Secondary metabolites of Hülle cells mediate protection of fungal reproductive and overwintering structures against fungivorous animals
Figures
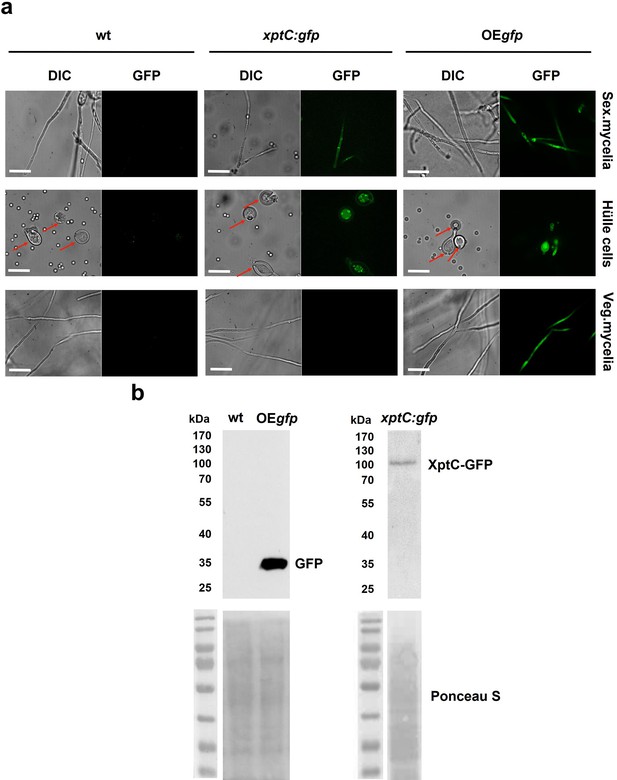
XptC is enriched in Aspergillus nidulans Hülle cells.
(a) Fluorescence microscopy of sexual mycelia, Hülle cells (after 3 days of incubation), and vegetative mycelium after 20 hr of incubation from xptC:gfp strain. A. nidulans wildtype (wt, AGB552) and constitutively expressed GFP strain (OEgfp, AGB596) were used as controls. Red arrows indicate Hülle cells. Scale bar = 20 µm. The fusion protein XptC-GFP is undetectable in vegetative mycelia. (b) Western hybridization of sexual (sex) tissues harvested 3 days after inoculation. α-GFP antibody was used. The fusion protein XptC-GFP was detected at approximately 95 kDa. A strain expressing GFP constitutively (OEgfp, AGB596) and wildtype (wt, AGB552) served as controls. Ponceau S staining was used as sample loading control.
-
Figure 1—source data 1
Raw data Western experiments.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig1-data1-v1.jpg
-
Figure 1—source data 2
Raw data Western experiments.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig1-data2-v1.jpg
-
Figure 1—source data 3
Labelled, uncropped wWestern experiments.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig1-data3-v1.jpg
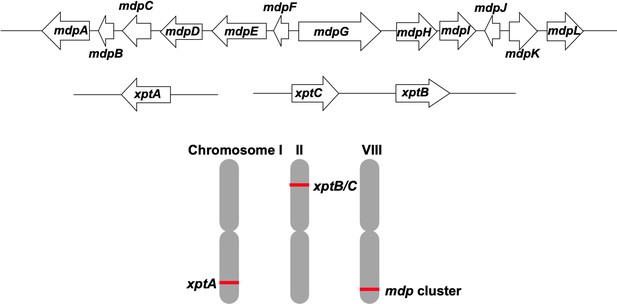
The epi-/shamixanthone-producing mdp and xpt genes are localized on different Aspergillus nidulans chromosomes.
The 12 mdp genes are clustered and located on chromosome VIII. The gene xptA is located on chromosome I, whereas xptB and xptC are located on chromosome II.
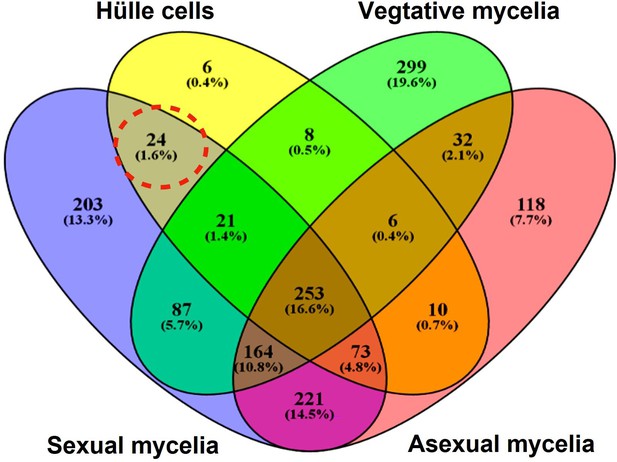
Mdp/Xpt proteins are expressed in sexual tissues of Aspergillus nidulans.
Venn diagram with 1525 identified proteins from different tissues (vegetative, asexual, sexual mycelia, and enriched Hülle cells) of A. nidulans A4. Vegetative mycelia were cultivated for 20 hr in liquid medium. Asexual and sexual cultures were grown for 3, 5, and 7 days on solid plates. Hülle cells were enriched from sexual tissues. Proteins that could be identified in two or more biological replicates and with two or more peptides per protein by LC-MS were used for the analysis. The dashed, red circle represents the 24 proteins (Supplementary file 1) that were exclusively identified from sexual mycelium and Hülle cell extracts.
