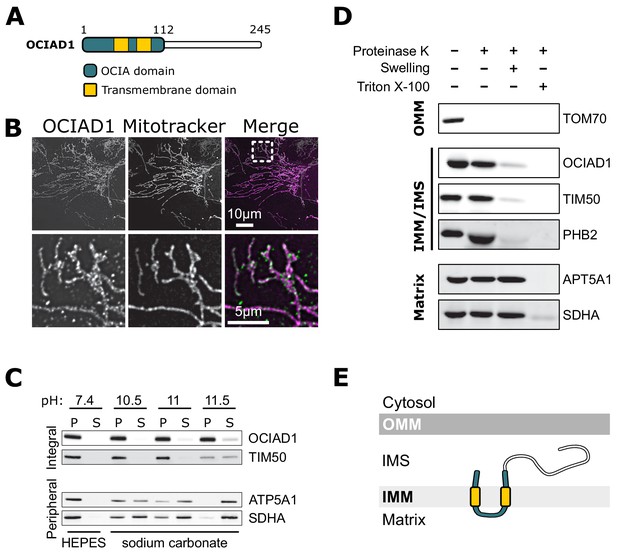
Ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) is an inner mitochondrial membrane protein.
(A) Schematic illustration of OCIAD1 domain organization. (B) Representative images of fixed U2OS cells stained with Mitotracker (magenta) and immunolabeled using anti-OCIAD1 antibodies (green). Lower panel is a magnification of the inset shown in the upper panel. (C) OCIAD1 is an integral membrane protein. Sodium carbonate extraction fractions (pH 10.5–11.5) immunoblotted with anti-OCIAD1, anti-TIM50, anti-ATP5A1, and anti-SDHA antibodies. P and S indicate pellet and soluble fractions, respectively. (D) OCIAD1 localizes to the inner membrane. Protease protection assay fractions immunoblotted with anti-OCIAD1, anti-prohibitin 2 (PHB2), anti-TIM50, anti-ATP5A1, and anti-SDHA antibodies. (OMM: outer mitochondrial membrane, IMM: inner mitochondrial membrane, IMS: intermembrane space). (E) Schematic illustration of OCIAD1 topology within the inner membrane.
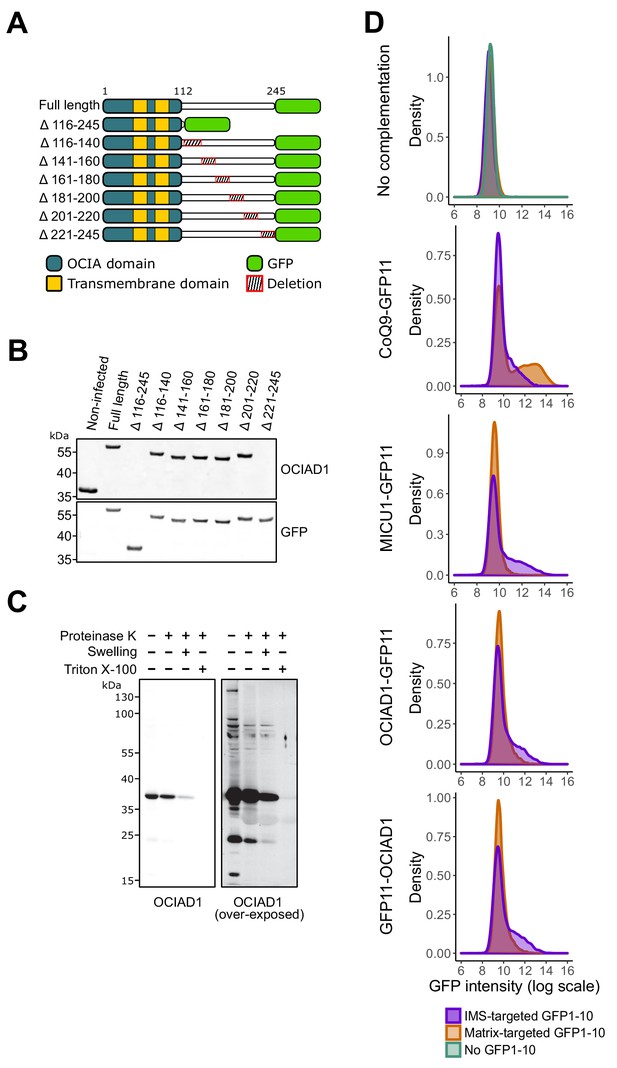
Ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) termini are localized in the mitochondrial intermembrane space.
