A chimeric nuclease substitutes a phage CRISPR-Cas system to provide sequence-specific immunity against subviral parasites
Figures
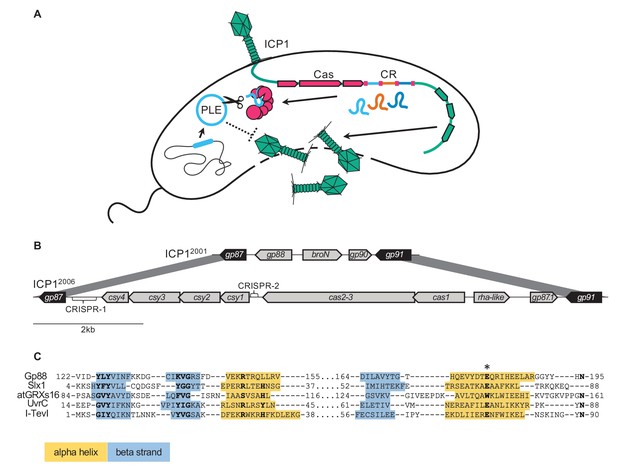
Some ICP1 isolates encode a free-standing nuclease in place of CRISPR-Cas.
(A) A model of ICP1 interference of phage-inducible chromosomal island-like elements (PLEs) via CRISPR. When ICP1 infects a PLE(+) V. cholerae cell, ICP1 is able to overcome PLE restriction and reproduce if it possesses a CRISPR-Cas system with complementary spacers to the PLE. Cas and CR refer to the CRISPR associated genes and CRISPR array respectively. (B) Schematics of the region between gp87 and gp91 as it appears in ICP12001 (top) and ICP12006 (bottom). Genes represented by black arrows are conserved in all ICP1 isolates, while genes represented with gray arrows covary with gp88 or CRISPR-Cas. (C) An alignment between the T5orf172 domain of Gp88 and the GIY-YIG domains of several structurally resolved endonucleases. Secondary structure for Gp88 was predicted using HHPRED (Zimmermann et al., 2018). Alpha helices are shown in yellow shading, and beta strands are shown in blue shading. Key residues of the GIY-YIG motif are bolded. We included an atypical GIY-YIG endonuclease domain from a chloroplast-encoded glutoredoxin atGRXs16 to demonstrate the potential for alternative residues at core motif positions. A conserved glutamate that was previously found to be required for catalysis in I-TevI is denoted by an asterisk (and corresponds to the E180A mutation in Gp88 in subsequent experiments).
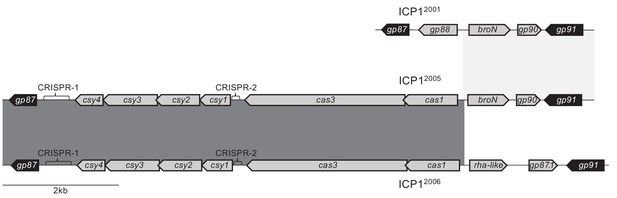
ICP12005 has an atypical CRISPR-Cas arrangement.
Genetic organization of the region between gp87 and gp91 in different ICP1 isolates (ICP12001[top], ICP12006[bottom], ICP12005[middle]). Other than ICP12005, all sequenced ICP1 isolates (Angermeyer et al., 2018) share their organization with ICP12001 or ICP12006.
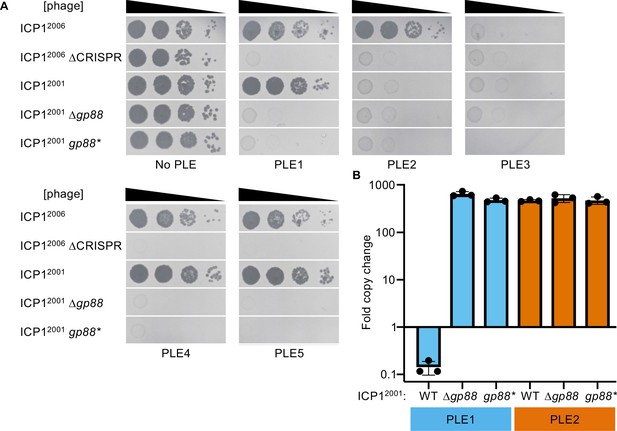
The alternative nuclease Gp88 controls ICP1 host range in a natural isolate that lacks CRISPR-Cas.
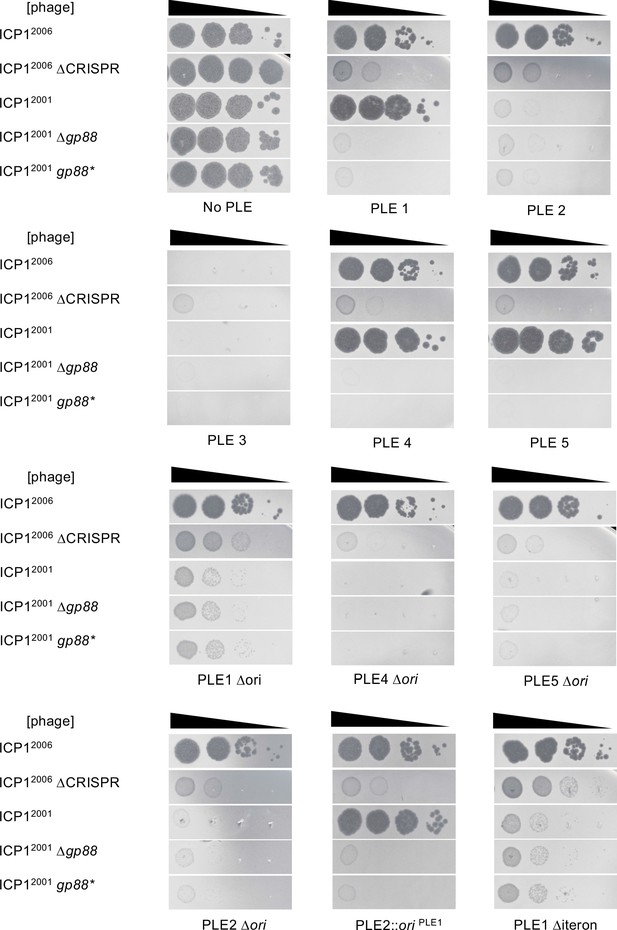
(A) Tenfold dilutions of the phage isolate or mutant derivative indicated spotted on V. cholerae with the PLE indicated (bacterial lawns in gray, zones of killing are shown in black). Gp88* possess a single amino acid substitution (E180A) predicted to abolish nuclease activity. Spot assays were performed in biological triplicate, and a single representative image is shown. Replicate spot assays are shown in Figure 2—figure supplement 1 and Figure 2—figure supplement 2. (B) Replication of PLE1 and PLE2 in V. cholerae host strains calculated as the fold change in PLE DNA copy 20 minutes post infection with the ICP1 variant indicated.