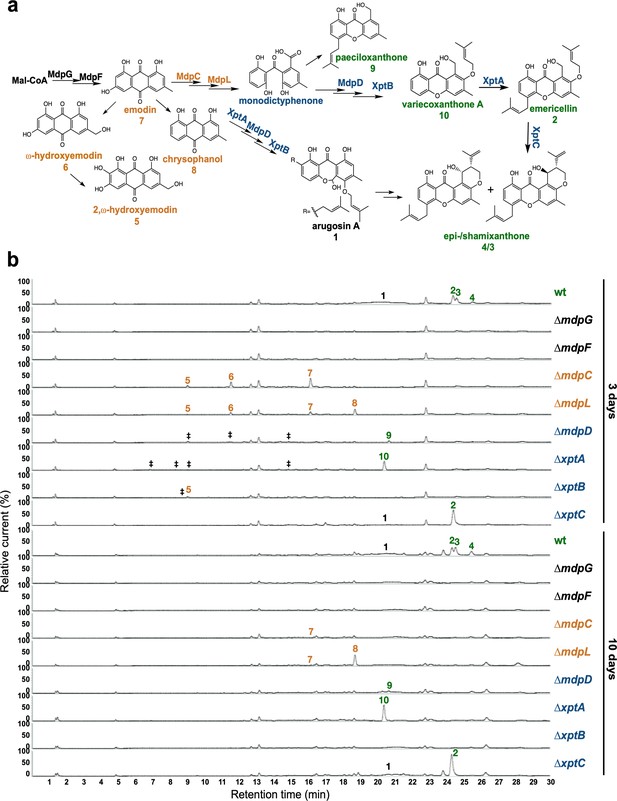
The mdp/xpt cluster metabolites are produced after 3 days of sexual growth in Aspergillus nidulans.
(a) Simplified biosynthetic pathway of epi-/shamixanthone in A. nidulans (Chiang et al., 2010; Pockrandt et al., 2012; Sanchez et al., 2011). Black enzymes are localized at the early steps of the biosynthetic pathway. Orange enzymes are localized at the middle of the biosynthetic pathway. Blue enzymes are localized at the late steps of the biosynthetic pathway. The cluster products are classified as four groups: anthraquinones are orange, benzophenones are blue, xanthones are green, arugosin A is black. (b) Chromatograms of secondary metabolites (SMs) of A. nidulans wildtype (wt, AGB552) and mdp/xpt deletion strains (∆). Conidia of A. nidulans wt and mdp/xpt mutant strains were point-inoculated on minimal medium (MM) and grown under conditions inducing the sexual cycle for 3 and 10 days. Extra- and intracellular metabolites were extracted and detected by LC-MS with a charged aerosol detector (CAD) in three independent experiments. Only CAD-detectable and identified SMs are shown with numbers: (1) arugosin A; (2) emericellin; (3) shamixanthone; (4) epishamixanthone; (5) 2,ω-dihydroxyemodin; (6) ω-hydroxyemodin; (7) emodin; (8) chrysophanol; (9) paeciloxanthone; (10) variecoxanthone A. ‡ marks unidentified compounds.
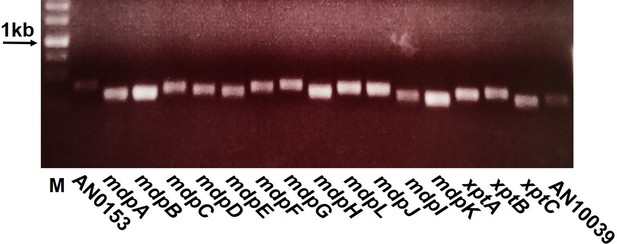
The mdp/xpt genes are expressed during fungal sexual development.
Aspergillus nidulans wildtype AGB552 conidia were point-inoculated on minimal medium (MM) agar plates and cultivated for 3 days under sexual conditions. RNA samples were harvested from three biological replicates and transcribed to cDNA. PCR was performed. AN0153 and AN10039 represent the genes localized at the border of the mdp cluster on chromosome VIII and were used as control. M = 1 kb DNA ladder (Thermo Fisher Scientific, SM0311).

Aspergillus nidulans developmental stages used for secondary metabolite analysis.
Conidia of A. nidulans AGB552 were point-inoculated on minimal medium (MM) agar plates and cultivated under sexual conditions. After 2 days, only conidiophores (Co) but no cleistothecia (Cl) or Hülle cells (Hc) were present. Young cleistothecia and Hülle cells were visible after 3 days and mature cleistothecia after 5 days. Scale bar = 200 µm.

Chromatograms of extracted secondary metabolites (SMs) from Aspergillus nidulans wildtype and mdp/xpt deletion strains.
Conidia of A. nidulans wildtype AGB552 and mdp/xpt deletion strains were point-inoculated on minimal medium (MM) agar plates and grown under sexual-inducing conditions for 2, 5, and 7 days. Extra- and intracellular metabolites were extracted and detected by LC-MS with a charged aerosol detector (CAD). Only CAD-detectable and identified SMs are shown with numbers: (1) arugosin A; (2) emericellin; (3) shamixanthone; (4) epishamixanthone; (5) 2,ω-dihydroxyemodin; (6) ω-hydroxyemodin; (7) emodin; (8) chrysophanol; (9) paeciloxanthone; (10) variecoxanthone A. ‡ marks unidentified compounds.
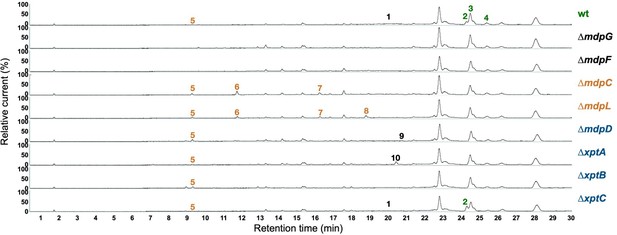
Chromatograms of secreted secondary metabolites (SMs) from Aspergillus nidulans wildtype and mdp/xpt deletion strains.