(A) Schematic illustrating the OCIAD1-GFP deletion constructs used for mapping the epitope of the anti-OCIAD1 polyclonal antibody. (B) Cell lysates from U2OS cells expressing either full-length or truncated OCIAD1-GFP were analyzed by Western blotting and immunoprobed using anti-OCIAD1 (Invitrogen, cat# PA5-20834) and anti-GFP antibodies. The anti-OCIAD1 polyclonal antibody recognizes an epitope located within the last 25 amino acids of OCIAD1 C-terminus. (C) Uncropped immunoblot for the OCIAD1 protease protection assay shown in Figure 3C alongside an over-exposed image of the same membrane. (D) U2OS cells stably expressing IMS- or matrix-targeted GFP1–10 were transiently transfected with various GFP11 constructs before assessing GFP complementation by flow cytometry analysis. No GFP1–10 and GFP1–10 alone (uppermost panel). CoQ9 tagged with C-terminal GFP11 expressed in matrix- or IMS-targeted GFP1–10 cells (second panel). MICUI tagged with C-terminal GFP11 expressed in matrix- or IMS-targeted GFP1–10 cells (third panel). N-terminal GFP11-tagged OCIAD1 construct (fourth panel) and C-terminal GFP11-tagged OCIAD1 construct (fifth panel) expressed in matrix- or IMS-targeted GFP1–10 cells.
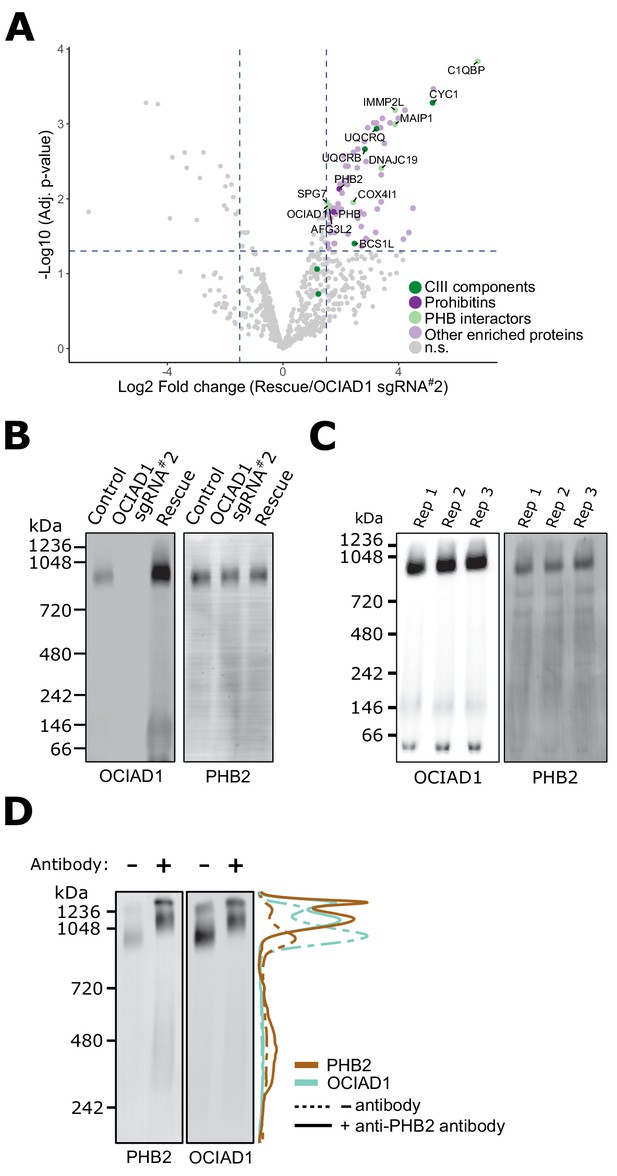
Ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) forms a complex with prohibitin supramolecular assemblies.
(A) Volcano plot showing the statistical significance (−log10 false discovery rate [FDR] adjusted p-value; y axis) versus log2 fold change (x axis) of proteins enriched in OCIAD1 pull-down performed on DSP-crosslinked K562 cell lysates from OCIAD1 knockdown cells and OCIAD1 knockdown cells rescued with wildtype OCIAD1. Proteins with a log2 fold change ≥ 1.5 and an adjusted p-value < 0.05 were considered significantly enriched (n = 3 biological replicates, n.s. = non significantly enriched). (B) Blue-native polyacrylamide gel electrophoresis (BN-PAGE) of lauryl maltose neopentyl glycol (LMNG) detergent-solubilized mitochondrial membranes isolated from U2OS control, OCIAD1 knockdown, and OCIAD1 knockdown cells rescued with wildtype OCIAD1. The membrane was immunoblotted with anti-OCIAD1 and anti-PHB2 antibodies. (C) BN-PAGE of LMNG detergent-solubilized mitochondrial membranes isolated from U2OS control cells (n = 3 biological replicates) and immunoblotted with anti-OCIAD1 and anti-PHB2 antibodies. Electrophoresis was stopped before elution of the migration front to calculate the fraction of OCIAD1 that associates with PHB2 assemblies (66.91 ± 0.35%). (D) Mitochondria from K562 cells solubilized with LMNG and pre-incubated with anti-Phb2 antibodies (solid line) or vehicle (dotted line) were analyzed by BN-PAGE and immunoblotted with anti-OCIAD1 and anti-prohibitin two antibodies. Line scan traces represent the distribution profile of P (brown) and OCIAD1 (light blue).
-
Figure 4—source data 1
Output of the limma statistical analysis of the OCIAD1 interactome in K562 OCIAD1 knockdown cells and K562 OCIAD1 knockdown cells rescued with wildtype OCIAD1.
- https://cdn.elifesciences.org/articles/67624/elife-67624-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Output of the limma statistical analysis of the whole proteome analysis by LC-MS/MS.