-
Figure 2—source data 1
Values for the graph in Figure 2B.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig2-data1-v1.xlsx
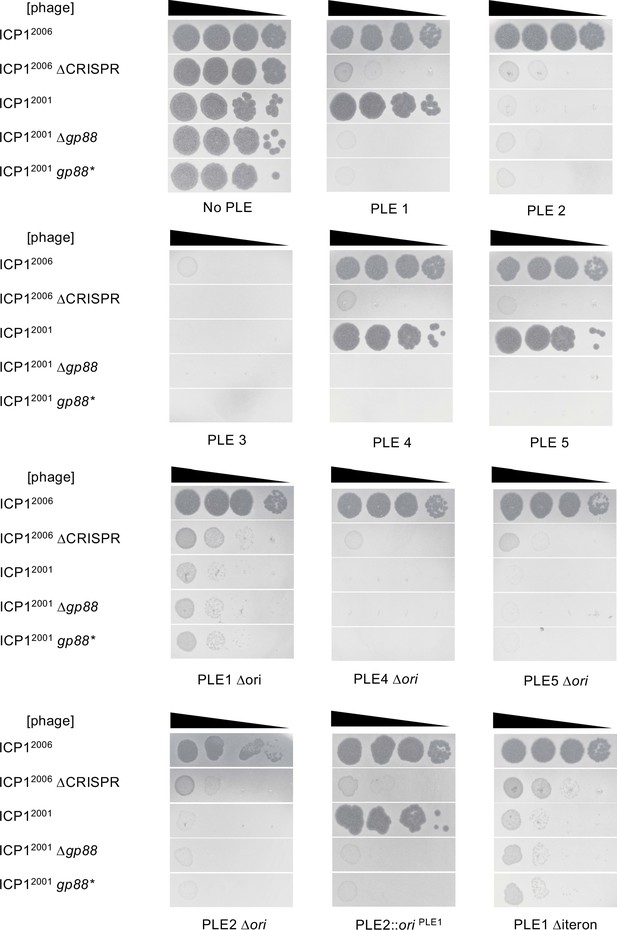
Biological replicate of spot assays.
Tenfold dilutions of the phage isolate or mutant derivative indicated spotted on V. cholerae with the PLE indicated (bacterial lawns in gray, zones of killing are shown in black). This figure and Figure 2—figure supplement 2 each represent one biological replicate, while a third replicate is divided across Figure 2B, Figure 4B, and Figure 5B. Strain backgrounds appear in the same order that they first appear in the main text. For each biological replicate, spot assays were performed in parallel for the different strains.
Biological replicate of spot assays.
Tenfold dilutions of the phage isolate or mutant derivative indicated spotted on V. cholerae with the PLE indicated (bacterial lawns in gray, zones of killing are shown in black). This figure and Figure 2—figure supplement 1 each represent one biological replicate, while a third replicate is divided across Figure 2B, Figure 4B, and Figure 5B. Strain backgrounds appear in the same order that they first appear in the main text. For each biological replicate, spot assays were performed in parallel for the different strains.
Phage-inducible chromosomal island-like element (PLE) replicons are modular and are composed of a compatible RepA initiation factor and origin of replication (ori).
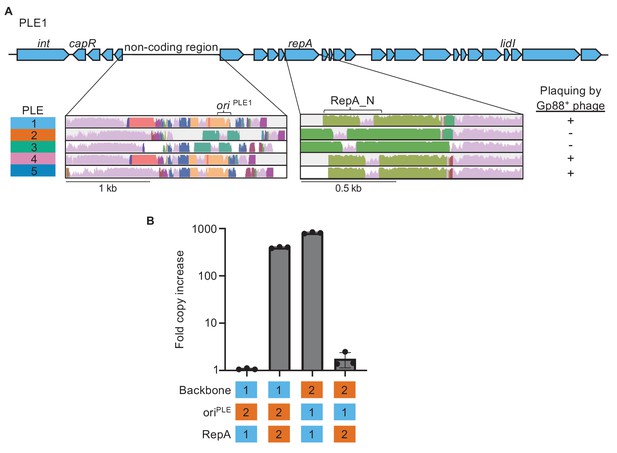
(A) Genomic organization of PLE1 with insets corresponding to the PLE noncoding region and repA. Previously characterized PLE genes are labeled. Insets are Mauve alignments showing sequence conservation of the denoted loci across the different PLEs. Shared color denotes sequence conservation, with the height of the histogram representing nucleotide sequence identity. The susceptibility of each PLE(+) V. cholerae host to plaquing by phage encoding Gp88 is indicated. (B) Replication of hybrid PLEs in V. cholerae calculated as the fold change in PLE DNA copy 20 minutes post infection with ICP12006 ∆CRISPR. Strains with PLE1 ∆repA (possessing the native oriPLE1 or ∆ori::oriPLE2) or PLE2 ∆repA (possessing the native oriPLE2 or ∆ori::oriPLE1) were complemented with a vector expressing the repA gene from PLE1 or PLE2. The backbone, identity of the ori, and RepA variant are indicated as being from PLE1 or PLE2.