Conidia of A. nidulans wildtype AGB552 and mdp/xpt deletion strains were point-inoculated on minimal medium (MM) agar plates and grown under sexual-inducing conditions for 3 days. Extracellular metabolites were extracted and detected by LC-MS with a charged aerosol detector (CAD). Only CAD-detectable and identified SMs are shown with numbers: (1) arugosin A; (2) emericellin; (3) shamixanthone; (4) epishamixanthone; (5) 2,ω-dihydroxyemodin; (6) ω-hydroxyemodin; (7) emodin; (8) chrysophanol; (9) paeciloxanthone; (10) variecoxanthone.
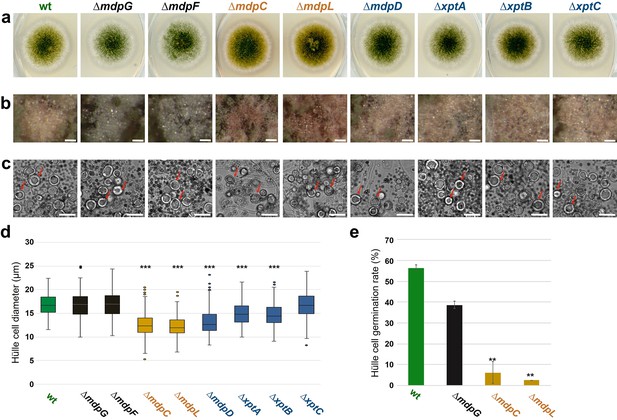
The mdp/xpt cluster metabolites are localized in Hülle cells.
(a) Colony phenotypes of Aspergillus nidulans wildtype (wt, AGB552) and mdp/xpt deletion strains (∆). Conidia were point-inoculated on minimal medium (MM) agar plates and cultivated 3 days under sexual conditions. (b) Photomicrographs of Hülle cells after 5 days. Scale bar = 50 µm. (c) Morphology of Hülle cells after 5 days. Red arrows indicate examples of Hülle cells. Scale bar = 25 µm. (d) Box plot of Hülle cells size after 5 days of sexual development (n ≥ 150). (e) Germination rate of Hülle cells. Detached Hülle cells were collected from cleistothecia surface after 5 days of sexual development and placed on fresh MM agar plates. The germination was monitored after 48 hr at 37°C. n = 40 (±1) with two biological replicates. All significance tests are in comparison to wildtype (wt), ***/**p < 0.005/0.05, two-tailed t-test.
-
Figure 3—source data 1
Hülle cells size after 5 days of sexual development.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Germination rate of Hülle cells.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig3-data2-v1.xlsx
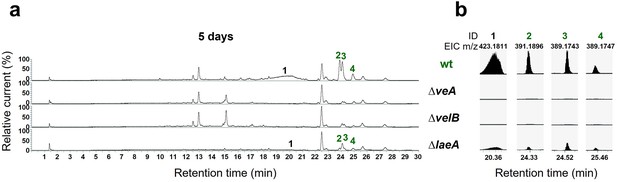
The velvet complex is required to produce mdp/xpt cluster metabolites.
(a) Chromatograms of secondary metabolites (SMs) of Aspergillus nidulans wildtype (wt, AGB552) and velvet complex gene deletion strains (∆). Conidia of A. nidulans strains were point-inoculated on minimal medium (MM) and sexually grown for 5 days. Extra- and intracellular metabolites were extracted and detected by LC-MS with a charged aerosol detector (CAD). Only CAD-detectable SMs of the mdp/xpt cluster are shown with numbers and were identified with MS and UV/VIS. (b) EICs (extracted ion chromatograms) of the compounds detected by CAD. m/z of 1 was detected in negative mode. m/z of 2, 3, and 4 was detected in positive mode. ID (compound number in this study): (1) arugosin A; (2) emericellin; (3) shamixanthone; (4) epishamixanthone.
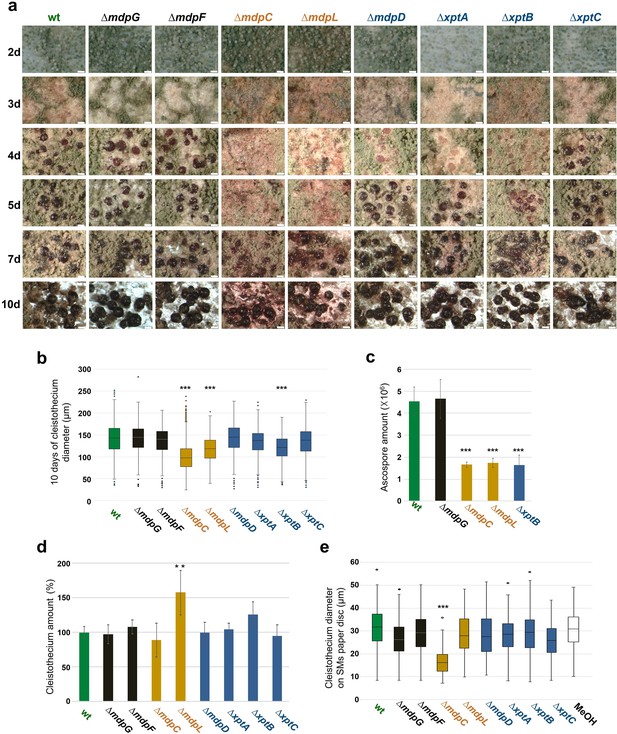
Intermediates of xanthone synthesis encoded by the mdp/xpt cluster repress sexual development of Aspergillus nidulans.