- https://cdn.elifesciences.org/articles/67624/elife-67624-fig4-data2-v1.xlsx
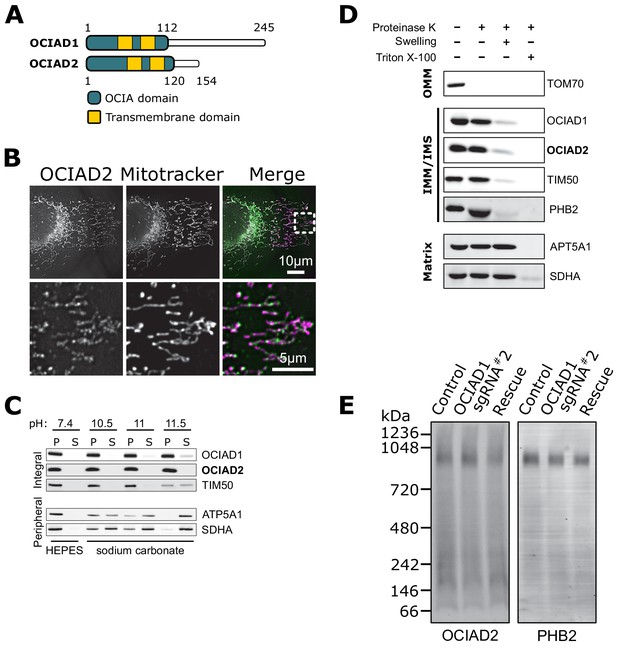
The ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) paralog, OCIAD2, localizes to the mitochondria inner membrane.
(A) Schematic of predicted OCIAD1 and OCIAD2 topologies. (B) Representative images of fixed U2OS cells stained with Mitotracker (magenta) and immunolabeled for OCIAD2 (green). The bottom panel is a magnification of the inset shown in the upper panel. (C) OCIAD2 is an integral membrane protein. Sodium carbonate extraction fractions (pH 10.5–11.5) immunoblotted with anti-OCIAD1, anti-OCIAD2, anti-TIM50, anti-ATP5A1, and anti-SDHA antibodies. P and S indicate pellet and soluble fractions, respectively. This panel, without the OCIAD2 blot, was shown in Figure 3C. (E) OCIAD2 localizes to the inner membrane. Protease protection assay fractions immunoblotted with anti-OCIAD1, anti-OCIAD2, anti-prohibitin 2 (PHB2), anti-TIM50, anti-ATP5A1, and anti-SDHA antibodies (OMM: outer mitochondrial membrane, IMM: inner mitochondrial membrane, IMS: intermembrane space). This panel, without the OCIAD2 blot, was shown in Figure 3D. (F) Blue-native polyacrylamide gel electrophoresis (BN-PAGE) of lauryl maltose neopentyl glycol (LMNG) detergent-solubilized mitochondrial membranes isolated from U2OS cells and immunoblotted with anti-OCIAD2 and anti-prohibitin antibodies.
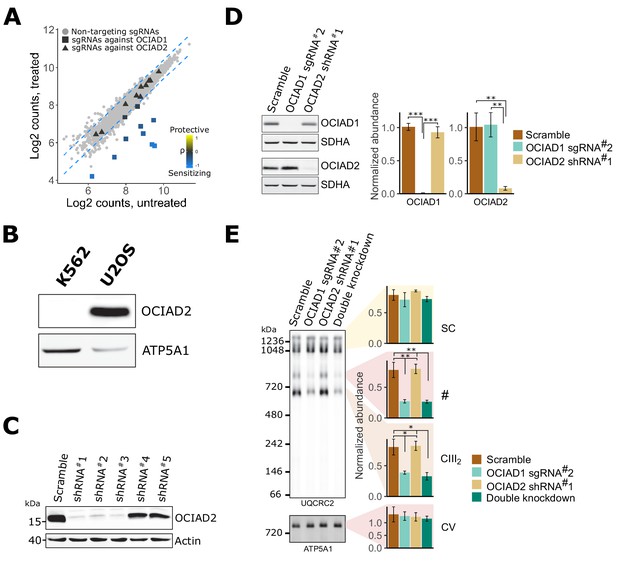
Ovarian carcinoma immunoreactive antigen domain-containing protein 1 ( OCIAD1) and OCIAD2 paralogs are functionally divergent.
(A) Read count distribution of all 10 sgRNAs targeting OCIAD1 (squares) and OCIAD2 (triangles) in untreated and antimycin-treated K562 cells. Gray circles represent non-targeting sgRNAs. Dashed blue lines represent the 95% prediction interval. (B) Western blot of U2OS and K562 cell extracts immunoblotted with anti-OCIAD2 and anti-ATP5A1 antibodies. (C) Western blot of cell extracts from U2OS cells expressing scramble or OCIAD2 shRNAs. The membrane was immunoblotted with anti-OCIAD2 and anti-β-actin antibodies. (D) Western blot of extracts from U2OS cells expressing OCIAD2 shRNA and OCIAD1gRNA. The membrane was immunoblotted with anti-OCIAD2 and anti-SDHA antibodies. SDHA was used as a loading control. (E) Blue-native polyacrylamide gel electrophoresis (BN-PAGE) analysis of digitonin-solubilized mitochondrial extracts from U2OS cells expressing OCIAD1 sgRNA#2 and OCIAD2 shRNA#1. ATP5A1 served as a loading control. Values represent normalized intensity ± SEM (n = 3 biological replicates). Asterisks (*p<0.05, **p<0.01, or ***p<0.001) correspond to the adjusted (false discovery rate [FDR]) p-values from the post-ANOVA (analysis of variance) pairwise t-test.