-
Figure 3—source data 1
Values for the graph in Figure 3B.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig3-data1-v1.xlsx
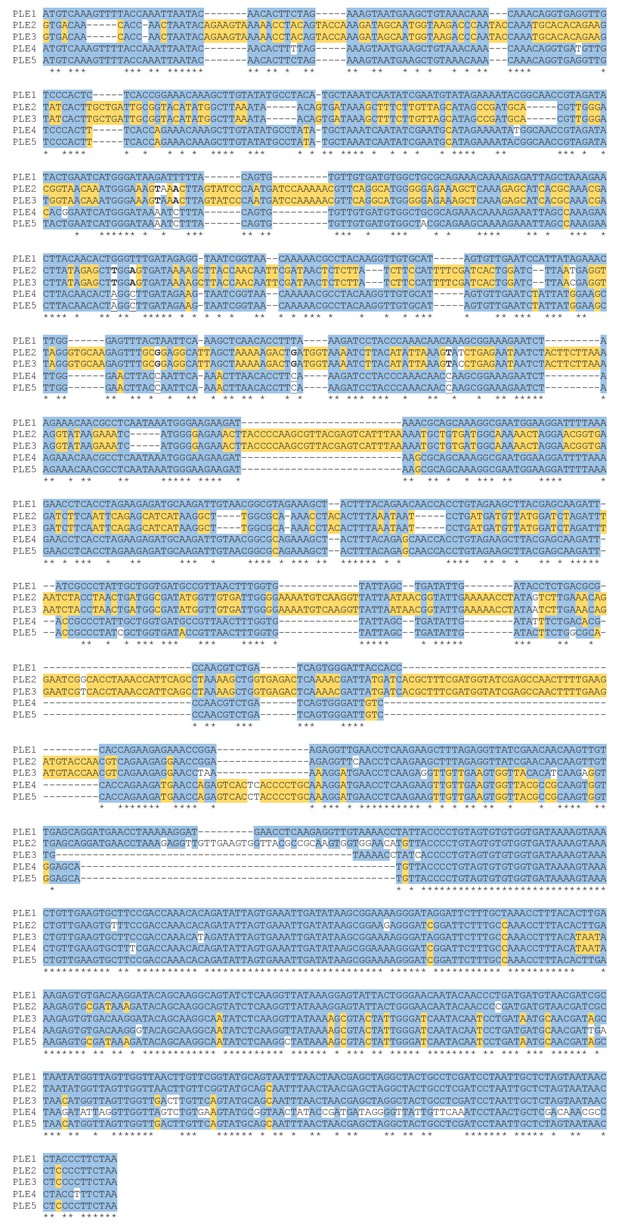
Alignment of repA from phaPLEs 1–5 encoding the conserved C-terminus.
The PLE1 sequence and identical nucleotides in the other PLE variants are shaded blue. Nucleotides that differ from PLE1 but co-occur in at least two PLE variants are shaded yellow. For positions where PLE1 is unique and both PLE2 and PLE3, and PLE4 and PLE5 have separate consensuses, the PLE2 and PLE3 consensus is bolded and unshaded, and the PLE4 and PLE5 consensus is underlined and unshaded. Nucleotides with no aligned matches are unshaded normal text.
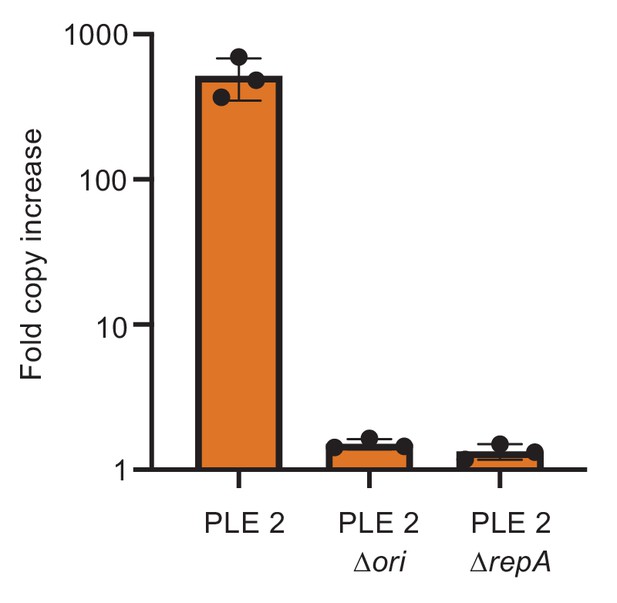
PLE2 requires RepA and a cognate origin of replication to replicate.
Replication of PLE2 in V. cholerae calculated as the fold change in PLE DNA copy 20 minutes post infection with ICP12006 ∆CRISPR.
-
Figure 3—figure supplement 2—source data 1
Values for the graph in Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig3-figsupp2-data1-v1.xlsx
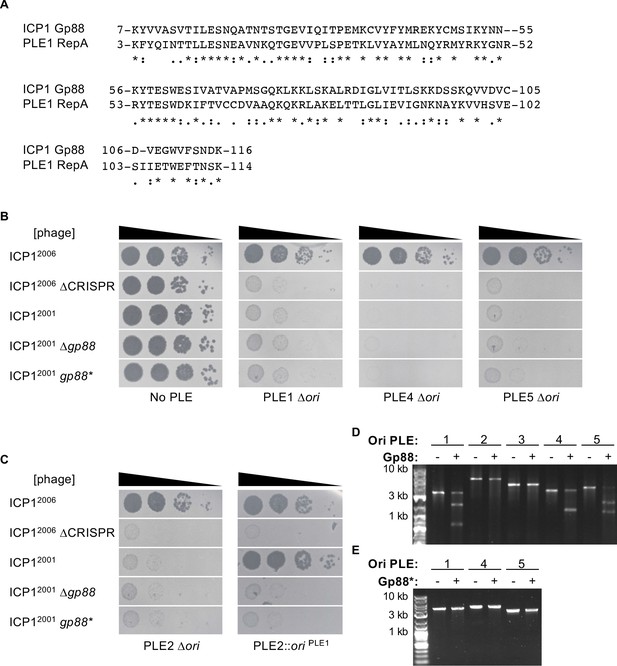
Gp88 is a PLE replication origin-directed nuclease.