(a) Photomicrographs of sexual structures of wildtype (wt, AGB552) and mdp/xpt deletion strains at different developmental stages. Scale bar = 100 µm. (b) Box plot of cleistothecia diameter after 10 days of sexual development (n ≥ 650). (c) Ascospore quantification after 10 days. Ten cleistothecia were broken in 100 µl 0.02% Tween buffer and released ascospores were quantified. Error bar indicates standard deviation with three biological and three technical replicates. (d) Cleistothecia quantification after 10 days. Error bar indicates standard deviation with five biological replicates; amount of cleistothecia of wt was set to 100%. (e) Box plot of cleistothecia diameter of A. nidulans wt grown on paper discs loaded with extracted secondary metabolites (SMs) of wt and mdp/xpt deletion strains. Strains were sexually grown for 5 days. SMs were extracted, solved in MeOH, and loaded on paper discs separately (pure MeOH was used as blank control). Paper discs were placed on agar plates inoculated with 1 × 105 conidia of A. nidulans wt. Cleistothecia on the paper discs were collected after 5 days of sexual growth (n ≥ 65). All significance tests are in comparison to wt, ***/**p < 0.005/0.05, two-tailed t-test.
-
Figure 5—source data 1
Cleistothecia diameter after 10 days of sexual development.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Ascospore quantification after 10 days.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig5-data2-v1.xlsx
-
Figure 5—source data 3
Cleistothecia quantification after 10 days.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig5-data3-v1.xlsx
-
Figure 5—source data 4
Cleistothecia diameter of Aspergillus nidulans wildtype grown on paper discs loaded with mdp/xpt deletion strain secondary metabolites (SMs).
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig5-data4-v1.xlsx
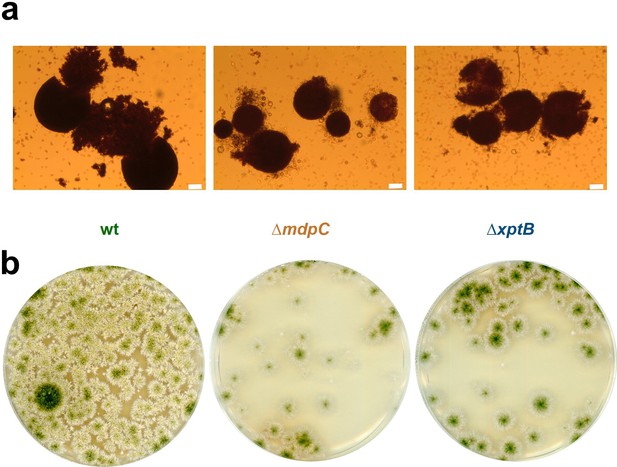
Ascospores from small cleistothecia of ΔmdpC and ΔxptB are viable.
Cleistothecia of Aspergillus nidulans wildtype AGB552, ΔmdpC and ΔxptB were harvested after 10 days of sexual development. (a) Manually opened cleistothecia and released ascospores. Scale bar = 20 µm. (b) One mature cleistothecium of each strain was opened manually in 100 µl Tween/NaCl solution and cultivated on an minimal medium (MM) agar plate for 3 days in light.
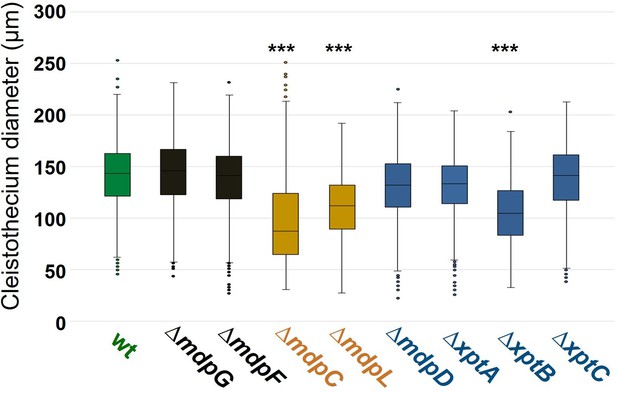
Box plot of cleistothecia size after 25 days of sexual growth.
Conidia of Aspergillus nidulans wildtype (wt) AGB552 and mdp/xpt deletion strains were point-inoculated on minimal medium (MM) agar plates and sexually grown for 25 days. Cleistothecia were collected and the diameters were measured. n ≥ 750, ***p < 0.005 (referenced to wt), two-tailed t-test.
-
Figure 5—figure supplement 2—source data 1
Cleistothecia size after 25 days of sexual growth.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig5-figsupp2-data1-v1.xlsx
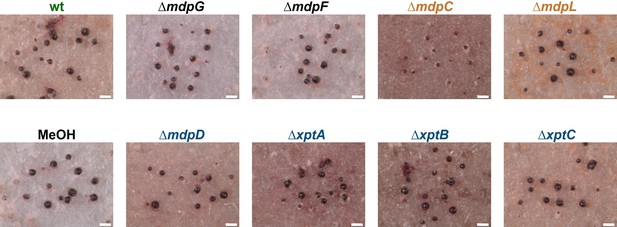
Secondary metabolites (SMs) of ∆mdpC affect the cleistothecia development of Aspergillus nidulans wildtype.