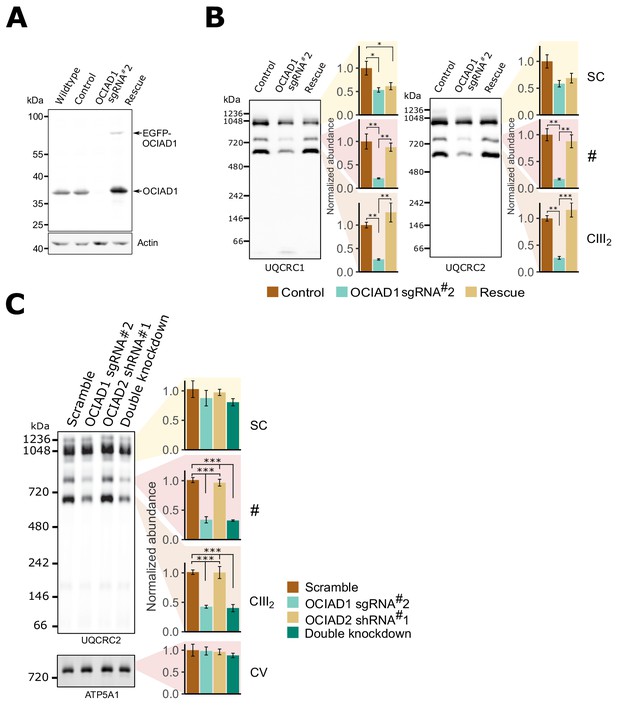
The role of ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) in CIII2 assembly is independent of cell type and glucose availability.
(A) Western blot of wildtype U2OS cells or U2OS cells expressing a non-targeting sgRNA, sgRNA#2 against OCIAD1, or sgRNA#2 and wildtype OCIAD1 rescued by lentivirus expression. The upper band (EGFP-OCIAD1) represents intact fusion gene product. The non-targeting sgRNA used in this study does not affect OCIAD1 expression (compare control and wildtype lanes). (B) Blue-native polyacrylamide gel electrophoresis (BN-PAGE) results using two CIII2 core subunits (UQCRC1, left and UQCRC2, right) showing that OCIAD1 is also required for CIII2 assembly in U2OS cells grown in glucose-containing media. (C) BN-PAGE indicating that silencing OCIAD1, but not OCIAD2, disrupts CIII2 assembly in U2OS cells grown in glucose-containing media. ATP5A1 served as a loading control. Values represent normalized intensity ± SEM (n = 3 biological replicates). Asterisks (*p<0.05, **p<0.01, or ***p<0.001) correspond to the adjusted (false discovery rate [FDR]) p-values from the post-ANOVA (analysis of variance) pairwise t-test.
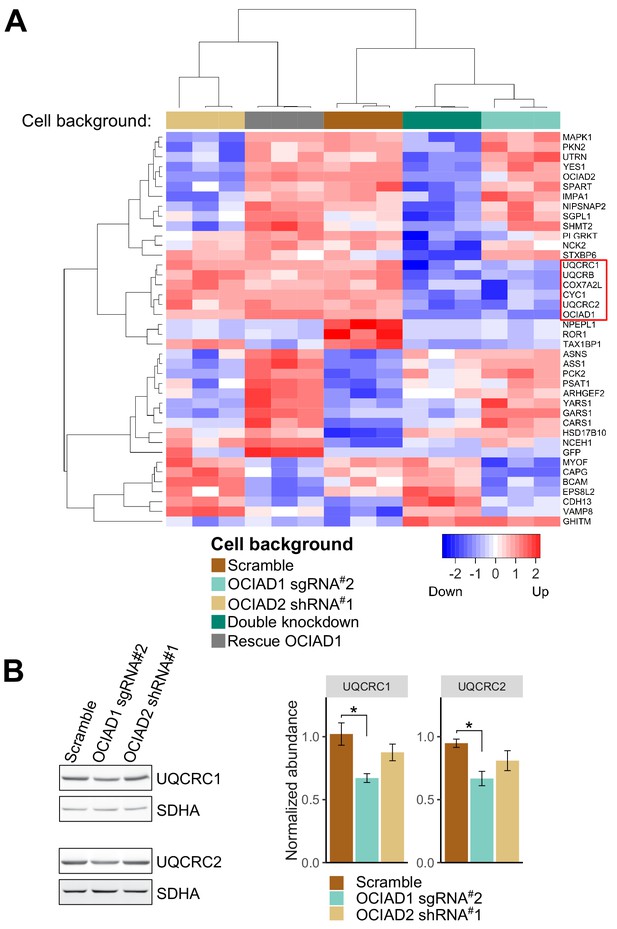
Ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) regulates steady-state levels of Complex III subunits.
