Science Forum: Author-sourced capture of pathway knowledge in computable form using Biofactoid
Figures
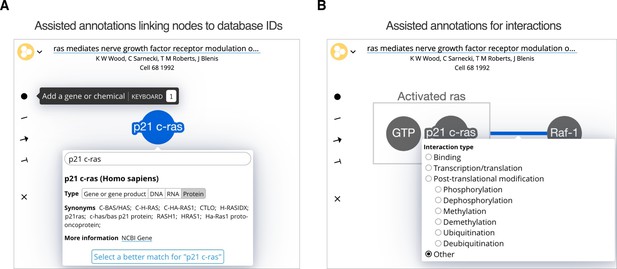
The Biofactoid curation tool.
Curation in Biofactoid involves drawing a network of relationships between genes or chemicals. (A) Genes and chemicals are represented by circles (nodes, highlighted in blue) where users provide a label, the type of gene product, and the organism. A search engine matches the label to a corresponding record from a database of genes or chemicals. (B) Relationships are represented by connecting genes, chemicals and/or rectangular complexes (shown in grey) with plain lines (when neither activation nor repression occur), arrows (to indicate activation), or ‘T-bars’ (to represent repression). Users select the mechanism that best describes the interaction. Complexes are represented as genes and/or chemicals enclosed by a box (e.g. the grey box labelled "Activated ras").
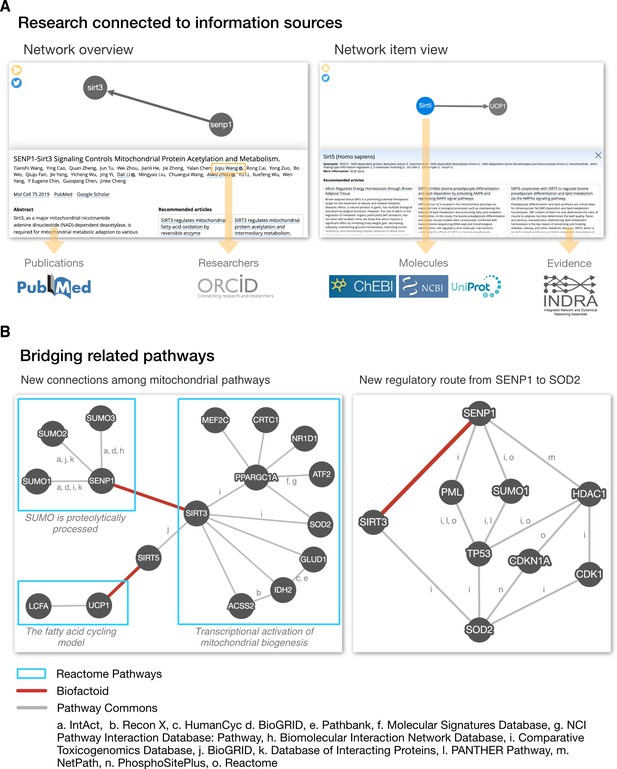
Biofactoid data is connected to information sources and establishes a bridge between related pathways.
(A) The Biofactoid Explorer is an interactive web app that publicly presents each author-curated entry alongside their article. Yellow arrows indicate how curated information is connected to outside knowledge bases. A “Network overview” (left) displays information about the article and pathway as a whole; a “Network item view” (right) displays information for a selected item (e.g. interaction, protein). (B) Biofactoid data establishes a bridge between related pathways described by structured biological knowledge from distinct data sources. Two author-curated interactions submitted to Biofactoid (red edges) bridge previously distinct pathways from the Reactome Pathway Database involved in mitochondrial biogenesis (left) and provide a new, more direct regulatory route between two mitochondrial genes (right). Pathway and interaction information was provided by Pathway Commons (pathwaycommons.org), a web resource that provides a single point of access for multiple public interaction and pathway databases. Details regarding the generation of these networks can be found in the section “Visualization of network data across sources” in Materials and methods.

Biofactoid pilot study.
A three-phase pilot tested the feasibility of Biofactoid and involved journal editors and authors of research articles. Phase I and II involved editors and authors whose articles were recently published. In Phase III, 2,065 published articles were screened and authors of suitable articles were invited to Biofactoid. The articles screened were from 16 journals (Table 1).
Tables
Prevalence of articles with pathway knowledge suitable for Biofactoid.
| ISSN | Journal* | Coverage² [Vol. (Issue)] | Articles screened | Hits | % Hits |
|---|---|---|---|---|---|
| 2211–1247 | Cell Reports | 30(1) - 32(11) | 953 | 109 | 10.3 |
| 1097–4164 | Molecular Cell | 73(1) - 79(6) | 725 | 85 | 10.5 |
| 1549–5477 | Genes & Development | 34(1-2) - 34(17-18) | 93 | 15 | 13.9 |
| 1476–4679 | Nature Cell Biology | 22(4) - 22(9) | 84 | 10 | 10.6 |
| 1083–351 X | Journal of Biological Chemistry | 295(31) - 295(37) | 210 | 21 | 9.1 |
| Total | - | - | 2065 | 240 | - |
| Weighted Average | - | - | - | - | 10.4 |
-
*
Only journals in which at least 80 ‘hits’ were identified were included. A ‘hit’ is an article that provides direct evidence for a molecular interaction that can be captured by Biofactoid. The ability of Biofactoid to capture an interaction depends upon the type of bioentities, the relationship types and organisms described in the article. In total, articles from 16 journals were screened: EMBO; Molecular and Cellular Biology; Cell; Cancer Cell; iScience; J Biol Chem; Cell Metabolism; Science; Nature Genetics; Science Signaling; Science Advances; Immunity; Cell Reports; Molecular Cell; Genes & Development; Nature Cell Biology. ²Coverage indicates the span of journal issues that were included. Only primary research articles from each issue were screened.
Comparison of non-centrally curated biocuration projects.
Comparison of projects that support community curation of pathway and interaction knowledge as their primary concern.
| Project | Scope | Source | Integrated curation tool | Automatic entity recognition | Single-article oriented | Ref. |
|---|---|---|---|---|---|---|
| Biofactoid | Pathway | Author | ✓ | ✓ | ✓ | This study. |
| Structured Digital Abstract (FEBS Letters) | Protein-Protein | Author | - | - | ✓ | Ceol et al., 2008; Leitner et al., 2010; Gerstein et al., 2007 |
| WikiPathways | Pathway | Anyone | - | - | - | Slenter et al., 2018 |
| SourceData | Figure | Author | ✓ | - | ✓ | Liechti et al., 2017 |