(A) Sequence alignment of the N-terminal portion of Gp88 with the RepA_N domain from PLE1 RepA. Identical residues are denoted with a ‘*.’ Strong residue similarity is denoted by ‘:’, and weak similarity is denoted by ‘•.’ (B, C) Tenfold dilutions of the phage isolate or mutant derivative indicated spotted on V. cholerae with the PLE indicated (bacterial lawns in gray, zones of killing are shown in black). Spot assays were performed in parallel with those in Figure 2, and images labeled with the same PLE background are the same image. Spot assays were performed in biological triplicate, and a single representative image is shown. Biological replicates are shown in Figure 2—figure supplement 1. Gp88* possess a single amino acid substitution (E180A) predicted to abolish nuclease activity. (B) shows phage susceptibility of V. cholerae with PLE1, PLE4, and PLE5 ∆ori derivatives as compared to a strain without PLE. (C) shows phage susceptibility for V. cholerae with PLE2 ∆ori and PLE2 ∆ori::oriPLE1. (D) Nuclease assay showing the integrity of a PCR product amplified from the noncoding region containing the ori from the PLE variant indicated (numbers) treated with (+) and without (–) 500 nM of purified Gp88. Nuclease assays were performed in triplicate, replicates are presented in Figure 4—figure supplement 3. (E) Nuclease assay showing the integrity of a PCR product amplified from the noncoding region containing the ori from the PLE variant indicated (numbers) treated with (+) and without (–) 500 nM of purified Gp88*. Nuclease assays were performed in triplicate, replicates are presented in Figure 4—figure supplement 4.
-
Figure 4—source data 1
Original uncropped gels for Figure 4D (top) and Figure 4E (bottom).
The cropped images shown in the figure are indicated by the orange boxes.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig4-data1-v1.pdf
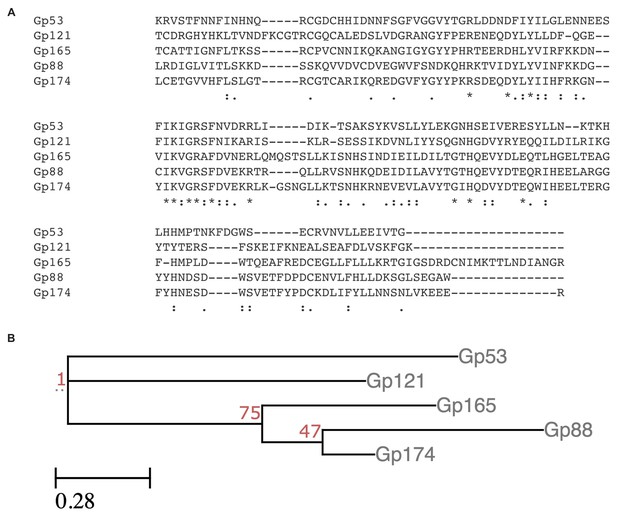
The nuclease domain of Gp88 is similar to those of putative homing endonucleases in ICP1.
(A) A multiple sequence alignment between the C-terminal portion of Gp88 and several homologous sequences found in putative homing endonucleases in the ICP1 genome. Identical residues are denoted with a ‘*.’ Strong residue similarity is denoted by ‘:,’ and weak similarity is denoted by ‘•.’ (B) A phylogenetic tree constructed from the multiple sequence alignment in (A).
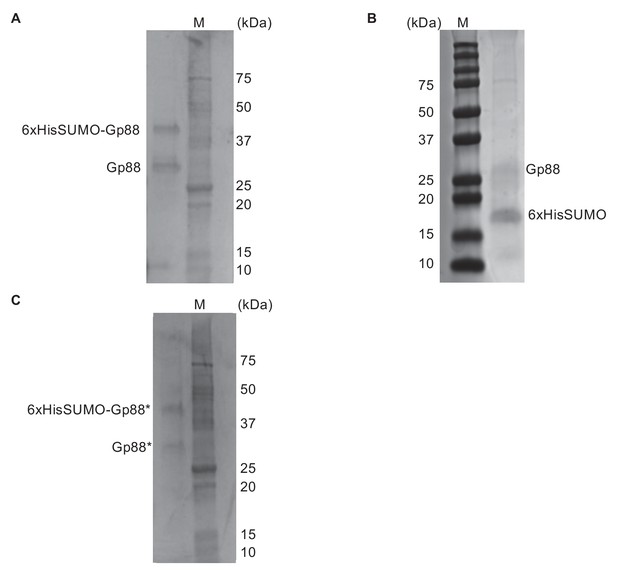
Protein preparations of Gp88 (Odn) and Gp88* (Odn*) used for in vitro assays.
Protein preparations were separated by SDS-PAGE and visualized by Coomassie staining. The marker (M) is indicated; the predicted molecular weight of untagged Gp88 is 26 kDa. (A) Gel showing wild-type Gp88 protein used for all cleavage assays except for those depicted in Figure 4—figure supplement 4 and Figure 6. (B) Gel showing wild-type Gp88 protein used for cleavage assays depicted in Figure 4—figure supplement 4 and Figure 6. (C) Gel showing Gp88* protein used for all in vitro assays that utilized Gp88*. Note that protein preparations (A) and (C) were performed in parallel; the preparation in (B) was completed separately.
-
Figure 4—figure supplement 2—source data 1
Original uncropped gels for Figure 4—figure supplement 2.