(A) Hierarchical clustering of unbiased proteomic analysis performed on whole-cell lysate from U2OS control cells, OCIAD1 knockdown cells, OCIAD2 knockdown cells, OCIAD1 and OCIAD2 double knockdown cells, and OCIAD1 knockdown cells rescued with wildtype OCIAD1. The analysis identified a small cluster (red square) enriched for Complex III proteins selectively downregulated in the OCIAD1 and OCIAD1/OCIAD2 knockdown cells. (B) Western blot analysis showing that two Complex III subunits (UQCRC1 and UQCRC2) are downregulated in mitochondria isolated from OCIAD1 knockdown U2OS cells but not from OCIAD2 knockdown U2OS cells. SDHA served as a loading control.
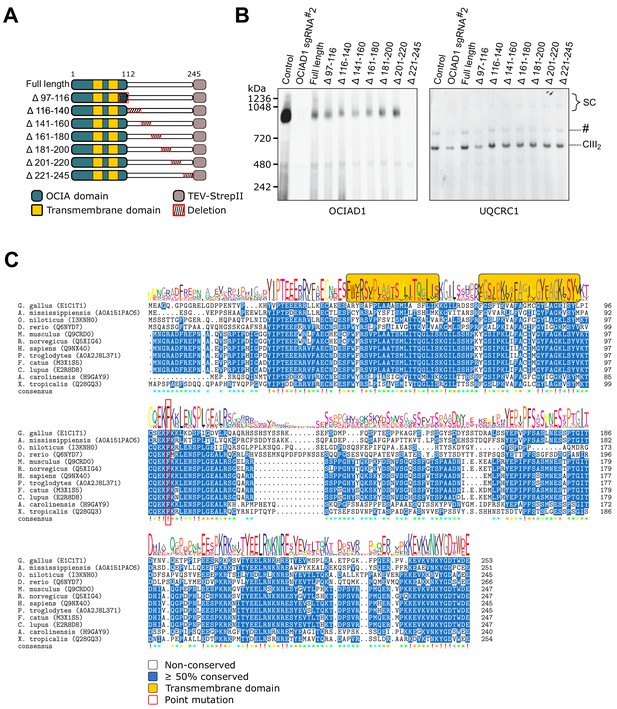
The distal region of the ovarian carcinoma immunoreactive antigen (OCIA) domain is essential for the function of OCIA domain-containing protein 1 (OCIAD1) in CIII2 assembly.
(A) Schematic showing the topology of the OCIAD1-TEV-StrepII isoforms with C-terminal tiled deletions. (B) Mitochondria were isolated from non-infected control and OCIAD1 knockdown cells, or OCIAD1 knockdown cells infected with a lentivirus expressing full-length or truncated OCIAD1 isoforms. Blue-native polyacrylamide gel electrophoresis (BN-PAGE) followed by Western blot analysis for OCIAD1 and UQCRC1 identified a small portion of the OCIA domain (a.a. 97–116) as putatively essential for CIII2 assembly. (C) Multiple sequence alignment of OCIAD1 protein sequences using Clustal Omega. Blue shading indicates over 50% of identical amino acids in all sequences. The red box indicates the location of the mutated residue.
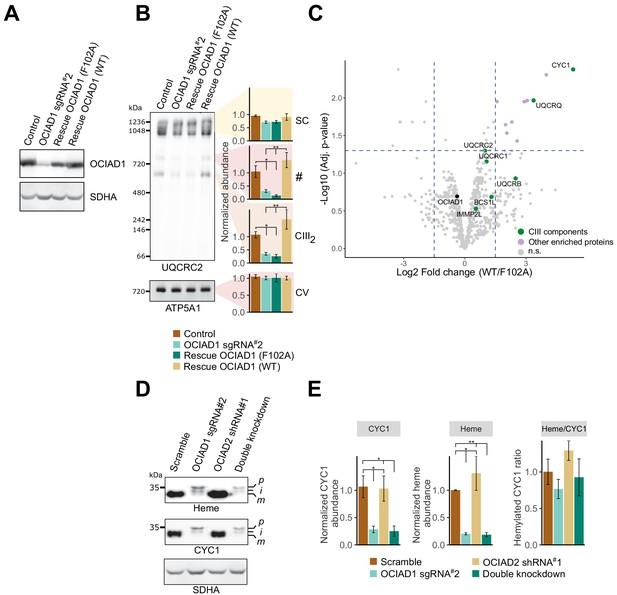
Ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) regulates the maturation of cytochrome c1 (CYC1).