A. nidulans wildtype (wt) AGB552 and mdp/xpt deletion strains were sexually grown for 5 days. SMs were extracted, solved in MeOH, and loaded onto filter discs. Pure MeOH was used as blank control. Filter discs were placed on agar plates inoculated with 1 × 105 conidia of A. nidulans wildtype. After 5 days of sexual development, the cleistothecia on each paper disc were monitored. Scale bars = 200 µm.
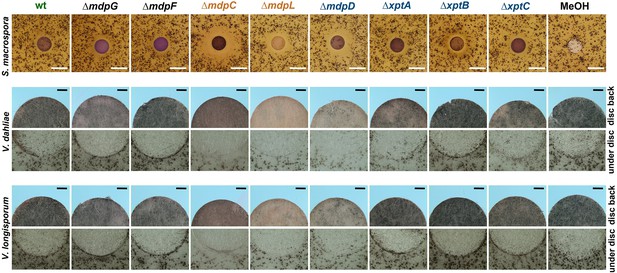
Intermediates of the mdp/xpt cluster repress fungal reproduction and resting structure formation.
Microphotographs of fungal reproduction and resting structures exposed to extracted secondary metabolites (SMs) of Aspergillus nidulans wildtype (wt) and mdp/xpt deletion strains. Conidia of A. nidulans strains were point-inoculated on minimal medium (MM) and sexually grown for 5 days. SMs were extracted, solved in MeOH, and loaded onto paper discs (MeOH solvent served as control). Paper discs were placed on agar plates inoculated with spores of the tested fungi. For Sordaria macrospora, BMM agar plates were inoculated with 2 × 105 spores and cultivated 7 days at 27°C. White bar = 1 cm. For Verticillium spp., simulated xylem medium (SXM) agar plates were inoculated with 1 × 105 Verticillium dahliae or Verticillium longisporum spores and cultivated 10 days at 25°C. The upper panel shows pictures taken from the back of the paper disc and the lower panel shows the agar under the paper disc. Black bar = 1 mm.
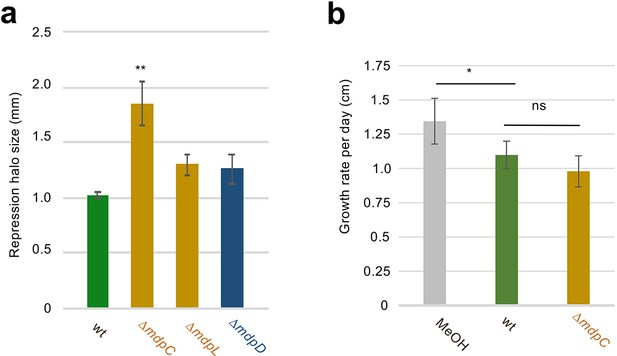
Effect of secondary metabolites (SMs) on perithecium formation and vegetative growth of Sordaria macrospora.
(a) Conidia of Aspergillus nidulans wildtype (wt) AGB552, ΔmdpC, ΔmdpL, and ΔmdpD were point-inoculated on minimal medium (MM) agar plates and sexually grown for 5 days. SMs were extracted and loaded onto filter discs. Filter discs were placed on BMM agar plates inoculated with spores of S. macrospora. Repression halo of S. macrospora perithecia formation was measured after 7 days of incubation at 27°C. Error bar indicates standard deviation with two technical and biological replicates. **p < 0.05 (referenced to wt), two-tailed t-test. (b) Race tube experiments with S. macrospora on BMM medium. SM extracts of Aspergillus nidulans AGB552 and ΔmdpC diluted in MeOH were used. As control only methanol (MeOH) was used. Growth rates were determined in cm/day. Experiment was carried out for 7 days at 27°C. *p < 0.05, two-tailed t-test. ns means not significant. Error bar indicates standard deviation with three biological replicates.
-
Figure 6—figure supplement 1—source data 1
Effect of secondary metabolites on perithecium formation.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig6-figsupp1-data1-v1.xlsx
-
Figure 6—figure supplement 1—source data 2
Effect of secondary metabolites on vegetative growth of Sordaria macrospora.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig6-figsupp1-data2-v1.xlsx
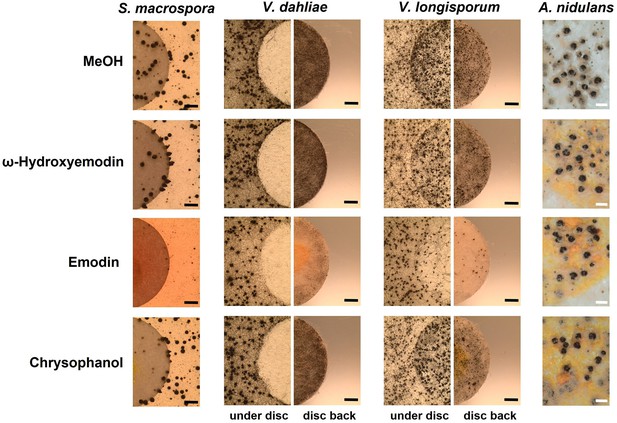
Emodin represses fungal reproduction and resting structure formation.
Pure emodin, ω-hydroxyemodin, and chrysophanol were dissolved in MeOH and loaded onto paper discs (final amount 75 µg per disc). MeOH served as blank control. Paper discs were placed on agar plates inoculated with spores of the tested fungi. For Sordaria macrospora, BMM agar plates were inoculated with 2 × 105 spores and cultivated 7 days at 27°C. For Verticillium spp., simulated xylem medium (SXM) agar plates were inoculated with 1 × 105 spores and cultivated 10 days at 25°C. Microsclerotia were monitored under the paper disc and at its back. Black bar = 1 mm. For Aspergillus nidulans, 1 × 105 conidia of wildtype were inoculated on minimal medium (MM) agar plates and sexually incubated for 5 days at 37°C. White bar = 200 µm.