The cropped images shown in the figure are indicated by the orange boxes.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig4-figsupp2-data1-v1.pdf
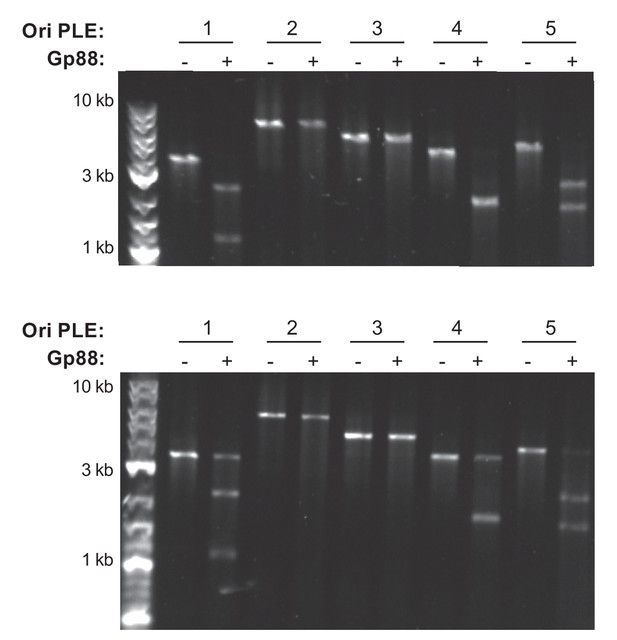
Replicates of Gp88 nuclease assay.
Two replicates of a nuclease assay showing the integrity of a PCR product amplified from the noncoding region containing the ori from the PLE variant indicated (numbers) treated with (+) and without (–) 500 nM of purified Gp88. A third replicate is depicted in Figure 4D.
-
Figure 4—figure supplement 3—source data 1
Original uncropped gels for Figure 4—figure supplement 3.
The cropped images shown in the figure are indicated by the orange boxes.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig4-figsupp3-data1-v1.pdf
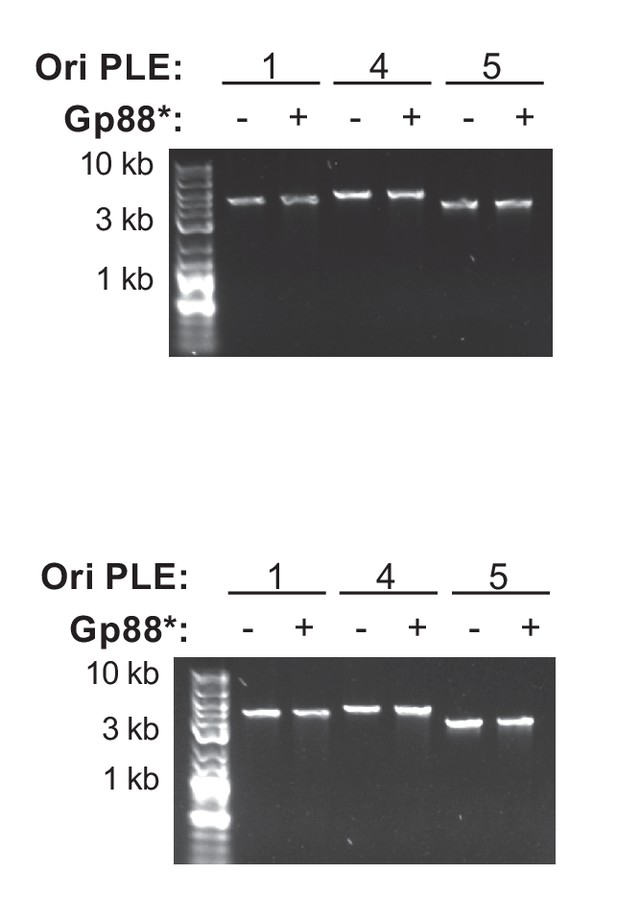
Replicates of Gp88* nuclease assay.
Two replicates of a nuclease assay showing the integrity of a PCR product amplified from the noncoding region containing the ori from the PLE variant indicated (numbers) treated with (+) and without (–) 500 nM of purified Gp88*. A third replicate is depicted in Figure 4E.
-
Figure 4—figure supplement 4—source data 1
Original uncropped gels for Figure 4—figure supplement 4.
The cropped images shown in the figure are indicated by the orange boxes.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig4-figsupp4-data1-v1.pdf
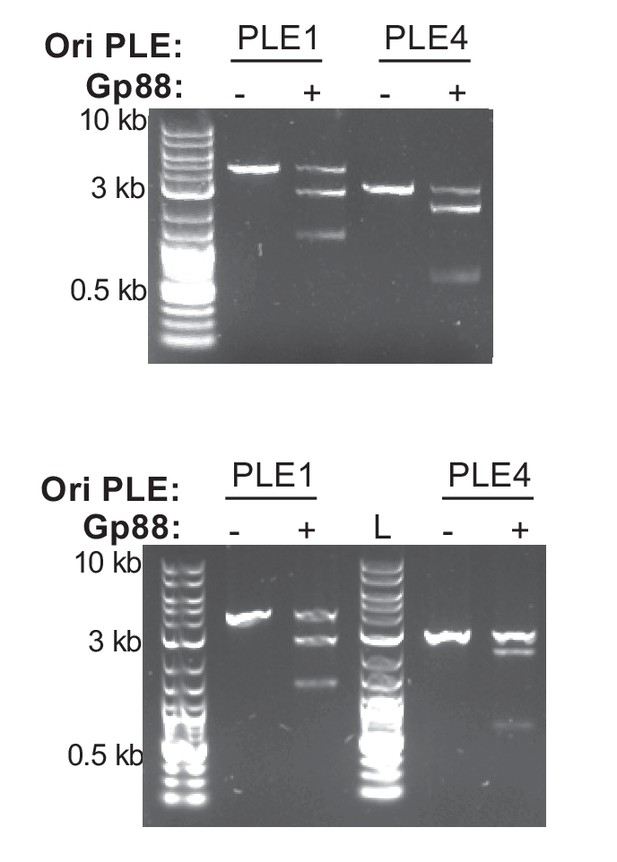
Cleavage of an altered PLE4 probe.