(A) Western blot showing OCIAD1 expression levels in K562 OCIAD1 knockdown cells rescued with either wildtype OCIAD1 or mutant (F102A) OCIAD1. (B) Blue-native polyacrylamide gel electrophoresis (PAGE) analysis showing that the F102A point mutant fails to rescue the CIII2 assembly defect. (C) Volcano plot showing proteins enriched in OCIAD1 pull-down performed in DSP-crosslinked cell lysate from K562 OCIAD1 knockdown cells rescued with either wildtype or F102A OCIAD1. Proteins with a log2 fold change ≥1.5 and an adjusted p-value<0.05 were considered significantly enriched (n = 3 biological replicates, n.s. = non-significantly enriched). (D) Western blot analysis of U2OS mitochondrial membranes solubilized in digitonin. Heme was detected by chemiluminescence before immunoblotting the membrane with the indicated antibodies. (E) Quantification of CYC1 (left) and heme (middle) levels from blot shown in (E). Right panel shows the proportion of CYC1 that is hemylated. Values represent normalized intensity ± SEM (n = 3 biological replicates). Asterisks (*p<0.05, **p<0.01, or ***p<0.001) correspond to the adjusted (false discovery rate [FDR]) p-values from the post-ANOVA (analysis of variance) pairwise t-test.
-
Figure 5—source data 1
Output of the limma statistical analysis of the ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) interactome in K562 OCIAD1 knockdown cells rescued with wildtype OCIAD1 or the OCIAD1 F102A mutant.
- https://cdn.elifesciences.org/articles/67624/elife-67624-fig5-data1-v1.xlsx
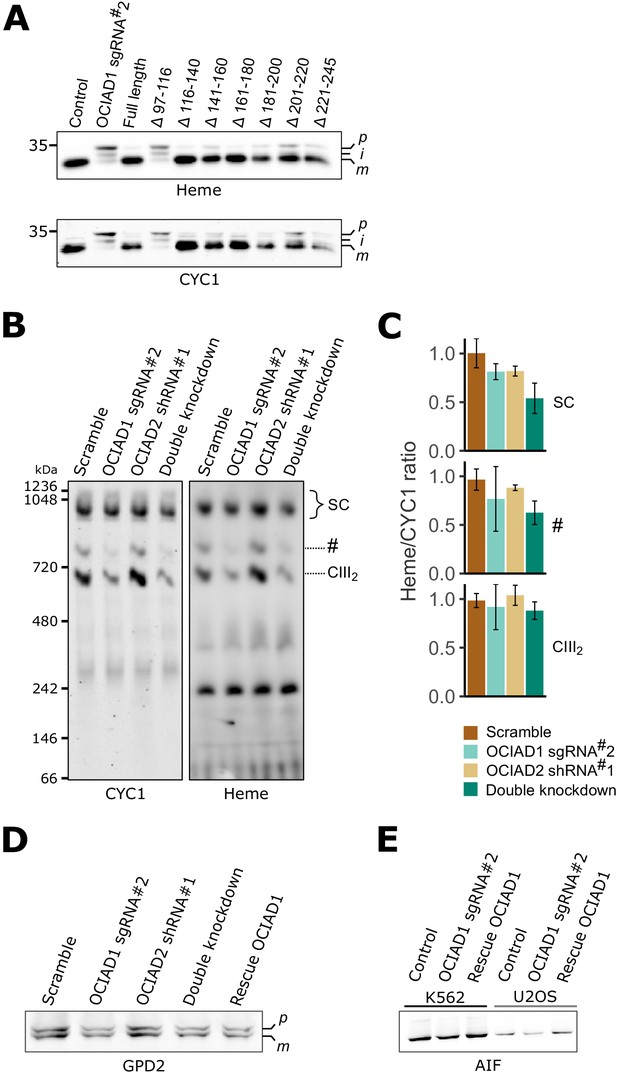
Mature CIII2 contains hemylated cytochrome c1 (CYC1) in ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) knockdown cells.
(A) Western blot analysis of mitochondria isolated from OCIAD1 knockdown cells rescued with truncated OCIAD1 isoforms shown in Figure 4—figure supplement 5A-B. Heme was detected by chemiluminescence before immunoblotting the membrane with an antibody against CYC1. Deleting the distal portion of the OCIA domain (Δ97–116) disrupted CYC1 maturation. (B) Clear-native polyacrylamide gel electrophoresis (PAGE) analysis of digitonin-solubilized mitochondrial membranes isolated from U2OS control cells, OCIAD1 knockdown cells, OCIAD2 knockdown cells, and OCIAD1 and OCIAD2 double knockdown cells. Heme was detected by chemiluminescence before immunoblotting the membrane with an antibody against CYC1. (C) Quantification of blots shown in B showing the proportion of hemylated CYC1 in CIII2 assemblies. Values represent normalized intensity ± SEM (n = 3 or 4 biological replicates). (D) Immunoblot analysis of the IMMP2L substrate GPD2 in mitochondria isolated from U2OS control cells, OCIAD1 knockdown cells, OCIAD2 knockdown cells, OCIAD1 and OCIAD2 double knockdown cells, and OCIAD1 knockdown cells rescued with wildtype OCIAD1. (E) Immunoblot analysis of the IMMP2L substrate AIF in mitochondria isolated from U2OS and K562 control cells, OCIAD1 knockdown cells, and OCIAD1 knockdown cells rescued with wildtype OCIAD1.