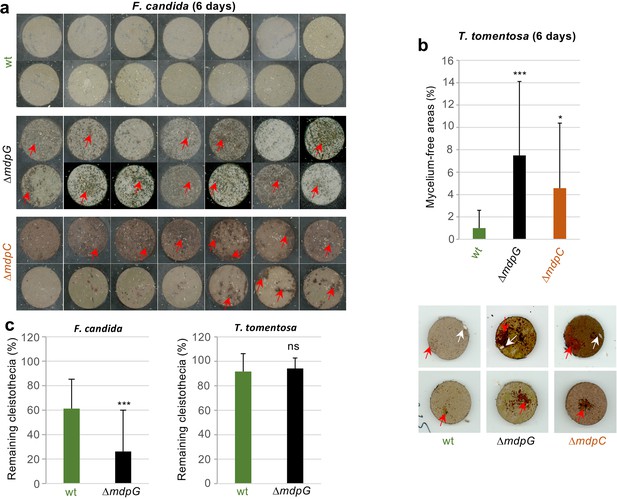
Metabolites of the mdp/xpt cluster in Aspergillus nidulans protect cleistothecia and sexual mycelium from fungal predators.
Approximately 30 springtail animals of Folsomia candida were placed on agar pieces covered with mycelium of fungal strains induced for 5 days under sexual conditions. Agar pieces with animals were incubated for 6 days at 22°C. For the two A. nidulans mutant strains ΔmdpG and ΔmdpC, mycelium-free areas could be observed (areas are indicated by red arrows). None of the agar pieces with mycelium of the wildtype showed large areas without mycelium. Two independent experiments were carried out. Each experiment contains at least six agar pieces from two different plates. (b) The same experimental setup as in (a) was used with the isopod Trichorhina tomentosa. Five animals were put on each agar piece. After 6 days, mycelium-free areas larger than 1% of the area of the agar piece were quantified using ImageJ software. Red arrows show mycelium-free areas. White arrows point on single T. tomentosa animals on the agar pieces. (c) Ten cleistothecia with Hülle cells of the indicated strains obtained from mycelium incubated for 4 days under sexual-inducing conditions were placed on water-agar plates and incubated with T. tomentosa (five animals) or F. candida (approximately 30 animals). Cleistothecia remaining after 24 hr were counted. Two biological replicates with T. tomentosa and four biological replicates with F. candida were performed. Each biological replicate contained six technical replicates. Statistical significance was determined using two-tailed t-test with *p < 0.05, ***p < 0.005 (referenced to wt). Error bar indicates the standard deviation.
-
Figure 8—source data 1
Eaten areas of sexual mycelium by Trichorhina tomentosa after 6 days.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig8-data1-v1.xlsx
-
Figure 8—source data 2
Eaten cleistothecia by the predators Trichorhina tomentosa and Folsomia candida.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig8-data2-v1.xlsx
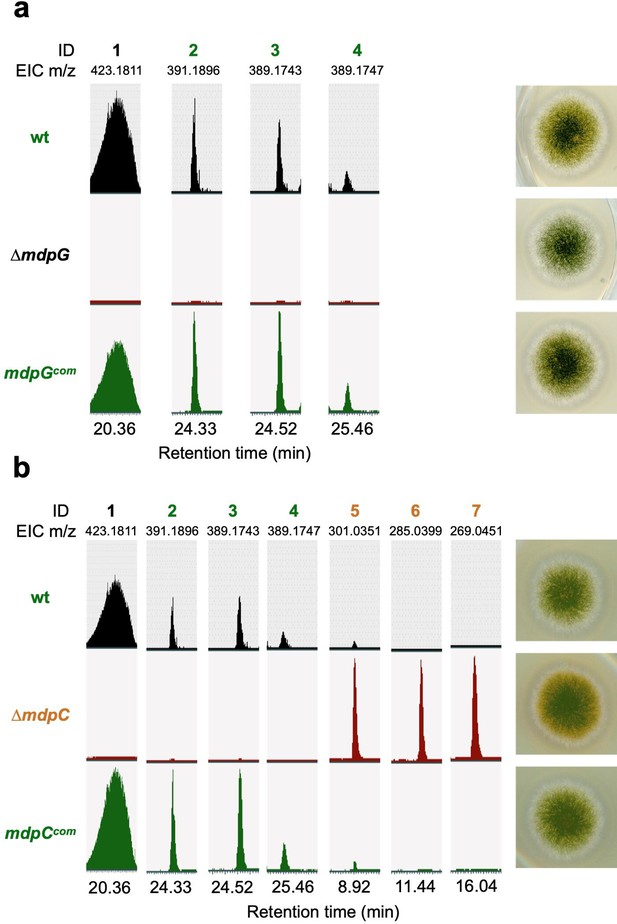
Complementation strains of mdpG and mdpC restored the phenotype and the secondary metabolite (SM) production.
The production of the mdp/xpt metabolites and the colony phenotype of (a) mdpG complementation (mdpGcom) and (b) mdpC complementation (mdpCcom) were restored to wildtype. Aspergillus nidulans conidiospores were point-inoculated on minimal medium (MM) agar plates and cultivated under sexual conditions for 3 days. Extra- and intracellular metabolites were extracted and detected by LC-MS. EICs (extracted ion chromatograms) of compounds are shown. m/z of 1, 5, 6, and 7 was detected in negative ion mode. m/z of 2, 3, and 4 was detected in positive ion mode. ID (compound number in this study): (1) arugosin A; (2) emericellin; (3) shamixanthone; (4) epishamixanthone; (5) 2,ω-dihydroxyemodin; (6) ω-hydroxyemodin; (7) emodin.