Two replicates of a nuclease assay showing the integrity of a PCR product amplified from the noncoding region containing the ori from the PLE1 or PLE4 treated with (+) and without (–) 500 nM of purified Gp88. The PLE4 probe used in this experiment is amplified with a different set of primers compared to the probe used in Figure 4D such that cleavage of the probe at or near the iterons would produce products that could be differentiated on the basis of size.
-
Figure 4—figure supplement 5—source data 1
Original uncropped gels for Figure 4—figure supplement 5.
The cropped images shown in the figure are indicated by the orange boxes.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig4-figsupp5-data1-v1.pdf
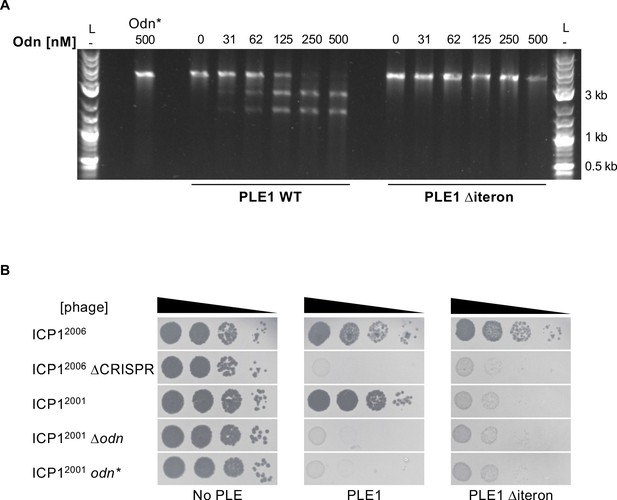
ICP1-encoded Odn (Gp88) requires the PLE iterons for cleavage.
(A) Nuclease assay showing the integrity of a PCR product amplified from the noncoding region containing the ori from wild-type (WT) PLE1 and the ∆iteron mutant, with purified Odn (31.25–500 nM) titrated in. 500 nM catalytically inactive Odn (Odn*) with a single amino acid substitution (E180A) with the WT PLE1 sequence was also included (far left). Nuclease assays were performed in triplicate and replicates are presented in Figure 5—figure supplement 1. (B) Tenfold dilutions of the phage isolate or mutant derivative indicated spotted on V. cholerae with the PLE indicated (bacterial lawns in gray, zones of killing are shown in black). Spot assays were performed in parallel with those in Figures 2 and 4, and images labeled with the same PLE background are the same image. Spot assays were performed in biological triplicate, and a single representative image is shown. Replicate assays are shown in Figure 2—figure supplement 1.
-
Figure 5—source data 1
Original uncropped gel for Figure 5A.
The cropped image shown in the figure is indicated by the orange box.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig5-data1-v1.pdf
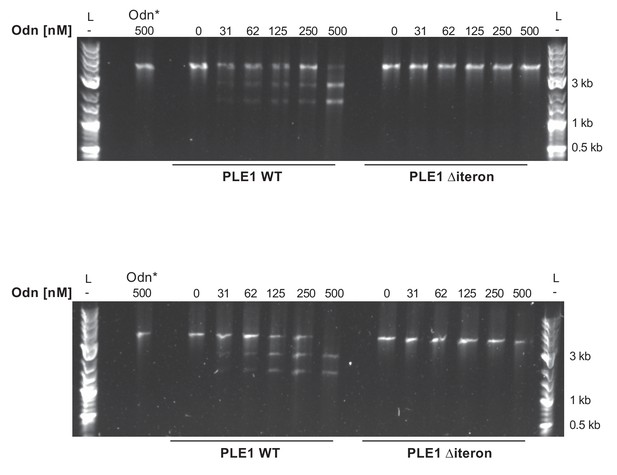
Replicates of nuclease assays.
Nuclease assays showing the integrity of a PCR product amplified from the noncoding region containing the ori from wild-type (WT) PLE1 and the ∆iteron mutant, with purified Odn (31.25–500 nM) titrated in. 500 nM catalytically inactive Odn (Odn*) with a single amino acid substitution (E180A) with the WT PLE1 sequence was also included. This assay was performed in triplicate, and two replicates are shown here. The third replicate is shown in Figure 5A.
-
Figure 5—figure supplement 1—source data 1
Original uncropped gels for Figure 5—figure supplement 1.
The cropped images shown in the figure are indicated by the orange boxes.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig5-figsupp1-data1-v1.pdf
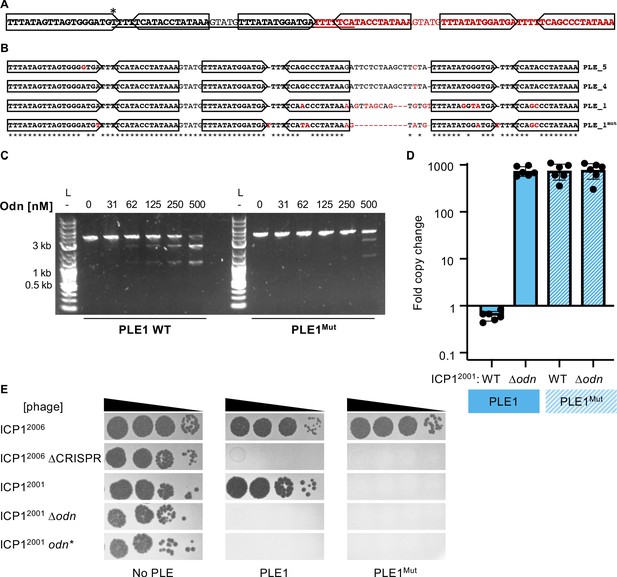
Mutations in PLE1 present in V. cholerae isolates from Pakistan render the PLE1 ori resistant to Odn-mediated cleavage.