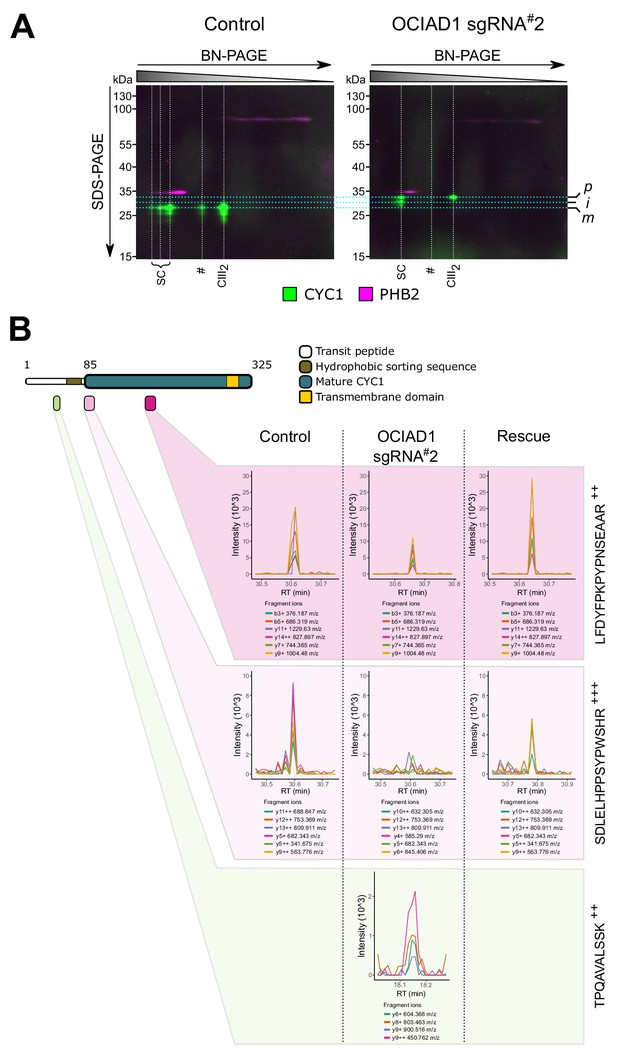
Ovarian carcinoma immunoreactive antigen domain-containing protein 1 (OCIAD1) regulates IMMP2L-dependent proteolytic processing of cytochrome c1 (CYC1).
(A) 2D-native/SDS-PAGE analysis of mitochondrial membranes isolated from K562 control and OCIAD1 knockdown cells and immunoblotted with CYC1 and PHB2 antibodies. CIII2 assemblies from OCIAD1 knockdown cells contained immature CYC1 of higher molecular weight. PHB2 staining served as an internal molecular size reference. Light blue horizontal lines represent the size of putative precursor (p), intermediate (i), and mature (m) CYC1. White vertical lines represent the different high-order CIII2 assemblies. (B) Extracted MS2 fragment ion chromatograms (XIC) for three diagnostic CYC1 peptides detected by diaPASEF mass spectrometry in blue-native polyacrylamide gel electrophoresis (BN-PAGE) gel slices excised from control cells, OCIAD1 knockdown cells, and OCIAD1 knockdown cells rescued with wildtype OCIAD1. Individual peptides displayed highly correlated fragment ion co-elution profiles strongly supportive of peptide identification. The TPQAVALSSK++ peptide (bottom panel), located at the N-terminus of the CYC1 hydrophobic sorting sequence, was only identified in CIII2 assemblies from OCIAD1 knockdown cells. Conversely, the SDLELHPPSYPWSHR+++ peptide (middle panel), which uniquely identifies the N-terminus of mature CYC1 but is not present in the tryptic digest of the CYC1 precursor, was reliably detected in CIII2 assemblies from control and OCIAD1 knockdown cells rescued with wildtype OCIAD1, but not from OCIAD1 knockdown cells. An internal peptide (LFDYFPKPYPNSEAAR+++, top panel) common to all CYC1 species (precursor, intermediate, mature) was detected in all cell lines, albeit at lower levels in OCIAD1 cells as expected.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | K562 dCas9-KRAB | Gilbert et al., 2014 doi:10.1016/j.cell.2014.09.029 | ||
| Cell line (Homo sapiens) | K562 control | This paper | K562 dCas9-KRAB transduced with pControlsgRNA | |
| Cell line (Homo sapiens) | K562 OCIAD1 sgRNA#2 | This paper | K562 dCas9-KRAB transduced with pOCIAD1sgRNA2 | |
| Cell line (Homo sapiens) | K562 OCIAD1 rescue | This paper | K562 OCIAD1 sgRNA#2 transduced with pUltra-OCIAD1 | |
| Cell line (Homo sapiens) | K562 OCIAD1 (F102A) rescue | This paper | K562 OCIAD1 sgRNA#2 transduced with pUltra-OCIAD1(F102A) | |
| Cell line (Homo sapiens) | U2OS dCas9-KRAB | This paper | U2OS transduced with pMH0006 (Addgene, cat# 135448; Chen et al., 2019) | |
| Cell line (Homo sapiens) | U2OS control | This paper | U2OS dCas9-KRAB transduced with pControlsgRNA | |
| Cell line (Homo sapiens) | U2OS OCIAD1 sgRNA #2 | This paper | U2OS dCas9-KRAB transduced with pOCIAD1sgRNA2 | |
| Cell line (Homo sapiens) | U2OS OCIAD1 rescue | This paper | U2OS OCIAD1 sgRNA#2 transduced with pUltra-OCIAD1 | |
| Cell line (Homo sapiens) | U2OS scramble | This paper | U2OS control cells transduced with pLKO.1-blast-Scramble (Addgene, cat# 26701) | |
| Cell line (Homo sapiens) | U2OS OCIAD2 shRNA#1 | This paper | U2OS control cells transduced with pLKO1-OCIAD2_shRNA1 | |
| Cell line (Homo sapiens) | U2OS double knockdown | This paper | U2OS OCIAD1 sgRNA#2 transduced with pLKO1-OCIAD2_shRNA1 | |