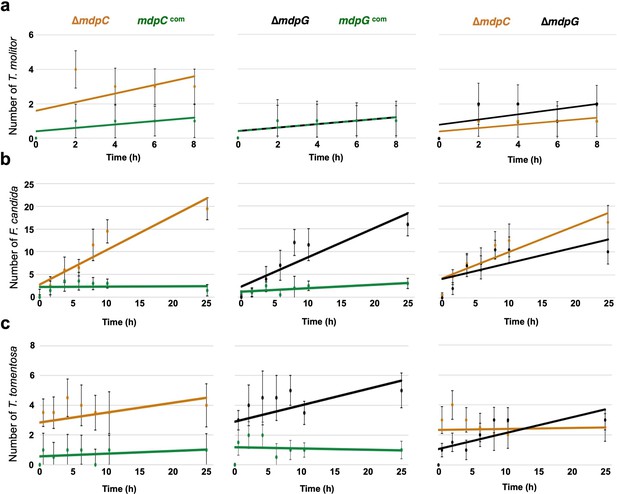
The final secondary metabolite (SM) products of the mdp/xpt cluster repel fungal predators.
Food preference test for Tenebrio molitor larvae (a), Folsomia candida (b), and Trichorhina tomentosa (c). Aspergillus nidulans spores were point-inoculated on minimal medium (MM) agar plates and incubated for 5 days under sexual conditions. A piece of the colony was cut out and used for animal feeding. Animals were placed into the center of the Petri dish containing fungal agar pieces on opposite sides. The number of animals on each side was counted. For T. molitor, 10 animals per plate were used. The experiment was performed with three biological and nine technical replicates and medians were calculated (the medians for the number of animals of ∆mdpG and mdpGcom were identical). For F. candida, approximately 20 animals per plate were used. For T. tomentosa, eight animals per plate were used. These experiments were performed with three biological and four technical replicates. Error bars represent 95% confidence interval; orange = ∆mdpC; black = ∆mdpG; green = complementation strains.
-
Figure 8—figure supplement 2—source data 1
The final secondary metabolite (SM) products of the mdp/xpt cluster repel fungal predators.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig8-figsupp2-data1-v1.xlsx
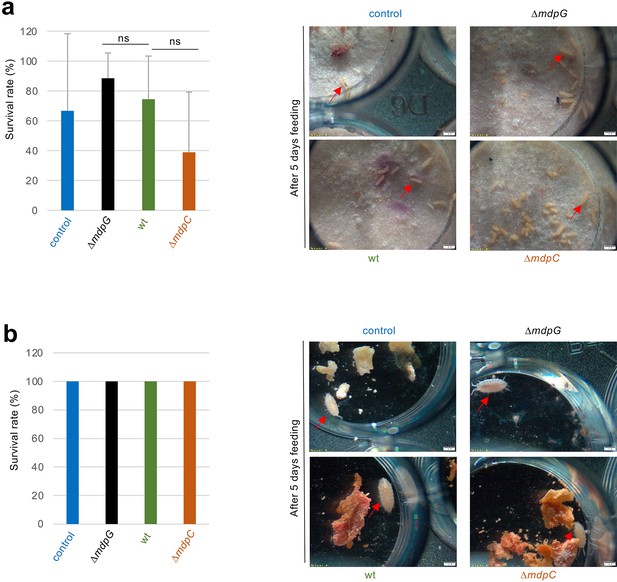
Final metabolite products of the mdp/xpt cluster in Aspergillus nidulans wildtype have no toxic effect on fungal predators.
Toxicity test of Folsomia candida (a) and Trichorhina tomentosa (b) fed with food containing secondary metabolites from the wildtype (AGB552), the ΔmdpG and the ΔmdpC strain. Food without secondary metabolites were used as control. (a) Left figure shows the survival rate of F. candida. Right images are photos from the toxicity assay after 5 days. (b) Same as (a) but for T. tomentosa. No significance could be observed in the survival rate of the wt in comparison to the mutant strains for both animals. T. tomentosa even shows a survival rate of 100% for all feeding conditions. Significance was calculated using the two-tailed t-test with p < 0.05 (referenced to wt). ns stands for not significant. Red arrows shows individual animals of F. candida (a) and T. tomentosa (b).
-
Figure 8—figure supplement 3—source data 1
Toxicity of secondary metabolites to Trichorhina tomentosa and Folsomia candida.
- https://cdn.elifesciences.org/articles/68058/elife-68058-fig8-figsupp3-data1-v1.xlsx
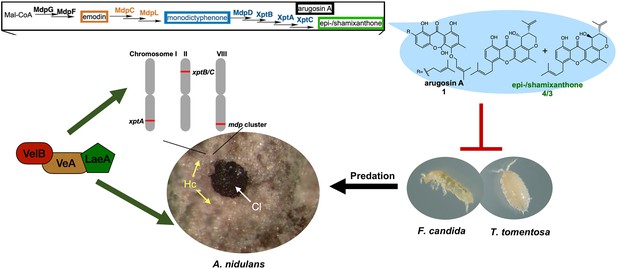
The mdp/xpt cluster metabolites establish a secure niche for Aspergillus nidulans.
The velvet complex VelB-VeA-LaeA regulates sexual development of A. nidulans and the expression of mdp/xpt genes. The resulting metabolites are accumulated in the sexual fruiting body (Cl) nursing Hülle cells (Hc). The final products arugosin A (1) and epi-/shamixanthone (4/3) protect sexual fruiting bodies from the animal predators Folsomia candida and Trichorhina tomentosa.