(A) The PLE1Mut iteron region. Iterons are bolded and sub-repeats are denoted with arrows. The underlined sequence is identical to the sequence in red. An asterisk (*) denotes the location of an A to T substitution. (B) An alignment of the iterons from PLE5, PLE4, PLE1, and PLE1Mut. Iterons are in bold with sub-repeats indicated with arrows. Sequence deviating from a consensus is shown in red. Regions with 100% conservation are denoted with an asterisk. (C) Nuclease assay showing the integrity of a PCR product amplified from the noncoding region containing the ori from wild-type (WT) PLE1 or PLE1Mut with purified Odn (31.25–500 nM) titrated in. Nuclease assays were performed in triplicate, and replicates are presented in Figure 6—figure supplement 1. (D) Replication of PLE1 WT and PLE1Mut in V. cholerae calculated as the fold change in PLE DNA copy 20 minutes post infection with the ICP1 variant indicated. (E) Tenfold dilutions of the phage isolate or mutant derivative indicated spotted on V. cholerae with the PLE indicated (bacterial lawns in gray, zones of killing are shown in black). Biological replicates of the spot assays are presented in Figure 6—figure supplement 2.
-
Figure 6—source data 1
Original uncropped gel for Figure 6C.
The cropped image shown in the figure is indicated by the orange box.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig6-data1-v1.pdf
-
Figure 6—source data 2
Values for the graph in Figure 6D.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig6-data2-v1.xlsx
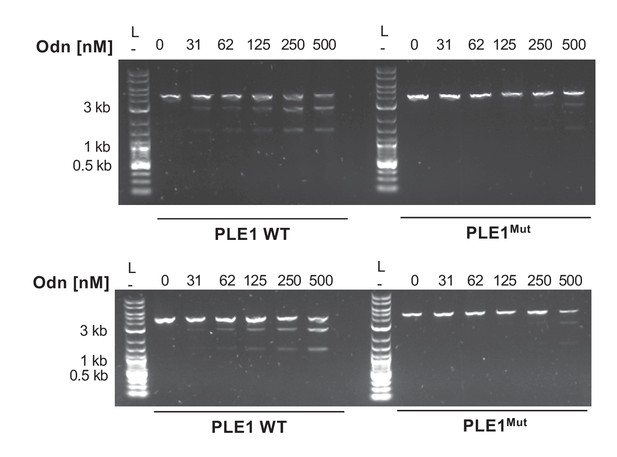
Replicates of nuclease assays.
Nuclease assays showing the integrity of a PCR product amplified from the noncoding region containing the ori from wild-type PLE1 (WT) or PLE1Mut with (+) and without (–) the addition of 500 nM oOdn. Nuclease assays were performed in triplicate. Two replicates are shown here, and the third is depicted in Figure 6C.
-
Figure 6—figure supplement 1—source data 1
Original uncropped gels for Figure 6—figure supplement 1.
The cropped images shown in the figure are indicated by the orange boxes.
- https://cdn.elifesciences.org/articles/68339/elife-68339-fig6-figsupp1-data1-v1.pdf
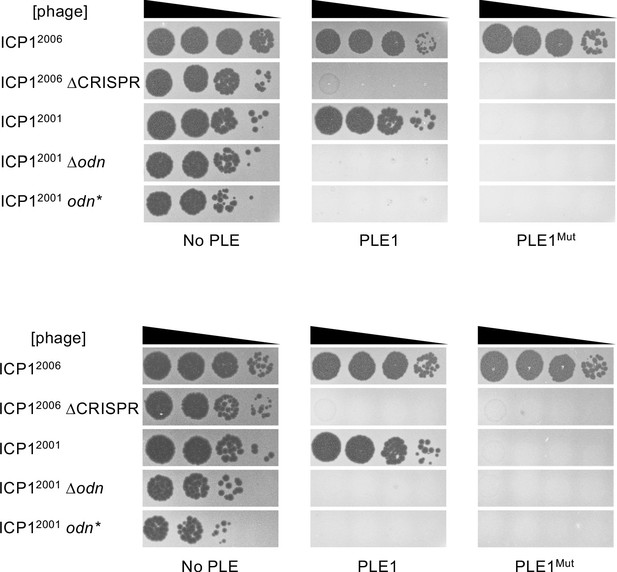
Biological replicates of PLE1Mut spot assays.