| Antibody | rabbit polyclonal anti-OCIAD1 | Invitrogen | PA5-20834 | RRID:AB_11155625 (1:2000-1:5000) |
| Antibody | mouse monoclonal anti-OCIAD1 | Proteintech | 66698–1-Ig | RRID:AB_2882051 (1:5000) |
| Antibody | Rabbit polyclonal anti-OCIAD2 | Invitrogen | PA5-59375 | RRID:AB_2644946 (1:500-1:5000) |
| Antibody | Mouse monoclonal anti-ATP5A1 | Proteintech | 66037–1-Ig | RRID:AB_11044196 (1:2000-1:5000) |
| Antibody | Rabbit polyclonal anti-NDUFB8 | Proteintech | 14794–1-AP | RRID:AB_2150970 (1:2000) |
| Antibody | Mouse monoclonal anti-SDHA | SantaCruz Biotechnology | sc-166947 | RRID:AB_10610526 (1:2000-1:5000) |
| Antibody | Rabbit polyclonal anti-UQCRC2 | Proteintech | 14742–1-AP | RRID:AB_2241442 (1:2000-1:5000) |
| Antibody | Mouse monoclonal anti-UQCRC1 | Invitrogen | 459140 | RRID:AB_2532227 (1:2000) |
| Antibody | Rabbit polyclonal anti-CYC1 | Proteintech | 10242–1-AP | RRID:AB_2090144 (1:1000) |
| Antibody | Mouse monoclonal anti-COXIV | Proteintech | 66110–1-1g | RRID:AB_2881509 (1:2000) |
| Antibody | Mouse monoclonal anti-PHB2 | Proteintech | 66424–1-Ig | RRID:AB_2811041 (1:5000) |
| Antibody | Rabbit polyclonal anti-TIM50 | Proteintech | 22229–1-AP | RRID:AB_2879039 (1:1000) |
| Antibody | Rabbit polyclonal anti-TOM70 | Proteintech | 14528–1-AP | RRID:AB_2303727 (1:1000) |
| Antibody | Mouse monoclonal anti-GFP | Proteintech | 66002–1-Ig | RRID:AB_11182611 (1:2000) |
| Antibody | Mouse monoclonal anti-β-actin | Proteintech | 66009–1-1g | RRID:AB_2687938 (1:10000) |
| Antibody | Rabbit polyclonal anti-GPD2 | Proteintech | 17219–1-AP | RRID:AB_2112476 (1:1000) |
| Antibody | Rabbit polyclonal anti-AIF | Proteintech | 17984–1-AP | RRID:AB_2224539 (1:1000) |
| Commercial assay or kit | SuperSignal West Femto | Thermo Scientific | 34094 | |
| Chemical compound, drug | Digitonin | Calbiochem | 300410 | |
| Chemical compound, drug | Lauryl maltose neopentyl glycol (LMNG) | Anatrace | NG310 | |
| Chemical compound, drug | Dithiobis(succinimidyl propionate) (DSP) | Life Technologies | 22585 | |
| Chemical compound, drug | LysC/Trypsin | Promega | V5071 | |
| Chemical compound, drug | ProteaseMAX | Promega | V2071 | |
| Other | μMACS protein A beads | Miltenyi Biotec | 130-071-001 | |
| Other | μ Columns | Miltenyi Biotec | 130-042-701 | |
| Other | μMACS Separator | Miltenyi Biotec | 130-042-602 | |
| Other | ZipTip with 0.6 µL C18 resin | Millipore Sigma | ZTC18S096 | |
| Other | Disposable gel cutter grids | The Gel Company | MEE2-7-25 | |
| Software, algorithm | DIA-NN | Version 1.7.12 and 1.7.13 (beta 1) | https://github.com/vdemichev/DiaNN | |
| Software, algorithm | ScreenProcessing | https://github.com/mhorlbeck/ScreenProcessing |
Additional files
-
Supplementary file 1
List of sgRNA guides used in this study.
Read counts and phenotypes for individual sgRNAs as well as gene-level phenotypes.
- https://cdn.elifesciences.org/articles/67624/elife-67624-supp1-v1.xlsx
-
Supplementary file 2
Results from the HHpred analysis.
- https://cdn.elifesciences.org/articles/67624/elife-67624-supp2-v1.zip
-
Supplementary file 3
List of primers and cloning strategy used in this study.
- https://cdn.elifesciences.org/articles/67624/elife-67624-supp3-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67624/elife-67624-transrepform-v1.docx