Tables
Five Mdp/Xpt proteins are found in sexual mycelia and Hülle cells.
| Gene ID (protein) | Putative function (Szwalbe et al., 2019) | 3 days | 5 days | 7 days | 20 hr | |||
|---|---|---|---|---|---|---|---|---|
| Sexual mycelia | Hülle cells | Sexual mycelia | Hülle cells | Sexual mycelia | Hülle cells | Vegetative mycelia | ||
| AN10023 (MdpL) | Baeyer-Villiger monooxygenase | 121 | 25 | 98 | – | 34 | 42 | 9 |
| AN7998 (XptC) | Reductase | 54 | 7 | 48 | 6 | 26 | – | 1 |
| AN12402 (XptB) | Prenyltransferase | 52 | 5 | 19 | – | – | 8 | – |
| AN10022 (MdpH) | Anthrone oxidase, decarboxylase | 27 | – | 2 | 5 | 27 | 6 | – |
| AN0150 (MdpG) | Polyketide synthase | 12 | 4 | – | 3 | – | – | – |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Aspergillus nidulans) | FGSC A4 | FGSC | Wildtype isolate (veA+) | |
| Strain, strain background (Aspergillus nidulans) | AGB552 | Bayram et al., 2012a | pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB596 | Bayram and Braus, 2012b | pgpdA:sgfp:phleoR; pabaA1; yA2; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1073 | This paper | ∆laeA::six; pabaA1; ∆nkuA::argB, veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1088 | This paper | xptC:gfp; pabaA1; yA2;ΔnkuA::argB, veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1236 | This paper | ∆mdpG::six; pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1237 | This paper | ∆mdpF::six; pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1238 | This paper | ∆mdpC::six; pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1239 | This paper | ∆mdpL::six; pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1240 | This paper | ∆mdpD::six; pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1241 | This paper | ∆xptA::six; pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1242 | This paper | ∆xptB::six; pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1243 | This paper | ∆xptC::six; pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1248 | This paper | ∆mdpG::six, mdpG:six::six; pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1249 | This paper | ∆mdpC::six, mdpC:six::six; pabaA1; ∆nkuA::argB; veA+ | |
| Strain, strain background (Aspergillus nidulans) | AGB1310 | This paper | ∆veA::six; pabaA1; ∆nkuA::argB | |
| Strain, strain background (Aspergillus nidulans) | AGB1311 | This paper | ∆velB::six; pabaA1; ∆nkuA::argB, veA+ | |
| Strain, strain background (Sordaria macrospora) | Taxid5147 | Nowrousian et al., 2010 | Wildtype isolate | |
| Strain, strain background (Verticillium longisporum) | VL43 | Zeise and Tiedemann, 2001 | Wildtype isolate | |
| Strain, strain background (Verticillium dahliae) | JR2 | Fradin et al., 2009 | Wildtype isolate | |
| Strain, strain background (Trichorhina tomentosa) | b.t.b.e. Insektenzucht GmbH | Wildtype | ||
| Strain, strain background (Folsomia candida) | Institute of Zoology (University of Göttingen, Germany) | Wildtype | ||
| Strain, strain background (Tenebrio molitor) | Zoo & Co. Zoo-Busch | Wildtype | ||
| Commercial assay or kit | GeneArt Seamless Cloning and Assembly Kit | Thermo Fisher Scientific | A13288 | |
| Commercial assay or kit | GeneArt Seamless Cloning and Assembly Enzyme Mix | Thermo Fisher Scientific | A14606 | |
| Commercial assay or kit | RNeasy Plant Mini Kit | Qiagen | Cat. No./ID: 74,904 | |
| Commercial assay or kit | QuantiTect Reverse Transcription Kit | Qiagen | Cat. No./ID: 205,311 | |
| Antibody | α-GFP antibody sc-9996 | Santa Cruz Biotechnology | RRID:AB_627695 | |
| Antibody | α-Mouse antibody G21234 | Invitrogen AG | ||
| Software, algorithm | ImageJ | Schneider et al., 2012 | RRID:SCR_003070 | |
| Software, algorithm | Proteome Discoverer 1.4 | Thermo Scientific | RRID:SCR_014477 | |
| Software, algorithm | Xcalibur (FreeStyle 1.4) | Thermo Scientific | RRID:SCR_014593 |
Additional files
-
Supplementary file 1
LC-MS analysis revealed 24 proteins that were exclusively produced in both sexual mycelia and Hülle cells.
- https://cdn.elifesciences.org/articles/68058/elife-68058-supp1-v1.docx
-
Supplementary file 2
Secondary metabolites produced by the mdp/xpt genes in Aspergillus nidulans sexual development identified by LC-MS.
- https://cdn.elifesciences.org/articles/68058/elife-68058-supp2-v1.docx
-
Supplementary file 3
Fungal strains used in this study.
- https://cdn.elifesciences.org/articles/68058/elife-68058-supp3-v1.docx
-
Supplementary file 4
Plasmids employed in this study.
- https://cdn.elifesciences.org/articles/68058/elife-68058-supp4-v1.docx
-
Supplementary file 5
Primers for DNA sequence amplification and plasmid construction.
- https://cdn.elifesciences.org/articles/68058/elife-68058-supp5-v1.docx
-
Supplementary file 6
Primers for semi-quantification of the mdp/xpt cluster genes.
- https://cdn.elifesciences.org/articles/68058/elife-68058-supp6-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68058/elife-68058-transrepform1-v1.docx