Tenfold dilutions of the phage isolate or mutant derivative indicated spotted on V. cholerae with the PLE indicated (bacterial lawns in gray, zones of killing are shown in black). A representative replicate is also shown in Figure 6E.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Vibrio cholerae) | RepAPLE1(PLE1 ORF11) | Barth et al., 2020b | WP_002040284.1 | |
| Gene (Vibrio cholerae) | RepAPLE2(PLE2 ORF14) | This paper | AGG36643.1 | |
| Gene (Bacteriophage ICP1) | Odn (ICP1_2001_Dha_0 gp88) | This paper | YP_004251029 | |
| Gene (Bacteriophage ICP1) | Odn* (ICP1_2001_Dha_0 gp88E180A) | This paper | The E180A mutation is predicted to abolish catalytic activity | |
| Recombinant DNA reagent | Ptac-repAPLE1(plasmid) | Barth et al., 2020b | pZKB129 | Inducible RepA from PLE1 |
| Recombinant DNA reagent | Ptac-repAPLE2(plasmid) | This paper | pKS2159 | Inducible RepA from PLE2 |
| Recombinant DNA reagent | pE-SUMO-Odn (plasmid) | This paper | pKS2187 | Vector to express 6xHisSumo-fusion protein, fused to N-terminus of Odn (Gp88) |
| Recombinant DNA reagent | pE-SUMO-Odn* (plasmid) | This paper | pKS2189 | Vector to express 6xHisSumo-fusion protein, fused to N-terminus of Odn* (Gp88E180A) |
| Strain, strain background (Vibrio cholerae) | PLE V. cholerae (E7946) | Levine et al., 1982 | KDS6 | |
| Strain, strain background (Vibrio cholerae) | PLE1 V. cholerae (PLE1 E7946) | O'Hara et al., 2017 | KDS36 | |
| Strain, strain background (Vibrio cholerae) | PLE2 V. cholerae (PLE2 E7946) | O'Hara et al., 2017 | KDS37 | |
| Strain, strain background (Vibrio cholerae) | PLE3 V. cholerae (PLE3 E7946) | O'Hara et al., 2017 | KDS38 | |
| Strain, strain background (Vibrio cholerae) | PLE4 V. cholerae (PLE4 E7946) | O'Hara et al., 2017 | KDS39 | |
| Strain, strain background (Vibrio cholerae) | PLE5 V. cholerae (PLE5 E7946) | O'Hara et al., 2017 | KDS40 | |
| Strain, strain background (Vibrio cholerae) | PLE1 ∆ori V. cholerae (PLE1 E7946) | This paper | KDS297 | Used for all spot assays |
| Strain, strain background (Vibrio cholerae) | PLE2 ∆ori V. cholerae (PLE2 E7946) | This paper | KDS298 | Figure 3—figure supplement 2 |
| Strain, strain background (Vibrio cholerae) | PLE2 ∆repA V. cholerae (PLE2 E7946) | This paper | KDS299 | Figure 3—figure supplement 2 |
| Strain, strain background (Vibrio cholerae) | PLE1 ∆repA ∆ori::oriPLE2; Ptac-repAPLE1 V. cholerae E7946 | This paper | KDS300 | Figure 3B |
| Strain, strain background (Vibrio cholerae) | PLE1 ∆repA ∆ori::oriPLE2; Ptac-repAPLE2 V. cholerae E7946 | This paper | KDS301 | Figure 3B |
| Strain, strain background (Vibrio cholerae) | PLE2 ∆repA ∆ori::oriPLE1; Ptac-repAPLE1 V. cholerae E7946 | This paper | KDS302 | Figure 3B |
| Strain, strain background (Vibrio cholerae) | PLE2 ∆repA ∆ori::oriPLE1; Ptac-repAPLE2 V. cholerae E7946 | This paper | KDS303 | Figure 3B |
| Strain, strain background (Vibrio cholerae) | PLE4 ∆ori V. cholerae (PLE4 E7946) | This paper | KDS304 | Used for all spot assays |
| Strain, strain background (Vibrio cholerae) | PLE5 ∆ori V. cholerae (PLE5 E7946) | This paper | KDS305 | Used for all spot assays |
| Strain, strain background (Vibrio cholerae) | PLE1 ∆iterons V. cholerae (PLE1 E7946) | Barth et al., 2020b | KDS263 | Used for all spot assays |
| Strain, strain background (Vibrio cholerae) | PLE2 ∆ori::ori PLE1V. cholerae (PLE2 E7946) | This paper | KDS306 | Used for all spot assays |
| Strain, strain background (Vibrio cholerae) | PLE1∆ori::oriMut∆lacZ::KanR V. cholerae E7946 (referred to as PLE1Mut) | This paper | KDS319 | Ori engineered to match what is observed in PLE1(+) strains from Pakistan: biosample accession numbers SAMN08979118, SAMN08979175, SAMN08979185, SAMN08979188, and SAMN08979253 |
| Strain, strain background (Escherichia coli) | pE-SUMO-Odn E. coli BL21 | This paper | KDS307 | Expression strain for Gp88/Odn |
| Strain, strain background (Escherichia coli) | pE-SUMO-Odn* E. coli BL21 | This paper | KDS308 | Expression strain for Gp88*/Odn* |
| Strain, strain background (Bacteriophage ICP1) | 2006 WT (ICP1_2006_Dha_E) | O'Hara et al., 2017 | MH310934 | |
| Strain, strain background (Bacteriophage ICP1) | 2006 ∆CR; ∆Cas2_3 (ICP1_2006_Dha_E) | McKitterick and Seed, 2018 | ||
| Strain, strain background (Bacteriophage ICP1) | 2001 WT (ICP1_2001_Dha_0) | Seed et al., 2011 | HQ641347 | |
| Strain, strain background (Bacteriophage ICP1) | 2001 ∆odn (ICP1_2001_Dha_0) | This paper | KSϕ93 | odn is gp88 |
| Strain, strain background (Bacteriophage ICP1) | 2001 odn* (ICP1_2001_Dha_0) | This paper | KSϕ134 | odn* isgp88E180A |
| Sequence-based reagent | 5'-AGGGTTTGAGTGCGATTACG-3' | O'Hara et al., 2017 | zac14 | qPCR primer targeting a conserved portion of the PLE noncoding region |
| Sequence-based reagent | 5'-TGAGGTTTTACCACCTTTTGC-3' | O'Hara et al., 2017 | zac15 | qPCR primer targeting a conserved portion of the PLE noncoding region |
| Sequence-based reagent | 5'-GTCATTTAACGCATCTTATCACC-3' | This paper | KS459 | F-primer used to amplify noncoding region probes for PLE1 and PLE5 |
| Sequence-based reagent | 5'-GGCTTAGCAACTGTCTACGG-3' | This paper | zac267 | F-primer used to amplify noncoding region probes for PLE2, PLE3, and PLE4 |
| Sequence-based reagent | 5'-GTTACGTCTGATTGCTGACG-3' | This paper | KS321 | R-primer used to amplify noncoding region probes for PLE1 |
| Sequence-based reagent | 5'-CCGCTTATATCAATTTCACTAATATCT-3' | This paper | zac269 | R-primer used to amplify noncoding region probes for PLE2 and PLE3 |
| Sequence-based reagent | 5'-GGACGGCTAAACCATTCTCG-3' | This paper | KS323 | R-primer used to amplify noncoding region probes for PLE4 and PLE5 |
| Sequence-based reagent | 5’-CATAAGGTTGGCTCCTCAATG-3’ | This paper | KS458 | R-primer used to amplify noncoding region probe for PLE4 in Figure 4—figure supplement 5 |





